Estimating the burden of α-thalassaemia in Thailand using a comprehensive prevalence database for Southeast Asia
Figures
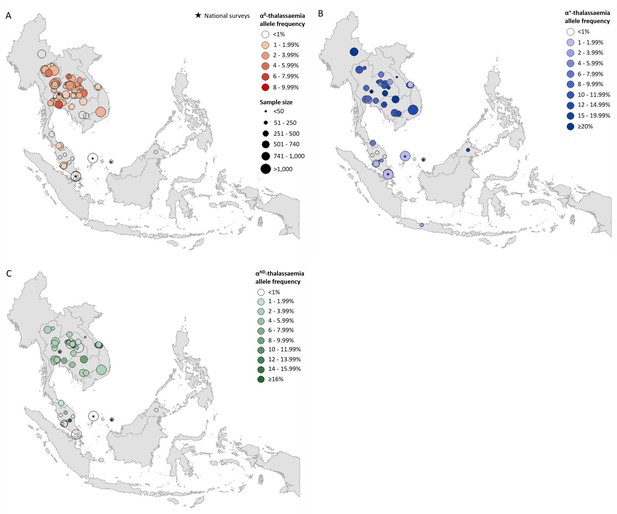
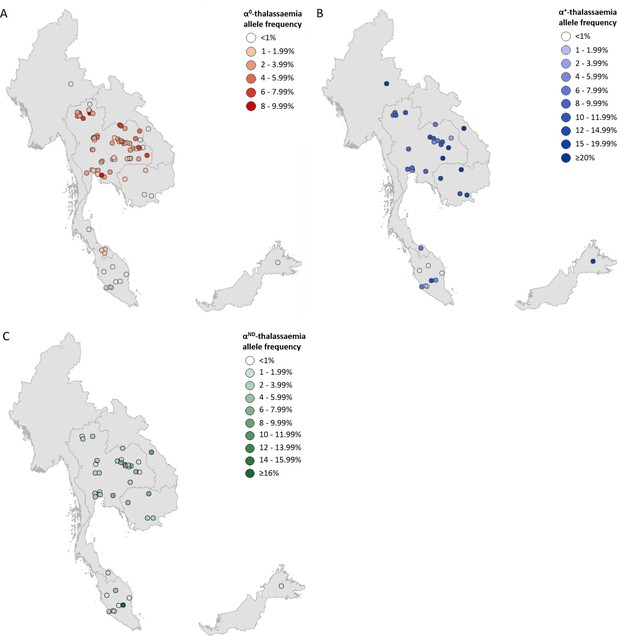
Descriptive maps of the observed allele frequencies in the database.
(A) α0-thalassaemia, (B) α+-thalassaemia and (C) αND-thalassaemia. A spatial jitter of up to 0.30 latitude and longitude decimal degree coordinates was applied to allow visualisation of spatially duplicated data points. Colour intensity indicates allele frequency; circle size represents the size of the survey size. Surveys that could only be mapped at the national level are indicated by a black star.
-
Figure 1—source data 1
Source data for Figure 1A,a map of the observed α0-thalassaemia allele frequencies in the database.
Data were obtained through a review of the published literature using a rigorous inclusion/exclusion protocol. In the figure, a spatial jitter of up to 0.30 latitude and longitude decimal degree coordinates was applied to allow visualisation of spatially duplicated data points.
- https://doi.org/10.7554/eLife.40580.006
-
Figure 1—source data 2
Source data for Figure 1B,a map of the observed α+-thalassaemia allele frequencies in the database.
Data were obtained through a review of the published literature using a rigorous inclusion/exclusion protocol. In the figure, a spatial jitter of up to 0.30 latitude and longitude decimal degree coordinates was applied to allow visualisation of spatially duplicated data points.
- https://doi.org/10.7554/eLife.40580.007
-
Figure 1—source data 3
Source data for Figure 1C,a map of the observed αND-thalassaemia allele frequencies in the database.
Data were obtained through a review of the published literature using a rigorous inclusion/exclusion protocol. In the figure, a spatial jitter of up to 0.30 latitude and longitude decimal degree coordinates was applied to allow visualisation of spatially duplicated data points.
