Rejection of immunogenic tumor clones is limited by clonal fraction
Figures
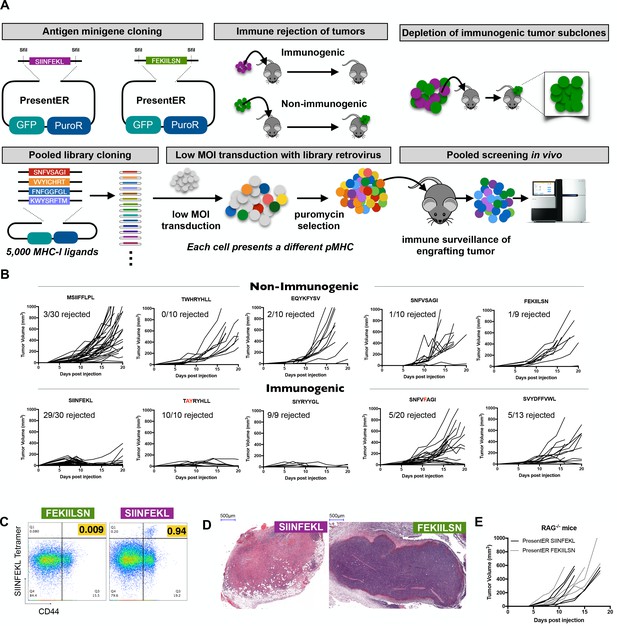
Cells expressing PresentER minigenes recapitulate the known immunogenicity of encoded antigens.
(A) A schematic of the cloning strategy of PresentER antigen minigenes along with the experiments performed in vivo in this manuscript. (B) 5 × 106 RMA/S cells expressing a PresentER minigene were injected subcutaneously into C57BL6/N mice and tumor size was monitored by caliper measurements. Top row: WT or non-binding peptide minigenes. Bottom row: mutated foreign peptide minigenes. All plots are compilations of several experiments. (C) Mice were injected with 5 × 106 RMA/S cells expressing PresentER-SIINFEKL or PresentER-FEKIILSN. Tumor draining lymph nodes were harvested 7 days later and stained with a SIINFEKL/H-2Kb tetramer and anti-CD44. (D) H and E staining of tumors expressing PresentER-SIINFEKL or PresentER-FEKIILSN. (E) RMA/S PresentER-SIINFEKL and PresentER-FEKIILSN cells were injected into Rag-/- mice and tumor growth was monitored.
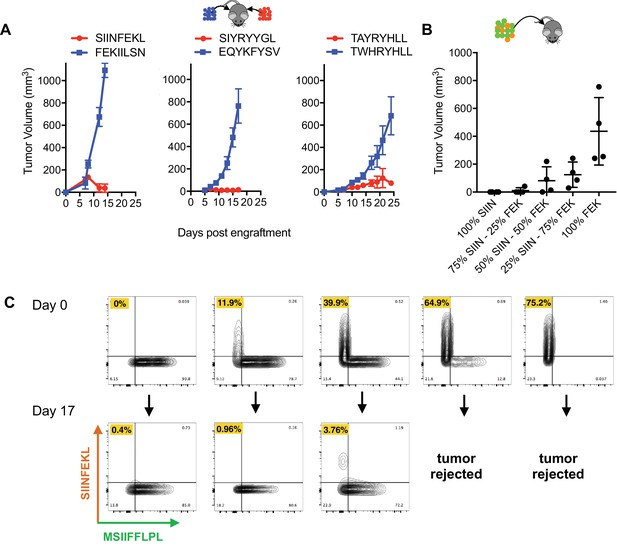
No abscopal effect and limited bystander killing of growing tumors in the RMA/S antigen minigene tumor model.
