Sox9+ messenger cells orchestrate large-scale skeletal regeneration in the mammalian rib
Figures
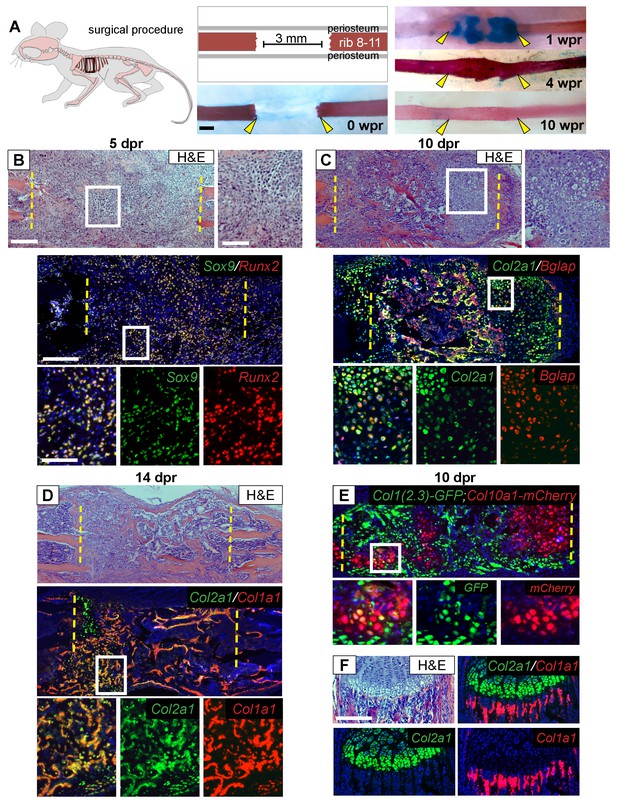
Regeneration involves skeletal cells with hybrid osteochondral properties.
(A) Schematic of the murine rib resection model. A 3 mm bone segment is resected from one rib (8-11), while the periosteum is carefully released and left in the mouse. Alizarin red and alcian blue whole mount staining indicates that repair occurs through a cartilage intermediate. The images show the outcome immediately after the resection at 0 wpr (weeks post resection, n = 2). At 1 wpr alcian blue positive material is evident between the cut ends (n = 3), by 4 wpr (weeks post resection) the lesion is fully-spanned by a mineralized callus (n = 2), while by 10 wpr remodeling has occurred (n = 2). (B–D) Histological sections stained with hematoxylin and eosin (H and E) (n > 5 for each time point) and near-adjacent double fluorescent RNA in situ hybridization (RNA-ISH) assays confirm the presence of a cartilage intermediate and show expression patterns in the repair callus. (B) At 5 dpr (days post resection) mesenchymal-like progenitor cells have moved into the resected region and are positive for the expression of both Sox9 (green) and Runx2 (red). The enlarged boxes with the separated color channels show co-expression in many of the cells within the resected region (overlap is yellow) (n = 3). (C) By 10 dpr bone and cartilage formation spans the resected region. Many cells that mediate repair express both the chondrocyte-associated gene Col2a1 (green), as well as the osteoblast-associated gene Bglap (red). The enlarged boxes show cells that have chondrocyte morphology expressing both Col2a1 and Bglap (n = 3). (D) At 14 dpr trabecular bone spans almost the entire resected region with only a small amount of cartilage at the cut ends; cells expressing both Col2a1 (green) and Col1a1 (red) are widespread. The enlarged boxes show the surface of newly formed trabecular bone where cells can be found that co-express Col1a1 and Col2a1 (n = 4). (E) At 10 dpr, animals double transgenic for Col1(2.3)-GFP;ColX-mCherry have mCherry (red) positive cells that are also expressing the osteoblast-specific reporter for Col1 (green). (F) Expression of Col2a1 (green) and Col1a1 (red) of the rib growth plate from an uninjured animal does not show a high degree of overlap (n = 5). Col2a1 is highly expressed in chondrocytes of the growth plate but not in osteoblasts forming new bone, while Col1a1 is highly expressed in the osteoblasts/cytes below the growth plate but not in cartilage cells. Scale bar for A = 500 microns, B-F = 200 microns, enlarged box H and E = 100 microns, enlarged box = 50 microns.