- https://doi.org/10.7554/eLife.40580.008

A map of the countries included in this study.
Here, we defined the Southeast Asian region according to the member states of the Association of Southeast Asian Nations (ASEAN) (http://asean.org/asean/asean-member-states/).
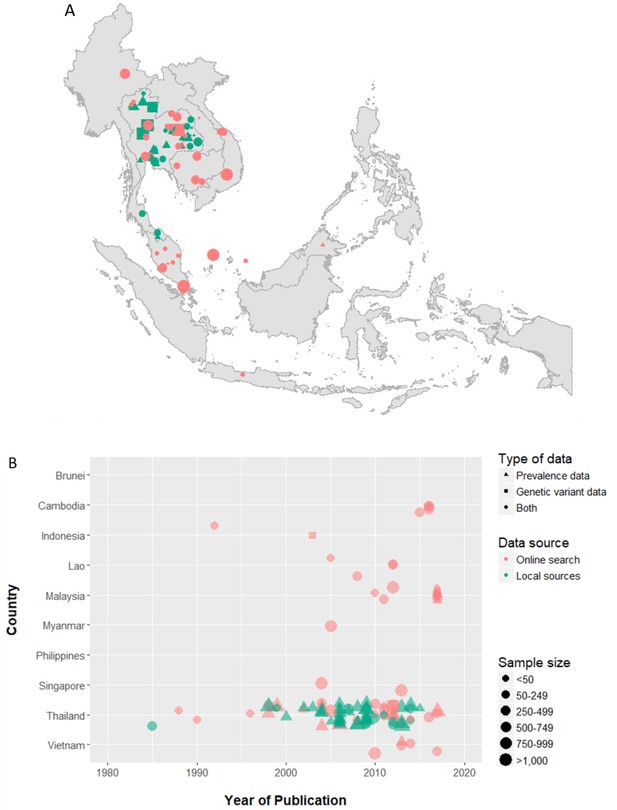
Spatial and temporal distributions of the α-thalassaemia surveys included in the final database.
In both panels, the shape of the data points indicates the type of data provided by the survey, the colour indicates whether the survey was found in our online literature search or in local journals, and size represents the sample size of the survey. In (A) a spatial jitter of up to 0.30 latitude and longitude decimal degree coordinates was applied to allow visualisation of spatially duplicated data points.
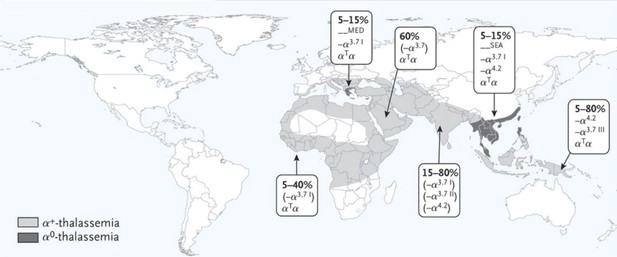
A map of our current knowledge of the global distribution, gene frequency and genetic diversity of α-thalassemia.
Only the most common variants for α+-thalassemia (-α3.7and -α4.2) and α0-thalassemia (--MED and --SEA) are shown for each region. The variants that appear in parentheses are those for which the data used to make this map are limited.
© 2014 Massachusetts Medical Society. From the New England Journal of Medicine, Piel FB and Weatherall DJ, ‘The α-Thalassaemias’, 371(20), 1912. Reprinted with permission from Massachusetts Medical Society.