(A) Mice were injected simultaneously with 5 × 106 RMA/S cells expressing an immunogenic PresentER minigene on one flank and a non-immunogenic minigene on the contralateral flank: SIINFEKL/FEKIILSN (left; n = 3), SIYRYYGL/EQYKFYSV (middle; n = 5) and TAYRYHLL/TWHRYHLL (right; n = 5). Tumor growth curves are shown. (B) Mixtures of 5 × 106 RMA/S PresentER-SIINFEKL and PresentER-FEKIILSN injected into wild type mice (n = 4 per condition). Tumor sizes at day 15 are presented because some animals had to be sacrificed on day 17. (C) CD45.1+ mice were injected with 5 mixtures of PresentER-SIINFEKL (mCherry) and PresentER-MSIIFFLPL (eGFP) cells at several ratios. The top row shows the percentage of SIINFEKL and MSIIFFLPL cells at time 0. Tumors were harvested at day 17, enzymatically disaggregated and the percentage of CD45.2+ eGFP+ and CD45.2 mCherry+ cells were quantified (bottom row). The percentage of PresentER-SIINFEKL cells in each pretumor and tumor sample is highlighted in yellow. The two tumors with the highest percentage of PresentER-SIINFEKL cells were complete rejected and no tumor cells could be recovered.
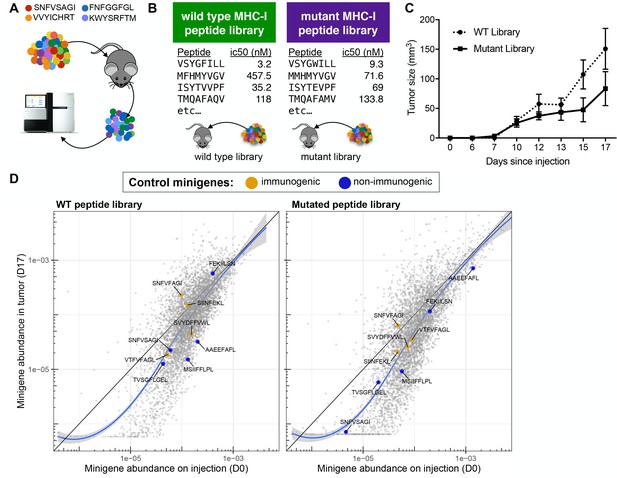
A drop-out screen for MHC-I peptide immunogenicity in immunocompetent mice.
(A) Schematic of the drop-out screen for MHC-I immunogenicity. Mice were injected with mixtures of RMA/S cells, each of which expresses a different peptide that also served as a genetic barcode for later deconvolution. (B) Two libraries of mouse MHC-I peptides were constructed. Left: Wild type peptides identified by searching the mouse proteome for peptides predicted to bind to MHC-I (NetMHCpan H-2Kb ic50 <500 nM). Right: single amino acid mutants of each of the wild type peptides. (C) C57BL6/N mice (n = 5 per group) were injected with 5 × 106 cells bearing libraries of wild type or mutant peptides and tumor growth monitored. (D) A scatter plot showing the average frequency of each minigene in the library before injection of the cells (x-axis) and after growth of the tumor in wild type mice (y-axis). The abundance of each minigene before injection (n = 3) is plotted on the x-axis while the abundance of each minigene after 17 days of growth in a wild type mouse is plotted on the y-axis (n = 5). Orange circles indicate positive control (immunogenic) minigenes; blue circles indicate negative control (non-immunogenic) minigenes. The straight black lines indicate x = y. LOESS (local best fit) lines are plotted in blue. The abundance of each minigene plotted is an average of all biological replicates.
-
Figure 3—source data 1
Abundance of each minigene in the pretumor and tumor samples.