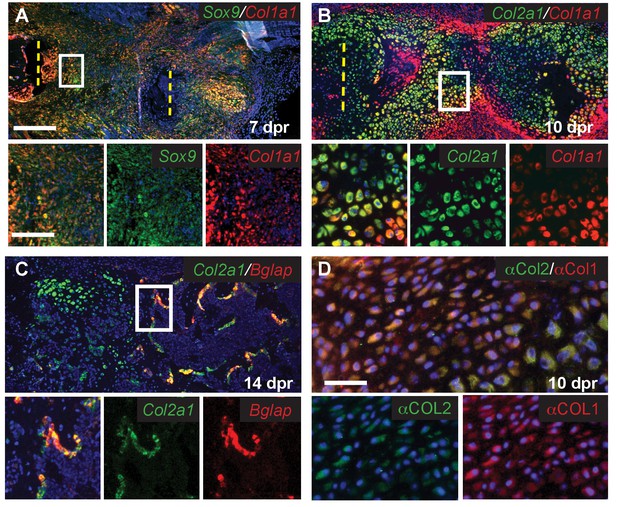
Analysis of hybrid skeletal cells.
(A) At 7 dpr the early progenitors not only express Sox9 (green) but also express the osteoblast-associated gene Col1a1 (red). (B) At 10 dpr, most of the callus cells co-express the chondrocyte-associated gene Col2a1 (green) and osteoblast-associated gene Col1a1 (red), especially those cells with chondrocyte morphology (n = 4). (C) At 14 dpr, cells in the newly formed trabecular bone co-express Col2a1 (green) and Bglap (red). (D) At 10 dpr, cells can be found that co-express COL1 (red) and COL2 (green) protein. Scale bar A-C = 200 microns, enlarged box = 50 microns; D = 50 microns.
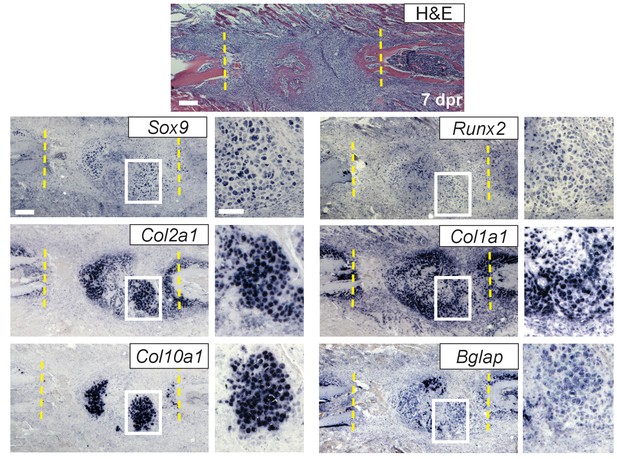
Colormetric RNA-ISH.
Near-adjacent sections of H and E staining and colormetric RNA-ISH of a 7 dpr repair callus suggests that similar cell populations express genes associated with cartilage (Sox9, Col2a1, Col10a1) as well as bone (Runx2, Col1a1, Bglap). The cartilage genes Sox9, Col2a1, and Col10a1 are strongly expressed in cells with chondrocyte morphology with Col10a1 expression most prevalent in the more mature, hypertrophic chondrocytes. The expression of Runx2, Bglap, and Col1a1 (genes associated with bone production) could also be found in these populations along with their expected expression in cells located in newly formed bone (n = 2 full series). Scale bar = 200 microns, enlarged box = 50 microns.
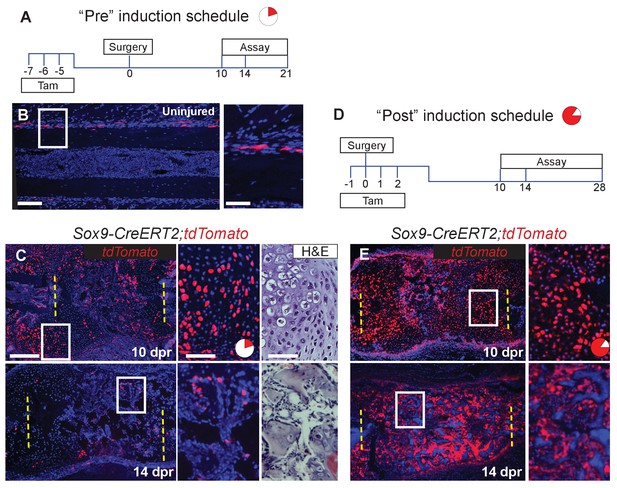
Sox9-CreERT2 marks cells that participate in repair.