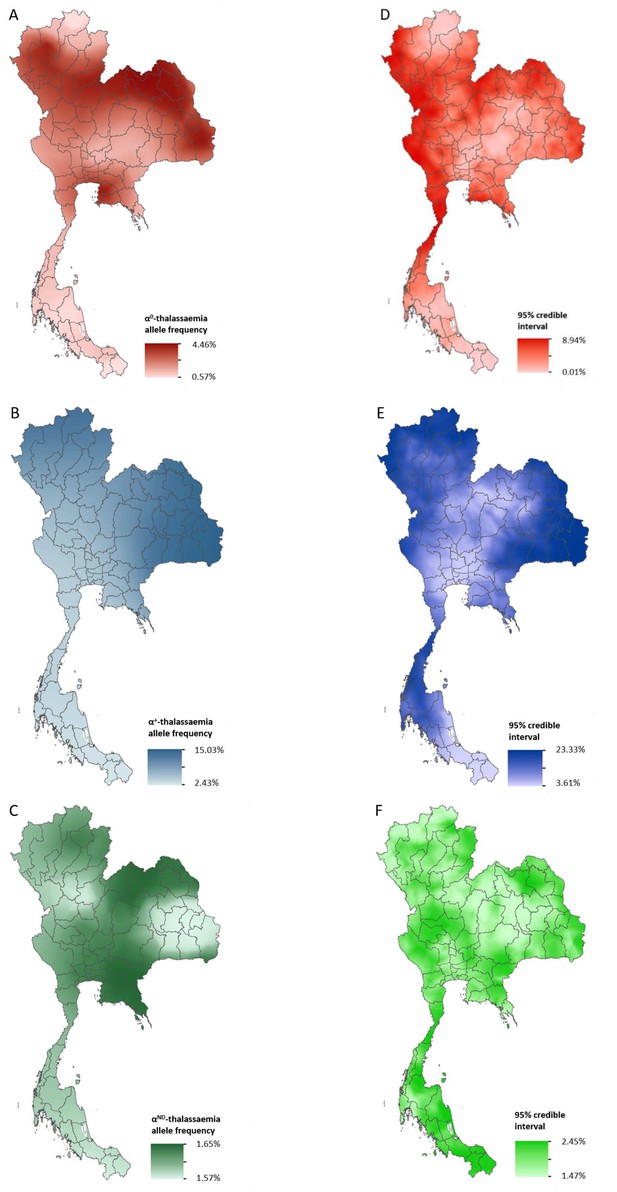
Maps of the mean of, and uncertainty in, the predicted α-thalassaemia allele frequencies in Thailand.
Panels A to C display the mean of the posterior predictive distribution (PPD) of 100 realisations of the geostatistical model. Panels D to F display the 95% credible interval of the PPD. Each row corresponds to a different α-thalassaemia form: α0-thalassaemia (A and D); α+-thalassaemia (B and E) and αND-thalassaemia (C and F). Figure 2—figure supplement 1 shows the observed data used to construct the models and Figure 2—figure supplement 2 displays the province names for reference.
Maps of the observed allele frequencies used to construct the models and generate the predicted continuous allele frequency maps for Thailand in Figure 2.
(A) α0-thalassaemia, (B) α+-thalassaemia and (C) αND-thalassaemia. A variable spatial jitter was applied to allow visualisation of spatially duplicated data points. Colour intensity indicates allele frequency.

A reference map of Thailand provinces.
The Bangkok Metropolitan Region, which includes Bangkok City and surrounding provinces, is shaded in red, with Bangkok City shaded a darker red.
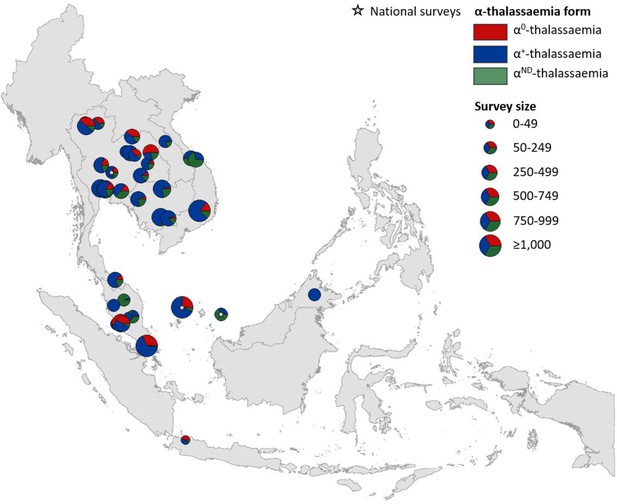
Map showing the proportions of α0-, α+- and αND-thalassaemia in Southeast Asia.
