The Mitotic Exit Network integrates temporal and spatial signals by distributing regulation across multiple components
Figures
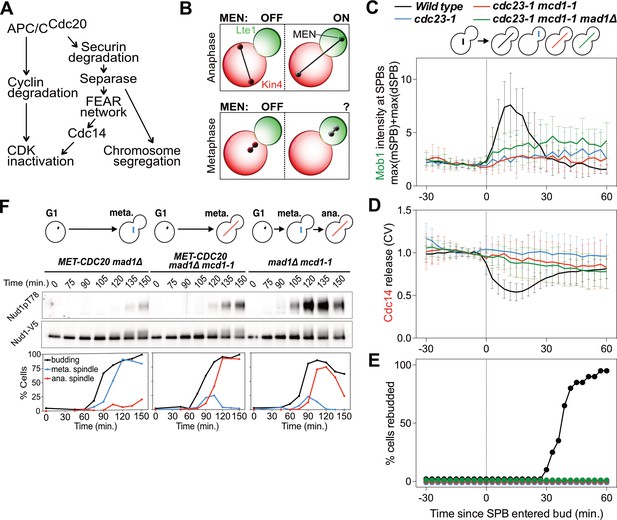
MEN activation requires APC/C activity.
(A) Control of the metaphase – anaphase transition. APC/CCdc20 has three roles in activating the MEN: cyclin degradation (1), activating the FEAR network (2), and triggering chromosome segregation (3). See text for more details. (B) The zone model explains spatial regulation of the MEN. Anaphase spindle elongation along the mother-bud axis drives one Tem1 bearing SPB into the bud. This removes the MEN GTPase from the inhibitory effect of Kin4 and brings it into contact with the MEN activator Lte1, activating the MEN. If spindle elongation fails to translocate a SPB into the bud the MEN does not become active. During metaphase SPBs can make excursions into the bud. We assayed MEN activity during these pre-anaphase excursions. (C, D, E) Wild type (A39323), cdc23-1 (A39374), cdc23-1 mcd1-1 (A40568), and cdc23-1 mcd1-1 mad1Δ (A39461) cells containing MOB1-eGFP, SPC42-mCherry, and CDC14-tdTomato were grown at 34°C and imaged every 3 min for 4–6 hr. Curves show mean and stdev. (n = 20 cells per condition; see Figure 1—figure supplement 2 for individual traces and sample images). Data were normalized to the average intensity measured during the 15 min prior to anaphase. Data were centered at the frame where at least one SPB had moved into the daughter for the first time. This centered timepoint was designated t = 3 min. (C) Mob1-eGFP intensity at mother and daughter cell SPBs [max(mSPB)+max(dSPB)]. (D) Cdc14 release from the nucleolus measured as coefficient of variation (CV = stdev/mean) of Cdc14 intensity within the cell. (E) Percent cells that exited mitosis and entered a new cell cycle as judged by their ability to form a new bud (rebudding). (F) MET-CDC20 mad1Δ (A37828), MET-CDC20 mad1Δ mcd1-1 (A37907), and mad1Δ mcd1-1 (A37739) cells containing NUD1-3V5 were arrested in G1 with α-factor (5 μg/ml) pheromone at room temperature in synthetic medium lacking methionine. After 3 hours cells were released into pheromone-free YEPD medium supplemented with 8 mM methionine at 37°C. Methionine was re-added every hour. The percentage of cells with buds (black), metaphase spindles (blue), and anaphase spindles (red) was determined at the indicated times. Nud1 T78 phosphorylation and total Nud1 levels were determined.
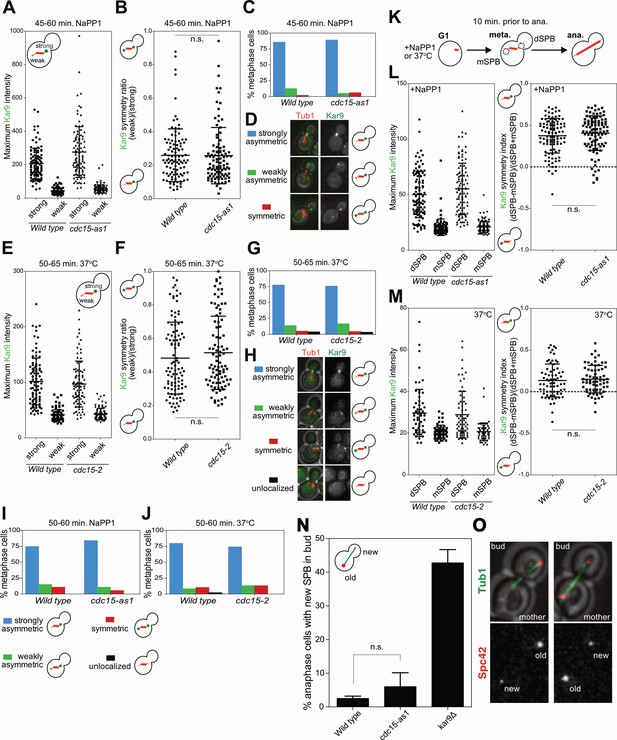
CDC15 mutants do not exhibit Kar9 localization and SPB inheritance defects.