(A) To target a sub-population of callus cells that arise from the periosteum, Sox9-CreERT2;tdTomato mice were injected for 3 consecutive days starting 7 days before analysis or before surgery. This tamoxifen induction schedule is referred to as ‘Pre’. The pie chart is a visual representation of number of cells affected in the callus at 10 dpr (red) and will be used throughout the figures as a reminder. (B) In uninjured Sox9-CreERT2;tdTomato mice, Sox9+ cells can be observed within the periosteum of the diaphysis by immunofluorescence (IF) for the tdTomato protein (red) (n = 3). They make up 6 ± 0.3% of the periosteal cells. (C) To determine if these cells participate in repair, rib resections were performed. At 10 dpr, lineage tracing of the Sox9+ cells show that the tdTomato+ expressing cells, 22 ± 1.3%, contribute to both cartilage and bone (n = 3) (IF for tdTomato). The enlarged images show chondrocytes, some of which are positive for tdTomato along with a near-adjacent section showing the histology of the area. At 14 dpr, tdTomato-expressing cells can be seen contributing to the trabecular bone. The enlarged image shows cells lining the trabeculae that are positive for tdTomato. (D) To activate Cre in a larger percentage of cells that build the repair callus, Sox9-CreERT2;tdTomato mice were injected with tamoxifen for 4 consecutive days, starting the day before surgery took place, the day of surgery, and for 2 more days. This tamoxifen induction schedule is referred to as ‘Post’. Another pie chart is used to visualize that a much greater portion of the callus is affected using this tamoxifen induction schedule at 10 dpr (85.5 ± 2.8%). (E) At 10 dpr, tdTomato+ cells are present in developing cartilage and bone, as well as in the periosteum surrounding the callus (native tdTomato fluoresence from a cryosection). The enlarged panel shows cells with chondrocyte morphology that are tdTomato+. At 14 dpr, tdTomato+ cells can be seen building new trabecular bone. In comparison to the Pre induced mice, these mice have significantly more tdTomato+ cells within the repair callus (n > 2 for all time points). Scale bar B = 100 microns, enlarged box = 50 microns; C, E = 200 microns.
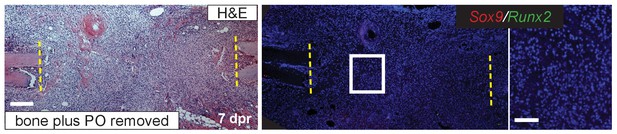
Periosteal removal.
Resections were performed in which both the bone and periosteum (PO) were removed. No cartilage is evident at 7 dpr. Sox9 (red) and Runx2 (green) expression is not readily detectable. Scale bar = 100 microns; enlarged box = 50 microns.
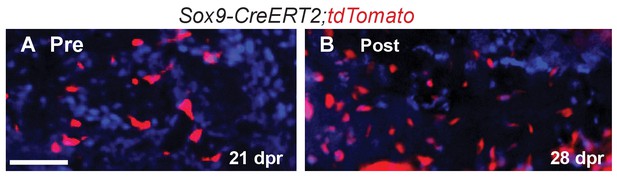
Lineage tracing at later time-points.
(A) Using the ‘Pre’ induction schedule, we observed tdTomato cells contributing to the trabecular bone during remodeling at 21 dpr. (B) Using the ‘Post’ induction schedule, tdTomato cells can still be observed at 28 dpr embedded in the remodeled bone. Scale bar = 50 microns.
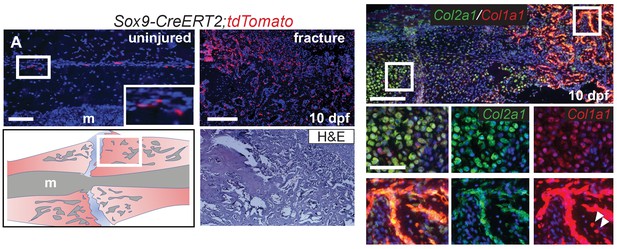
Femur fracture.