Three surveys were mapped at the national level (indicated by a white star). The size of the pie charts reflects survey size.
-
Figure 3—source data 1
Source data for Figure 3, a map showing the proportions of α0-, α+- and αND-thalassaemia in Southeast Asia.
Reported allele frequencies (converted to percentages here) and sample size were used to calculate the number of chromosomes bearing each form of α-thalassaemia. Sample size was multiplied by two to obtain the total number of chromosomes in the study sample. In total, 40 surveys included genetic diversity information for the three α-thalassaemia forms.
- https://doi.org/10.7554/eLife.40580.015
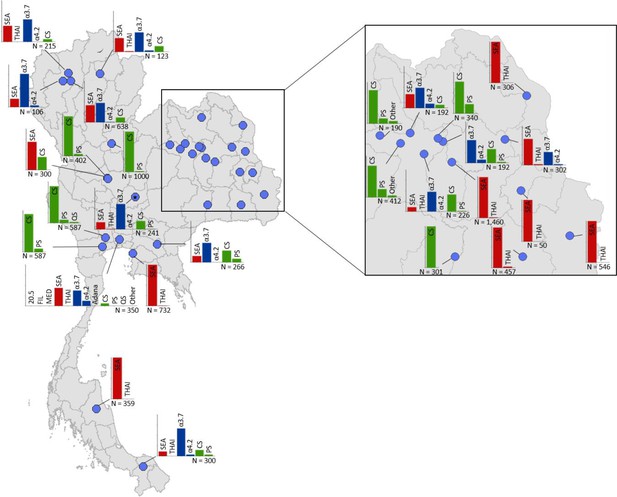
Map showing the allele frequencies of specific α-thalassaemia variants in Thailand.
Given the high number of surveys in northeast Thailand, this region has been magnified. The y-axis scale is the same across all bar charts, ranging from 0 to 1. The variants that were tested for in each survey are indicated above each bar. α0-thalassaemia mutations are shown in red, α+-thalassaemia mutations in blue and αND-thalassaemia mutations in green. Empty spaces along the x-axis indicate an absence of the corresponding mutation in the survey sample. The sample size of the survey is given under each plot. Bar charts are connected to their spatial location by a black line.
-
Figure 4—source data 1
Source data for Figure 4, a map showing the proportions of specific α-thalassaemia variants in Thailand.
'NA' indicates those variants that were not tested for in the survey. In Thailand, 29 surveys included genetic diversity information for specific α-thalassaemia variants.
- https://doi.org/10.7554/eLife.40580.017
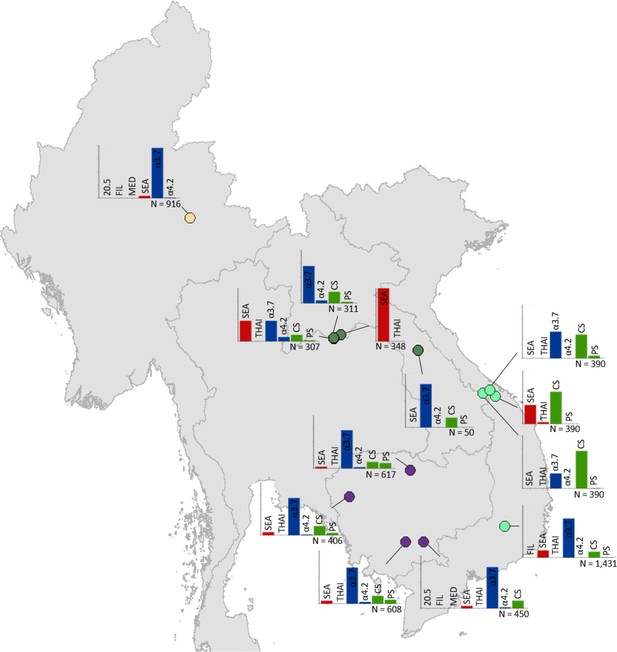
Map showing the allele frequencies of specific α-thalassaemia variants in Myanmar, Lao PDR, Cambodia and Vietnam.