- https://doi.org/10.7554/eLife.41090.006
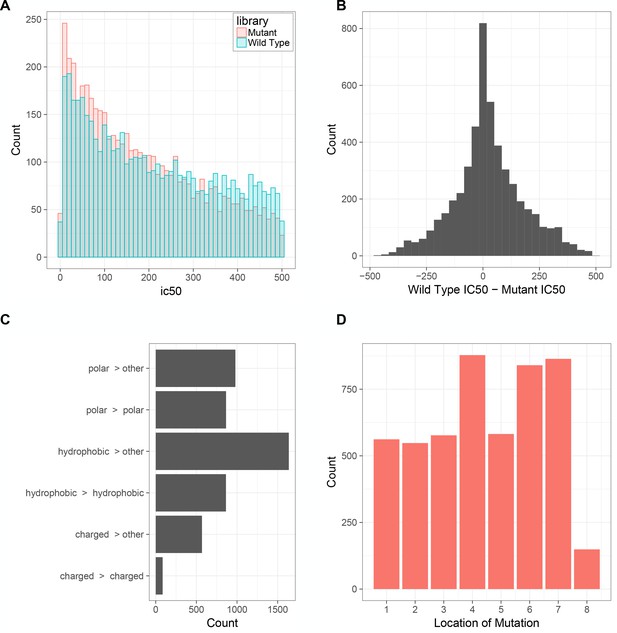
Characteristics of the wild-type and mutated antigen minigene libraries.
(A) Distribution of MHC-I affinities in the wild-type and mutated library. (B) Difference in NetMHCPan predicted H2-Kb affinity (ic50) between wild-type and mutant peptides (C) The type of amino acid changes in the mutant library (Polar: Q,N,H,S,T,Y,C,M,W; Charged: R,K,D,E; Hydrophobic: A,I,L,F,V,P,G). (D) The positions of the mutations in the mutant library with respect to the wild-type 8-mer.
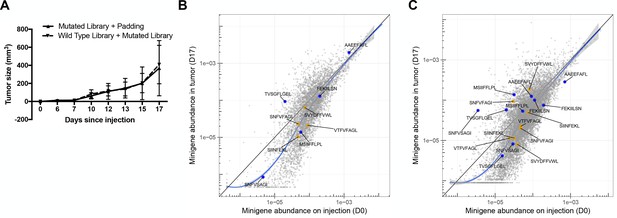
Five C57BL6/N mice were injected with 5 × 106 RMA/S cells bearing the mutated peptide library plus 1 × 106 untransduced RMA/S cells (‘padded’ mutant peptide library) to provide a buffer against bystander killing in the event that most of the cells in the tumor were immunogenic.
An additional five C57BL6/N mice were injected with a mixture of 107 wild type library and mutated library cells. (A) Growth curves for tumors bearing the ‘padded’ mutated peptide library and mutated peptide library cells. A scatter plot showing the average frequency of each minigene in the (B) ‘padded’ mutated peptide library or (C) mixed wild type and mutated peptide library before and after growth in wild type mice. The abundance of each minigene in culture (x-axis; n = 3) versus the abundance of each minigene after 17 days of growth in a wild type mouse (y-axis; n = 5). Orange circles indicate positive control (immunogenic) minigenes; blue circles indicate negative control (non-immunogenic) minigenes. The straight black lines indicate x = y. LOESS (local best fit) lines are plotted in blue.
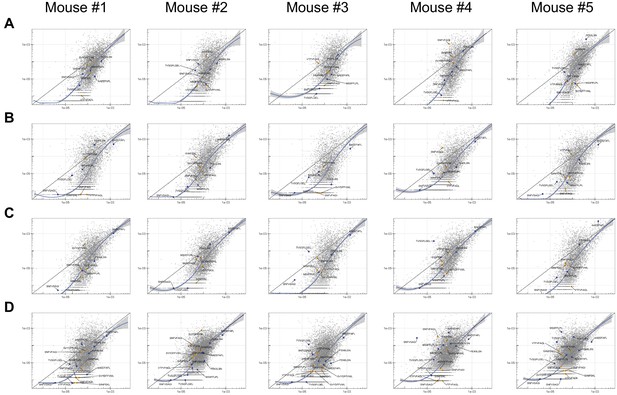
The abundance of each minigene in each mouse tumor after 17 days of growth in vivo (y-axis) compared to the abundance of each minigene in culture (x-axis) before injection across four groups of tumors: (A) wild type, (B) mutated, (C) ‘padded’ mutated and (D) mixed wild type and mutated peptide libraries.
The straight black lines indicate x = y. LOESS (local best fit) lines are plotted in blue.