(A) In the femur of Sox9-CreERT2;tdTomato mice, Sox9+ cells can be observed within the periosteum by immunofluorescence (IF) for the tdTomato protein (red) (n = 2). Femur fractures were performed on these mice under the Pre tamoxifen induction schedule and the indicated portion of the callus (diagram) is shown enlarged at 10 days post fracture (dpf). Lineage tracing of the Sox9+ periosteal subpopulation shows that they participate in femur fracture repair. A near adjacent H and E stained section shows that many of the lineage traced cells participate in making new trabecular bone ((n = 2), m: marrow cavity). (B) Double RNA-ISH of Col1a1 (red) and Col2a1 (green) shows high co-expression in the femur fracture callus at 10 dpf. The enlarged boxes show that both chondrocytes and trabecular bone in the callus have strong co-expression of both cartilage and bone associated genes (n = 4). White arrowheads indicate osteocytes within bone matrix that still express Col1a1. Scale bar A, B = 200; enlarged boxes = 50 microns.
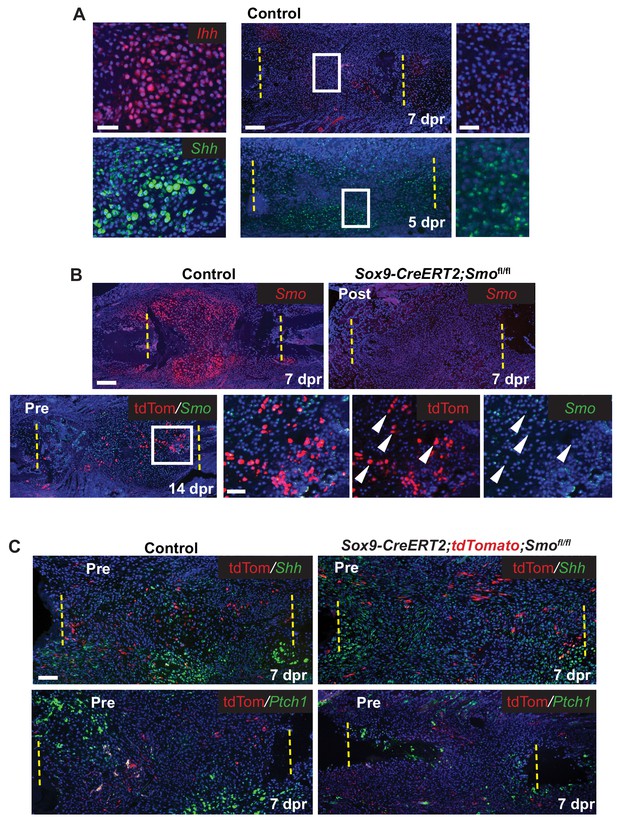
Hh signaling during rib repair.
(A) Expression of Ihh (red) and Shh (green) is evident in differentiating cartilage cells. At earlier stages, prior to cartilage formation, Ihh is hard to detect even at 7 dpr, while Shh is expressed at 5 dpr in many cells across the lesion. (B) Many cells express Smo (red) in the control callus at 7 dpr, while in the Post KO, Smo expression is not detectable. Fluorescent RNA-ISH for Smo (green) combined with IF for tdTomato (red) shows that when using the Pre induction schedule, most Tdtomato+ cells are negative for Smo expression, while many non-Sox9-positive lineage cells still express Smo. White arrow heads indicate tdTomato+ cells that are negative for Smo expression. (C) IF for the tdTomato protein (red) in combination with RNA-ISH for Shh and Ptch1 (green) at 7 dpr. In control mice, Shh and Ptch1 expression can be seen in many cells across the callus but most strongly in the chondrocytes. Cells neighboring tdTomato+ cells do not have strong Ptch1 expression. In Pre KO mice, tdTomato+ cells can be seen throughout the callus. Ptch1 expression is strongest in the small regions of cartilage that form at the cut ends. Scale bar A = 200 microns, enlarged boxes = 50 microns; B = 200 microns, enlarged boxes = 50 microns; C = 100 microns.