The y-axis scale is the same across all bar charts, ranging from 0 to 1. The variants that were tested for in each survey are indicated above each bar. α0-thalassaemia mutations are shown in red, α+-thalassaemia mutations in blue and αND-thalassaemia mutations in green. Empty spaces along the x-axis indicate an absence of the corresponding mutation in the survey sample. The sample size of the survey is given under each plot. Bar charts are connected to their spatial location by a black line. Data points are coloured by country, using the same colour scale as that in Figure 1—figure supplement 1.
-
Figure 5—source data 1
Source data for Figure 5, a map showing the proportions of specific α-thalassaemia variants in Cambodia, Lao PDR, Myanmar and Vietnam.
'NA' indicates those variants that were not tested for in the survey. In total, 13 surveys included genetic diversity information for specific α-thalassaemia variants in these countries.
- https://doi.org/10.7554/eLife.40580.013
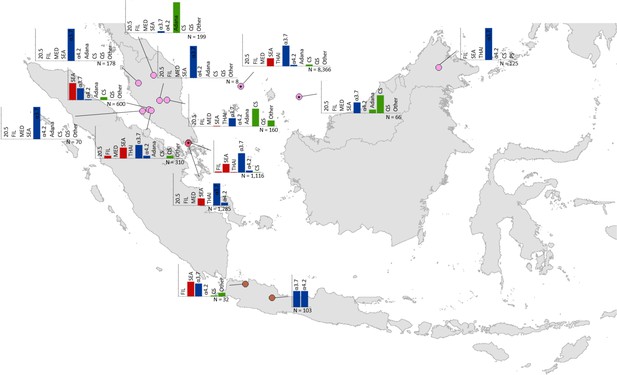
Map showing the allele frequencies of specific α-thalassaemia variants in Malaysia, Singapore and Indonesia.
The y-axis scale is the same across all bar charts, ranging from 0 to 1. The variants that were tested for in each survey are indicated above each bar. α0-thalassaemia mutations are shown in red, α+-thalassaemia mutations in blue and αND-thalassaemia mutations in green. Empty spaces along the x-axis indicate an absence of the corresponding mutation in the survey sample. The sample size of the survey is given under each plot. Bar charts are connected to their spatial location by a black line. Data points are coloured by country, using the same colour scale as that in Figure 1—figure supplement 1.
-
Figure 6—source data 1
Source data for Figure 6, a map showing the proportions of specific α-thalassaemia variants in Indonesia, Malaysia and Singapore.
'NA' indicates those variants that were not tested for in the survey. In total, 14 surveys included genetic diversity information for specific α-thalassaemia variants in these countries.
- https://doi.org/10.7554/eLife.40580.019
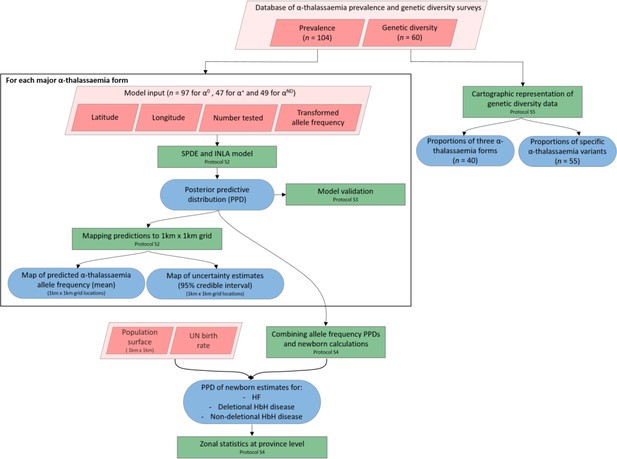
A schematic overview of the methodology used in this study and a breakdown of the data types analysed.
Pink diamonds indicate the database and input data; green boxes denote model processes and data visualisation steps; blue rods represent study outputs. .