(A–D, I, L) Wild type (A40512) and cdc15-as1 (A40524) cells containing an mCherry-TUB1 and KAR9-GFP fusion were treated with 10 μM 1-Na-PP1 for the indicated time period at 25°C on agar pads. (E–H, J, M) Wild type (A40530) and temperature sensitive cdc15-2 (A40529) cells containing the mCherry-TUB1 and KAR9-GFP fusions were heat shocked for the indicated period of time at 37°C on agar pads. (A, E) Cells were imaged and metaphase cells were identified by spindle morphology. Astral microtubule associated Kar9 intensity was determined for both SPBs in metaphase. The two SPBs in each metaphase cell were placed into one of two categories: SPB with high levels of Kar9 bound to emanating microtubules (strong) or the pole with low levels of Kar9 bound to emanating microtubules (weak). Mean and stdev are shown (n > 90 cells). (B, F) Quantifications from (A) and (E) were used to create a weak SPB/strong SPB localization ratio in (B) and (F), respectively. Mean, stdev, and results of t-test are shown. (C, G) Qualitative classification of Kar9 localization for cells quantified in (A) and (E). (D, H) Sample images of Kar9 localization categories quantified in (C) and (G). (I, J) Cells were imaged every 10 s for 100 s. Metaphase cells were identified by spindle morphology and Kar9 localization was quantified using the classifiers described in (H); n > 45 cells. (K–M) Cells were imaged every 5 m for 4 hr. G1 cells were followed until anaphase and Kar9 intensity was measured on astral microtubules 10 min prior to anaphase onset. Mother and daughter bound SPBs were identified during anaphase. Kar9 intensity and Kar9 symmetry index [(dSPB-mSPB)/(dSPB +mSPB)] was determined. Mean, stdev, and results of t-test are shown (n > 60 cells). (N, O) Wild type (A40559), cdc15-as1 (A40556), or kar9Δ (A40562) cells containing SPC42-mCherry and GFP-TUB1 were treated with 10 μM 1-Na-PP1 for 45 min at 25°C on agar pads. Cells were then imaged every 15 m for 3 hr. SPB inheritance was determined by the brightness of the Spc42-mCherry signal entering the bud (N); Mean and stdev of three experiments are shown, t-test. Sample images of Spc42-mCherry signal are shown in (O).
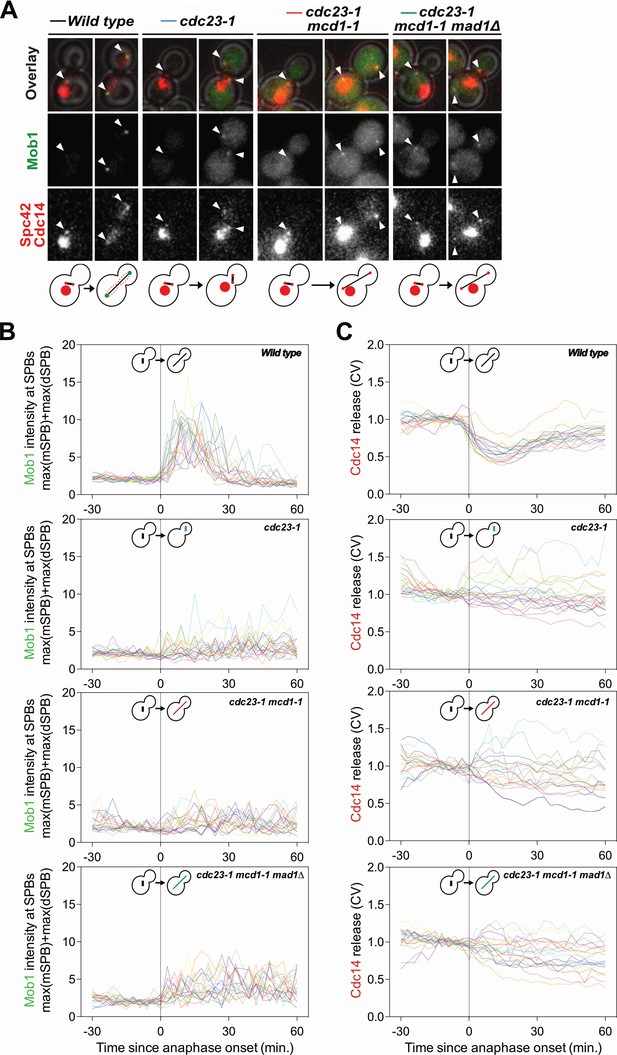
Mob1 localization to SPBs and release of Cdc14 from the nucleolus depends on the APC/C.