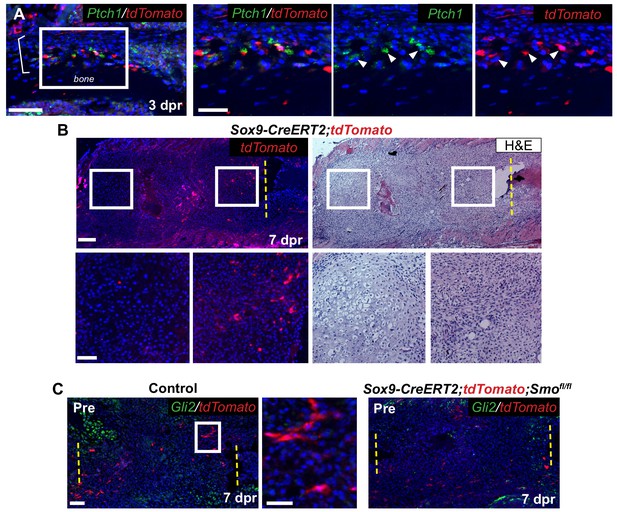
Characterization of Tdtomato+ cells during repair.
(A) RNA-ISH for Ptch1 (green) shows expression in tdTomato+ cells in the thickened periosteum (bracket) at 3 dpr close to the cut end. (B) Near-adjacent sections of IF for tdTomato (red) and H and E straining. At 7 dpr there are tdTomato+ cells seen throughout the callus, a majority of them have not yet differentiated into chondrocytes. The enlarged boxes of the IF and H and E on the left show a region of differentiated chondrocytes with no tdTomato+ cells. The enlarged boxes on the right show a region with many tdTomato+ cells that are less mature. (C) IF for the tdTomato protein (red) in combination with RNA-ISH for Gli2 (green) at 7 dpr. In control mice Gli2 expression can be seen most strongly in areas where chondrocytes are differentiating. Cells neighboring tdTomato+ cells do not have strong Gli2 expression. In Pre KO calluses, tdTomato+ cells can still be seen throughout the callus. Some Gli2 expression can be seen in the small regions of cartilage that form at the cut ends. Scale bar A = 100 microns, enlarged boxes = 50 microns; B = 200 microns, enlarged boxes = 100 microns; C = 100 microns, enlarged boxes = 25 microns.
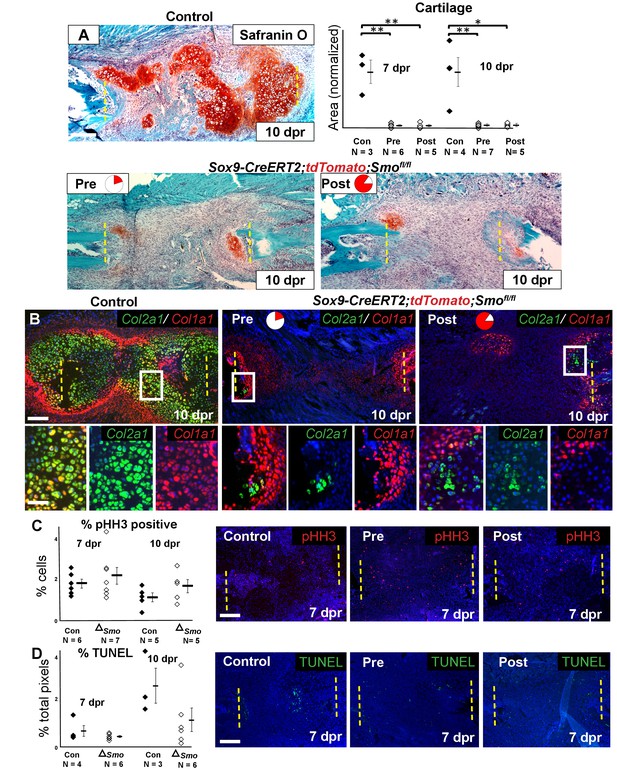
Requirement of Hh signaling for rib callus formation.