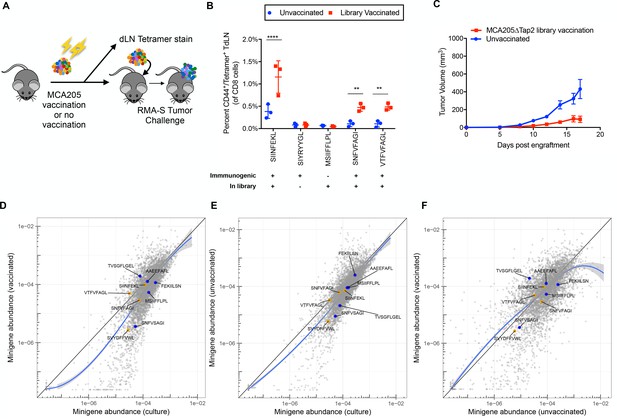
Vaccination of wild type mice with minigene library-expressing MCA205ΔTap2 cells leads to increased antigen-reactive T cells, but not increased immune surveillance
(A) A schematic of the vaccinations performed on C57BL/6N mice. 107 irradiated MCA205ΔTap2 cells expressing wild type library peptides were injected subcutaneously every six days (for a total of three vaccinations) into eight animals. On day 18, three mice from each group were sacrificed for tetramer analysis. Draining lymph nodes and splenocytes were stained with H-2Kb peptide tetramers. At day 18, the remaining five mice were challenged with 5 × 106 RMA-S cells expressing the library. (B) Splenocytes and draining lymph node cells from vaccinated animals were stained for CD8, CD44, and H-2Kb/peptide tetramers. Five control peptides were evaluated: four found in the library and one peptide not found in the library. The frequency of CD44/tetramer positive CD8 cells is reported. (C) Growth curves of RMA/S library tumors in in vaccinated or unvaccinated mice. (D-F) Average abundance of each minigene in cultured cells before injection into mice (x-axis) compared to minigene abundance in tumors harvested from vaccinated (n = 4; y-axis) (D) or non-vaccinated (n = 5; y-axis) (E) mice. Each circle is a minigene. Orange circles indicate positive control (immunogenic) minigenes; blue circles indicate negative control (non-immunogenic) minigenes. (F) Direct comparison of minigene abundance in tumors grown in vaccinated and non-vaccinated animals. The straight black lines indicate x = y. LOESS (local best fit) lines are plotted in blue.
-
Figure 4—source data 1
Abundance of each minigene in the tumors of vaccinated and non-vaccinated animals.
- https://doi.org/10.7554/eLife.41090.012
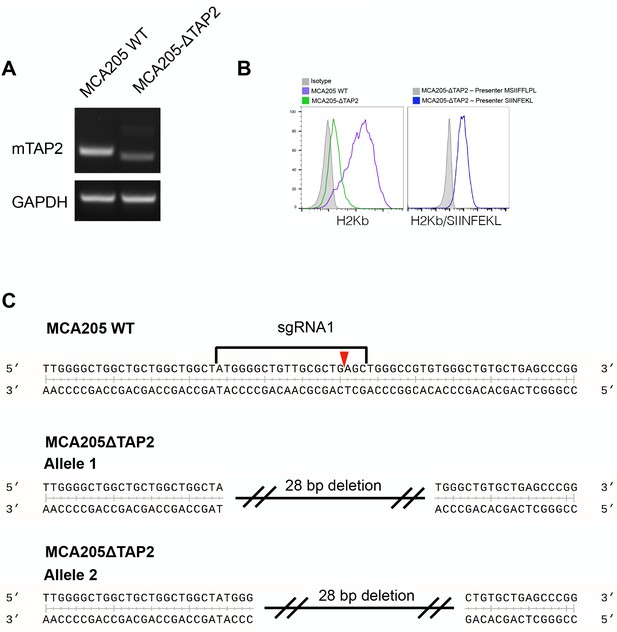
MCA205ΔTap2 cell line was generated by transient transfection of MCA205 cells with a plasmid encoding Cas9 and an sgRNA directed at Tap2.