Related to Figure 1C–E. (A) Sample images. SPBs are marked with white arrowheads. Mob1 localization is shown in green, Spc42 and Cdc14 in red. (B, C) Traces of 20 individual cells from Figure 1C and D, respectively.
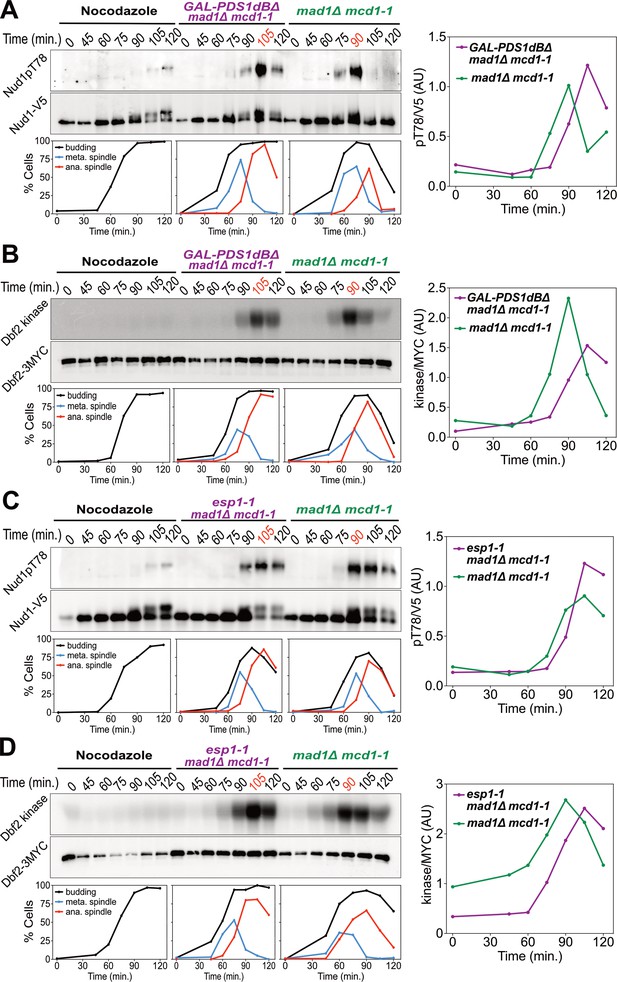
Securin degradation and Separase function promote MEN activation.
(A, B) GAL-PDS1dBΔ mad1Δ mcd1-1 and mad1Δ mcd1-1 cells containing (A) NUD1-3V5 (A37904 and A37739) or (B) DBF2-3MYC (A37497 and A37523) were arrested in G1 with α-factor pheromone at room temperature in YEP medium containing raffinose. After 3 hours cells were released into pheromone-free YEP medium containing raffinose and galactose at 34°C. A control sample was released from the G1 arrest into medium containing nocodazole (15 μg/ml) to arrest cells in metaphase. (C, D) esp1-1 mad1Δ mcd1-1 and mad1Δ mcd1-1 cells containing (C) NUD1-3V5 (A38079 and A37739) or (D) DBF2-3MYC (A37500 and A37523) were arrested in G1 with α-factor pheromone at room temperature in YEPD medium. After 3 hours cells were released into pheromone-free YEPD medium at 37°C. A control sample was released from the G1 arrest into medium containing nocodazole. (A–D) Percentage of cells with buds (black), metaphase spindles (blue), and anaphase spindles (red) was determined at the indicated times. (A, C) Nud1 T78 phosphorylation and total Nud1 levels were determined. (B, D) Dbf2 kinase activity and Dbf2 protein levels were determined.