(A) Safranin O staining was used to visualize cartilage formation. At 10 dpr, the control callus has significantly more cartilage then both the Pre KO and Post KO repair calluses. In the graph showing the quantification of cartilage based on Safranin O staining at both 7 and 10 dpr, data are presented as compared to the average of controls, normalized to 1. When comparing both the Pre and Post KO to the control, the difference in the amount of cartilage is statistically significant, but between the Pre and Post KO, there is no statistically significant difference. See Figure 4—source data 1. (B) Double fluorescent RNA-ISH of Col1a1 (red) and Col2a1 (green) expression shows that there are fewer hybrid cells that mediate large scale repair in the KO calluses when compared to the control. Most of the cells in the control callus express high levels of both Col1a1 and Col2a1, while in both Pre and Post KO calluses many of the chondrocytes only express Col2a1. The enlarged boxes show cells with chondrocyte morphology with color channels merged and separated. (C) IF against pHH3 was used to mark cells undergoing proliferation. The percentage of positive cells in the callus was calculated vs. the total number of callus cells. No statistically significant difference between the control (n = 6, 5) and the Sox9-CreERT2; Smofl/fl (n = 7, 5) mice at 7 or 10 dpr (p=0.460 and 0.210 respectively) was evident. Representative panels are shown. See Figure 4—source data 2. (D) TUNEL staining was used to detect apoptotic cell death at 7 and 10 dpr. The graph shows the percentage of green pixels, in comparison to total pixels in the callus area. There is no statistically significant difference between the control (n = 4,3) and Sox9-creERT2;Smofl/fl (n = 6,6) mice at 7 and 10 dpr (p=0.243 and 0.141 respectively). Representative panels are shown. See Figure 4—source data 3. Whisker bars are mean ± SEM. Statistical differences were determined using the unpaired t test. *p value < 0.005 **p value < 0.001; Scale bar A = 200 microns, B = 50 microns, enlarged box = 25 microns, C, D = 200 microns.
-
Figure 4—source data 1
Quantification of Safranin O.
Data collected for the quantification of Safranin O shown in Figure 4A.
- https://doi.org/10.7554/eLife.40715.015
-
Figure 4—source data 2
Quantification of pHH3.
Data collected for the quantification of pHH3 shown in Figure 4C.
- https://doi.org/10.7554/eLife.40715.016
-
Figure 4—source data 3
Quantification of TUNEL positivity.
Data collected for the quantification of TUNEL positivity shown in Figure 4D.
- https://doi.org/10.7554/eLife.40715.017
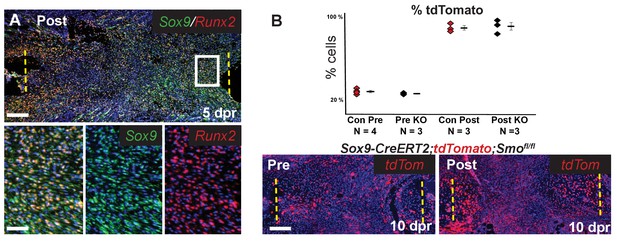
Characterization of Smo knock-out calluses.
(A) At 5 dpr, in Post KO mice, the progenitors that fill the resected region express both Sox9 (green) and Runx2 (red), as seen in the control animals. (B) The percentage of tdTomato+ cells within the callus in Pre KO and Post KO mice is compared to controls at 10 dpr. In Pre-induced control mice 22.8 ± 1.3% of the cells in the callus were tdTomato+ while in the Pre KO mice 20.6 ± 0.44% of the cells were tdTomato+. In Post induced control mice 85.5 ± 2.8% of the callus cells were tdTomato+ while in the Post KO 87 ± 3.9% of the cells were tdTomato+. There was no statistically significant difference between controls and KOs. (Pre-induction p=0.262 and Post-induction p=0.757). Panels show lineage tracing of tdTomato+ cells from Pre KO and Post KO mice demonstrating that the majority of cells null for Smo are still present and participate. See Figure 4—figure supplement 1—source data 1. Scale bar A, B = 200 microns, enlarged = 75 microns.
-
Figure 4—figure supplement 1—source data 1
Quantification of cells that are tdTomato+.
Data collected for the quantification of cells that are tdTomato+ shown in Figure 4—figure supplement 1B.