A single cell clone with an INDEL in both alleles of Tap2 was selected and expanded. (A) RT-PCR of genomic DNA shows a smaller band than wild-type cells. (B) Left: Loss of surface H2-Kb in the ΔTap2 line. Right: transduction of MCA205ΔTap2 with PresentER-SIINFEKL or PresentER-MSIIFFLPL and staining with SIINFEKL/H2-Kb TCR mimic antibody shows Tap independent presentation of the SIINFEKL antigen. (C) Next generation sequence of the MCA205ΔTap2 single cell clone shows two different 28 bp deletions in both Tap2 loci.
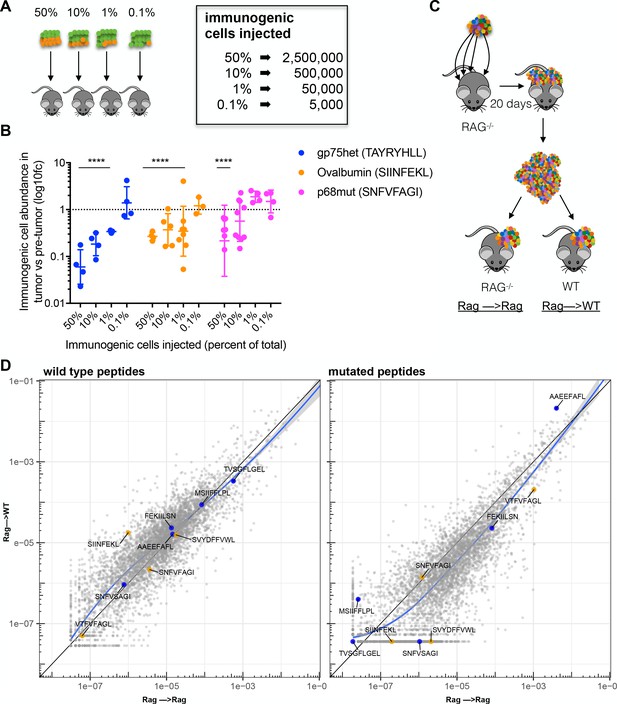
Immunosurveillance fails when tumor subclone frequency is low.
(A) Schematic and (B) results of experiments to detect antigen-specific immune surveillance thresholds. CD45.1+ C57BL/6N mice were injected with mixtures of immunogenic and non-immunogenic RMA/S cells. Mixtures were 50%, 90%, 99% or 99.9% non-immunogenic PresentER-MSIIFFLPL (mCherry) mixed with 50%, 10%, 1% or 0.1% cells expressing one of three different immunogenic minigenes (eGFP) noted in the inset. After 17 days in the mouse, tumors were enzymatically disaggregated, stained with CD45.2 and analyzed by flow cytometry. t-tests with the alternative hypothesis that log10 fold change is 0 were performed for each group. Some tumors were rejected entirely, and these were excluded from the presented depletion analysis. At least three tumors per group yielded sufficient numbers of cells to be confident that depletion had occurred. All groups marked with four stars reached a p-value of <0.001. (C) In order to overcome the hypothesized lack of immune surveillance at low minigene abundance, 2 Rag-/- mice were injected at 4 sites with 5 × 106 RMA/S library cells per site. Tumors were harvested after 20 days, minced and pooled. Approximately 1 mm3 of tumor fragments were implanted subcutaneously into the flank of either wild-type or Rag-/- mice and allowed to grow for 17 days. (D) The abundance of each minigene (average of 3 mice) in tumors transferred to WT mice (y-axis) compared to tumors transferred to Rag-/- mice (x-axis) is plotted. Each circle is a minigene. Orange circles indicate positive control (immunogenic) minigenes; blue circles indicate negative control (non-immunogenic) minigenes. The straight black lines indicate x = y. LOESS (local best fit) lines are plotted in blue.
-
Figure 5—source data 1
Abundance of each minigene in the tumors transferred from RAG to WT or RAG to RAG animals.