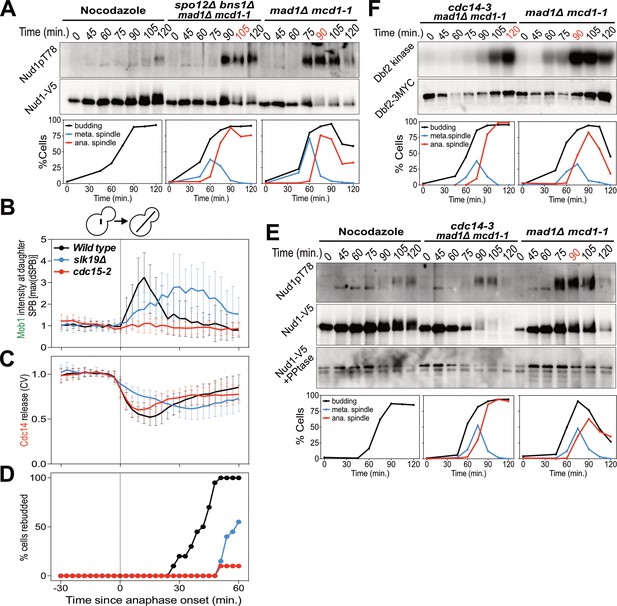
The FEAR network promotes MEN signaling by activating Cdc14.
(A) spo12Δ bns1Δ mad1Δ mcd1-1 (A38127) and mad1Δ mcd1-1 (A37739) cells containing NUD1-3V5 were arrested in G1 with α-factor pheromone at room temperature. (E, F) cdc14-3 mad1Δ mcd1-1 and mad1Δ mcd1-1 cells containing (E) NUD1-3V5 (A38112 and A37739) or (F) DBF2-3MYC (A37714 and A37523) were arrested in G1 with α-factor pheromone at room temperature. (A, E, F) After 3 hours cells were released into pheromone-free medium at 37°C. A control sample was released from the G1 arrest into medium containing nocodazole. The percentage of cells with buds (black), metaphase spindles (blue), and anaphase spindles (red) was determined at the indicated times. (A, E) Nud1 T78 phosphorylation and total Nud1 levels were determined. Note, Nud1 is hyperphosphorylated in cdc14-3 cells. Samples were therefore treated with Lambda Phosphatase to detect the protein (Nud1-V5 + PPtase, Western). (F) Dbf2 associated kinase activity and Dbf2 protein levels were determined. (B, C, D) Wild type (A39323), slk19Δ (A39378), and cdc15-2 (A39353) cells containing MOB1-eGFP, SPC42-mCherry, and CDC14-tdTomato were grown at 34°C and imaged every 3 min for 4 hr. Curves show mean and stdev. (n = 20 cells; see Figure 3—figure supplement 1 for individual traces). Data were normalized to the average intensity measured during the 15 min prior to anaphase. Data were centered at the first frame where anaphase onset (spindle >3 μm) was detected. This centered timepoint was designated t = 3 min. (B) Mob1-eGFP intensity at the daughter bound SPB[max(dSPB)]. (C) Cdc14 coefficient of variation (CV = stdev/mean) within the cell. (D) Percent cells that rebudded, indicating exit from mitosis and entry into the next cell cycle occurred.
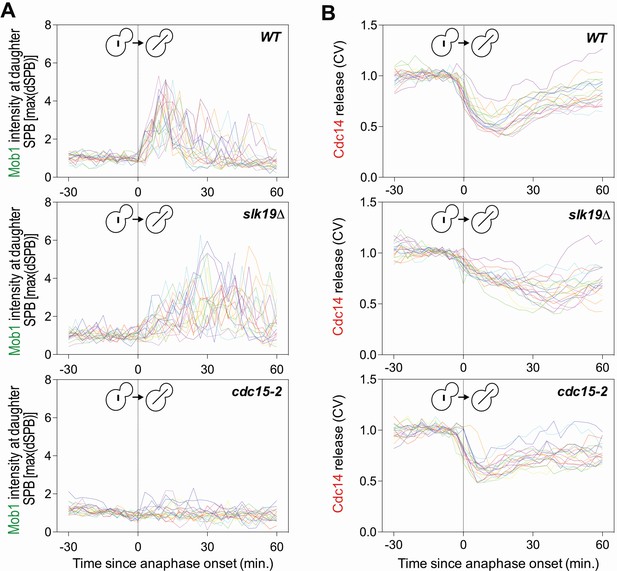
Deletion of SLK19 delays MEN activation and Cdc14 release from the nucleolus.