- https://doi.org/10.7554/eLife.40715.014
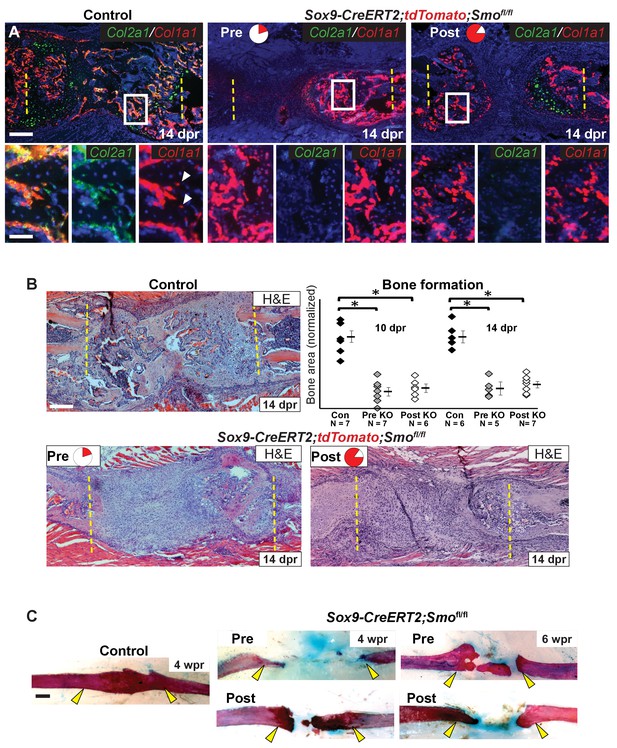
Hh signaling is required for bone formation.
(A) Double fluorescent RNA-ISH of Col1a1 (red) and Col2a1 (green). The enlarged boxes show an area of newly formed trabecular bone in separate channels. In both KO contexts, the cells building the trabecular bone express Col1a1 at high levels, but are largely negative for the expression of Col2a1. While, in the control, cells lining the trabecular bone express high levels of both. White arrowheads point to differentiating osteocytes that still express Cola1. (B) H and E staining at 14 dpr shows the histology of the repair callus in control, Pre, and Post KO animals. Both KO mice have much less bone than in controls and many of the cells that have entered the lesion have a progenitor-like morphology. Bone formation was quantified based on histology and the data is shown compared to the average of controls which has been normalized to one in the graph. At both 10 and 14 dpr, both the Pre KO and the Post KO have significantly less bone when compared to the controls. No statistically significant difference is seen when comparing the Pre to the Post KO. See Figure 5—source data 1. Whiskers show mean ± SEM. Statistical differences were determined using the unpaired t test. *p value < 0.0002 (C) Alizarin red and alcian blue whole mount staining show that at 4 wpr, the resected region is fully spanned by mineralized material in control mice (n = 3), while both the Pre KO and Post KO animals fail to heal. Similar results can still be seen at 6 wpr. Scale bar A, B = 200 microns, enlarged box = 50 microns, C = 500 microns.
-
Figure 5—source data 1
Quantification of bone.
Data collected for the quantification of bone shown in Figure 5B.
- https://doi.org/10.7554/eLife.40715.020
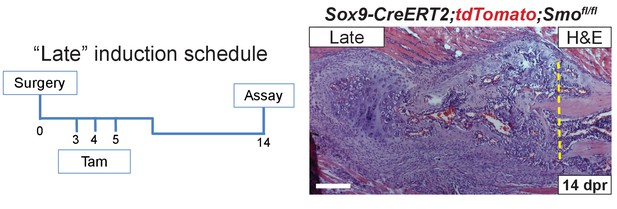
Late KO of Smo.
‘Late’ tamoxifen induction schedule: Sox9-CreERT2;tdTomato;Smofl/fl mice were injected with tamoxifen days 3–5 post resection. H and E staining at 14 dpr shows ample trabecular bone and cartilage formation. Although repair is delayed when compared to control mice, there is much more bone and cartilage formation when compared to either the Pre or Post KO calluses at 14 dpr (n = 3).