- https://doi.org/10.7554/eLife.41090.014
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (M musculus) | RMA/S | 10.1002/ijc. 2910470711 | RRID:CVCL_2180 | |
| Cell line (M musculus) | MCA-205 | PMID: 2303716 | MCA-205 | |
| Cell line (M musculus) | MCA205∆Tap2 | this paper | Generated using CRISPR/Cas9 described in materials/methods | |
| Recombinant DNA reagent | PresentER- SIINFEKL | this paper | Generated using procedure described in materials/methods | |
| Recombinant DNA reagent | PresentER- FEKIILSN | this paper | Generated using procedure described in materials/methods | |
| Recombinant DNA reagent | PresentER- MSIIFFLPL | this paper | Generated using procedure described in materials/methods | |
| Recombinant DNA reagent | PresentER- SNFVSAGI | this paper | Generated using procedure described in materials/methods | |
| Recombinant DNA reagent | PresentER- SNFVFAGI | this paper | Generated using procedure described in materials/methods | |
| Recombinant DNA reagent | PresentER- SIYRYYGL | this paper | Generated using procedure described in materials/methods | |
| Recombinant DNA reagent | PresentER- EQYKFYSV | this paper | Generated using procedure described in materials/methods | |
| Recombinant DNA reagent | PresentER- TWHRYHLL | this paper | Generated using procedure described in materials/methods | |
| Recombinant DNA reagent | PresentER- TAYRYHLL | this paper | Generated using procedure described in materials/methods | |
| Recombinant DNA reagent | LentiCRISPRv2 | Addgene | Addgene:52961 | |
| Strain, strain background (m musculus) | C57BL6/N | Envigo | Envigo:44 | |
| Strain, strain background (m musculus) | B6.SJL- Ptprca/BoyAiTac | Taconic Biosciences | Taconic:4007 F | |
| Strain, strain background (m musculus) | B6.129S6- Rag2tm1Fwa N12 | Taconic Biosciences | Taconic:RAGN12-F | |
| Antibody | anti-H-2Kb | Biolegend | Biolegend:116517 | used at 1:200 |
| Antibody | anti-SIINFEKL /H2kb | Biolegend | Biolegend:141605 | used at 1:200 |
| Antibody | anti-CD45.2 | eBioscience | eBioscience:17-0454-81 | used at 1:400 |
| Antibody | anti-CD8a | BD Pharmingen | BD Pharmingen:553030 | used at 1:200 |
| Antibody | anti-CD3 | BD Pharmingen | BD Pharmingen:553067 | used at 1:400 |
| Antibody | anti-CD44 | Biolegend | Biolegend:103012 | used at 1:400 |
| Other | H-2Kb SIINFEKL tetramer | NIH Tetramer Core | NIH Tetramer Core:K(b)/Ova.SIINFEKL | used at 10 nM |
| Other | H-2Kb SIYRYYGL tetramer | NIH Tetramer Core | NIH Tetramer Core:K(b)/SIYRYYGL | used at 10 nM |
| Other | H-2Kb MSIIFFLPL tetramer | NIH Tetramer Core | NIH Tetramer Core:custom | used at 10 nM |
| Other | H-2Kb SNFVFAGI tetramer | NIH Tetramer Core | NIH Tetramer Core:K(b)/mp68.SNFVFAGI | used at 10 nM |
| Other | H-2Kb VTFVFAGL tetramer | NIH Tetramer Core | NIH Tetramer Core:custom | used at 10 nM |
Additional files
-
Supplementary file 1
Tables of oligonucleotide sequences used in this manuscript, including qPCR primers, cloning, amplification and Illumina sequencing oligonucleotides.
- https://doi.org/10.7554/eLife.41090.015
-
Supplementary file 2
Metadata corresponding to the wild type and mutant PresentER minigene libraries: peptide, gene, NetMHCPan predicted H-2Kb ic50 and type of mutation.
- https://doi.org/10.7554/eLife.41090.016
-
Transparent reporting form
- https://doi.org/10.7554/eLife.41090.017