Tables
A breakdown of the genotypes for the three clinically important forms of α-thalassaemia – Hb Bart’s hydrops fetalis, deletional HbH disease and non-deletional HbH disease – and the Hardy-Weinberg equilibrium (HWE) proportions used for their calculation.
To compare our model output with previous newborn estimates for Hb Bart’s hydrops fetalis and deletional HbH disease, we paired our allele frequency maps with 2003 demographic and birth data and included a measure of consanguinity in our calculations.
| Genotype | Disorder | HWE proportions | Inclusion of population coefficient of consanguinity (F) |
|---|---|---|---|
| --/-- | Hb Bart’s hydrops fetalis | p2 | p2 + Fp(1 p) |
| -α/-- | Deletional HbH disease | 2pq | 2pq(1 F) |
| ααND/-- | Non-deletional HbH disease | 2pr | 2pr(1 F) |
Summary of the α-thalassaemia dataset characteristics according to the type of data provided (allele frequency data or genetic variant data), and overall.
Numbers correspond to individual surveys that met the study inclusion criteria. As some sources reported more than one survey from multiple locations or in multiple population groups, the number of surveys is greater than the number of references in the Supplementary files 1 and 2. Some surveys reported data on both α-thalassaemia prevalence and genetic diversity and are therefore included twice in these columns, but once in the overall column.
| Allele frequency data | Genetic diversity data | Overall | |
|---|---|---|---|
| Total surveys | 104 | 60 | 106 |
| Number of countries | 8 | 8 | 8 |
| Publication time | |||
| 1959–1969 | 0 | 0 | 0 |
| 1970–1979 | 0 | 0 | 0 |
| 1980–1989 | 2 | 2 | 2 |
| 1990–1999 | 8 | 5 | 8 |
| 2000–2009 | 46 | 15 | 47 |
| 2010–2017 | 47 | 38 | 48 |
| N/A | 1 | 0 | 1 |
| Spatial extent | |||
| Admin 0 centroids | 4 | 4 | 4 |
| Admin one centroids | 27 | 12 | 27 |
| Admin two centroids | 10 | 8 | 9 |
| Admin three centroids | 3 | 3 | 3 |
| Points | 46 | 22 | 47 |
| Multiple centroids/points | 14 | 11 | 16 |
| Total individuals sampled | 132,157 | 32,237 | 133,649 |
| Survey count by sample size | |||
| ≤50 | 3 | 2 | 4 |
| 51–250 | 22 | 18 | 22 |
| 251–500 | 38 | 23 | 38 |
| 501–750 | 21 | 10 | 21 |
| 751–1000 | 3 | 1 | 3 |
| >1000 | 17 | 6 | 18 |
Observed allele frequency ranges for different α-thalassaemia forms.
https://doi.org/10.7554/eLife.40580.031| Country | Allele frequency range (%) | ||
|---|---|---|---|
| α0-thalassaemia | α+-thalassaemia | αND-thalassaemia | |
| Brunei Darussalam | No surveys identified | No surveys identified | No surveys identified |
| Cambodia | 0.80–1.10 | 10.30–26.30 | 2.44–4.20 |
| Indonesia | No surveys identified | 2.91 | No surveys identified |
| Lao PDR | 0.00–6.19 | 4.60–40.00 | 2.28–9.00 |
| Malaysia | 0.00–1.92 | 0.00–16.80 | 0.00–16.25 |
| Myanmar | 0.93 | 20.58 | No surveys identified |
| Philippines | No surveys identified | No surveys identified | No surveys identified |
| Singapore | 0.86–0.90 | 1.88–3.04 | 0.04 |
| Thailand | 0.00–9.29 | 2.98–21.43 | 0.00–7.30 |
| Vietnam | 0.00–2.66 | 1.59–14.4 | 2.07–14.43 |
Additional files
-
Supplementary file 1
References for allele frequency data.
- https://doi.org/10.7554/eLife.40580.022
-
Supplementary file 2
References for genetic variant data.
- https://doi.org/10.7554/eLife.40580.023
-
Transparent reporting form
- https://doi.org/10.7554/eLife.40580.024