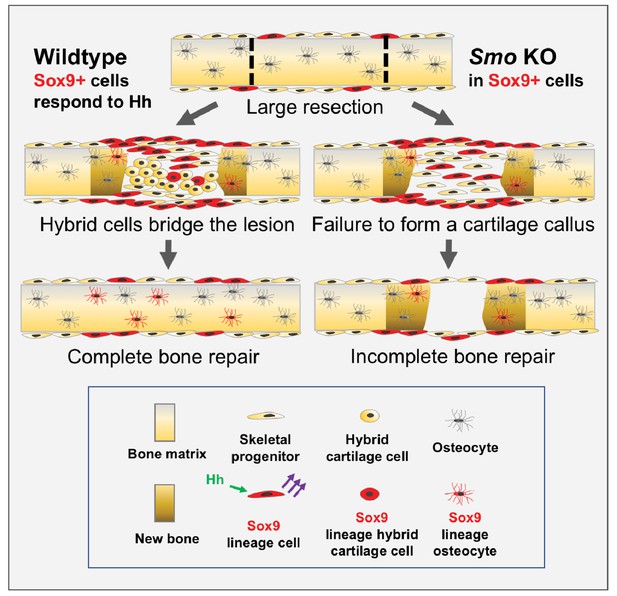
Model for large-scale bone repair.
In wildtype animals, represented by the left side of the diagram, extensive gaps in the mouse rib (as defined by the hatched lines) can naturally regenerate. A Sox9-expressing periosteal subpopulation (indicated in red) along with other skeletal progenitors (yellow) proliferate and migrate into the lesion. These Sox9+ lineage cells require Hh signaling (green arrows) to be able to signal via a yet-to-be identified mechanism to neighboring cells (purple arrows). This signal induces neighboring cells to differentiate into a reparative callus with hybrid osteochondral qualities, leading to complete bridging and bone repair. Sox9+ lineage cells ultimately contribute to the callus and regenerated bone (indicated in brown) although they are represented in the minority. The right side of the diagram represents the outcome of a Pre regimen KO of Smo (tamoxifen administered prior to injury). When Smo is removed from the Sox9+ periosteal subpopulation prior to resection, the Sox9+ lineage cells can still contribute but are not activated and therefore do not relay a differentiation signal to neighboring cells. Thus, the entire callus fails to differentiate into a hybrid osteochondral callus. While some bone forms via direct ossification, this is not sufficient and healing fails.
Tables
| Reagent type | Designation | Source/ Reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (M. musculus) | B6.Cg-Tg;Col1a1*2.3-GFP1Rowe/J | JAX 013134 | MGI: 151234 | |
| Genetic reagent (M. musculus) | Tg;Col10a1-mCherry3Pmay/J | Maye et al., 2011 JAX 017465 | MGI: 5428042 | P Maye |
| Genetic reagent (M. musculus) | Sox9tm1(cre/ERT2)Haak | Soeda et al., 2010 | MGI: 4867441 | H Akiyama |
| Genetic reagent (M. musculus) | B6;129S6-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J | JAX 007905 | MGI: 3809524 | |
| Genetic reagent (M. musculus) | Smotm2Amc/J | JAX 004526 | ||
| Antibody | Rabbit polyclonal anti-Collagen I | Abcam ab34710 | 1:250 | |
| Antibody | Goat polyclonal anti-Collagen II | Southern Biotech 1320–01 | 1:200 | |
| Antibody | Rabbit polyclonal anti-pHH3 | Millipore 06–570 | 1:200 | |
| Antibody | Chicken polyclonal anti-mCherry | Novus Biological NBP2-25158SS | 1:200 Also detects tdTomato | |
| Antibody | Alexa Fluor 488 Donkey polyclonal anti Goat IgG (H and L) | Abcam ab150129 | 1:500 | |
| Antibody | Alexa Fluor 568 Goat polyclonal anti-Rabbit IgG (H + L) | ThermoFisher A-11011 | 1:250 | |
| Antibody | Alexa Fluor 568 Goat polyclonal anti-Chicken IgY (H and L) | Abcam ab175477 | 1:500 | |
| Antibody | Alexa Fluor 488 Donkey anti-Goat IgG (H and L) | Abcam ab150129 | 1:500 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.40715.023