Related to Figure 3B–D. (A, B) Traces of 20 individual cells from Figure 3B and C, respectively. The FEAR network mutant slk19Δ delayed Mob1 localization to the daughter SPB (A) and delayed release of Cdc14 from the nucleolus (B). The MEN mutant cdc15-2 failed to localize Mob1 to the daughter SPB (A) and only transiently released Cdc14 from the nucleolus (B).
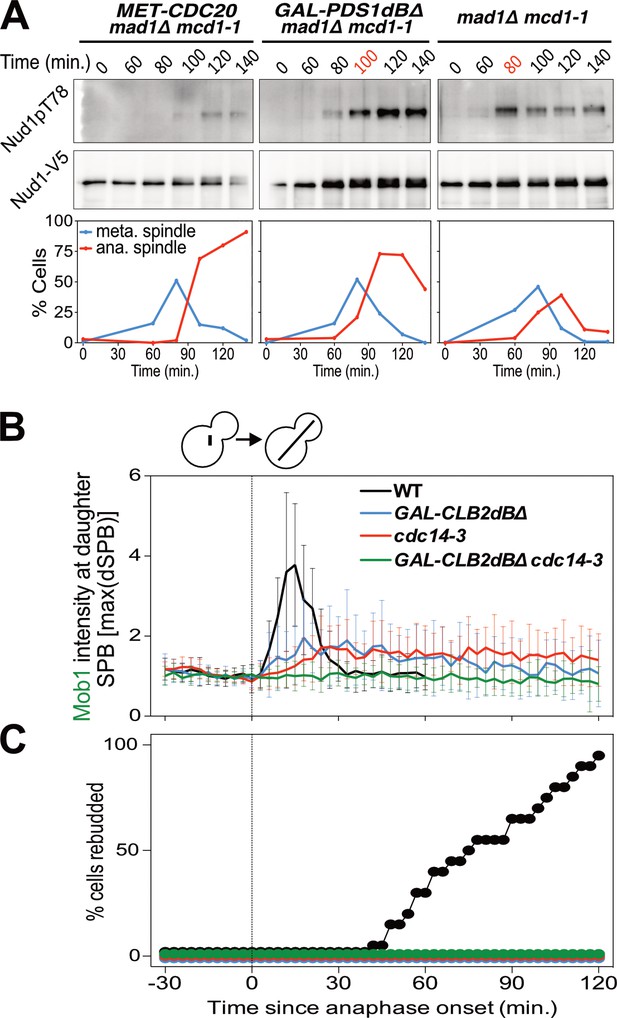
Mitotic CDKs antagonize the MEN.
(A) MET-CDC20 mad1Δ mcd1-1 (A37907), GAL-PDS1dBΔ mad1Δ mcd1-1 (A37904), and mad1Δ mcd1-1 (A37739) cells containing NUD1-3V5 were arrested in G1 with α-factor pheromone at room temperature in synthetic medium lacking methionine and supplemented with raffinose. After 3 hours cells were released into pheromone-free YEP medium containing raffinose, galactose and 8 mM methionine at 34°C. Methionine was re-added every hour. The percentage of cells with buds (black), metaphase spindles (blue), and anaphase spindles (red) as well as Nud1 T78 phosphorylation and total Nud1 levels were determined at the indicated times. (B,C) Wild type (A33715), GAL-CLB2dBΔ (A39542), cdc14-3 (A39541), and GAL-CLB2dBΔ cdc14-3 (A39540) cells containing MOB1-eGFP and SPC42-mCherry were grown at 34°C in SC medium containing raffinose and galactose and imaged every 3 min for 5 hr. Curves show mean and stdev. (n = 20 cells; see Figure 4—figure supplement 1 for individual traces). Data were normalized to the average intensity measured during the 15 min prior to anaphase. Data were centered at the first frame where anaphase onset (spindle >3 μm) was detected. This centered timepoint was designated t = 3 min. (B) Mob1-eGFP intensity at daughter bound SPB [max(dSPB)] was determined. (C) Percent cells that rebudded, indicating that exit from mitosis and entry into the next cell cycle had occurred.
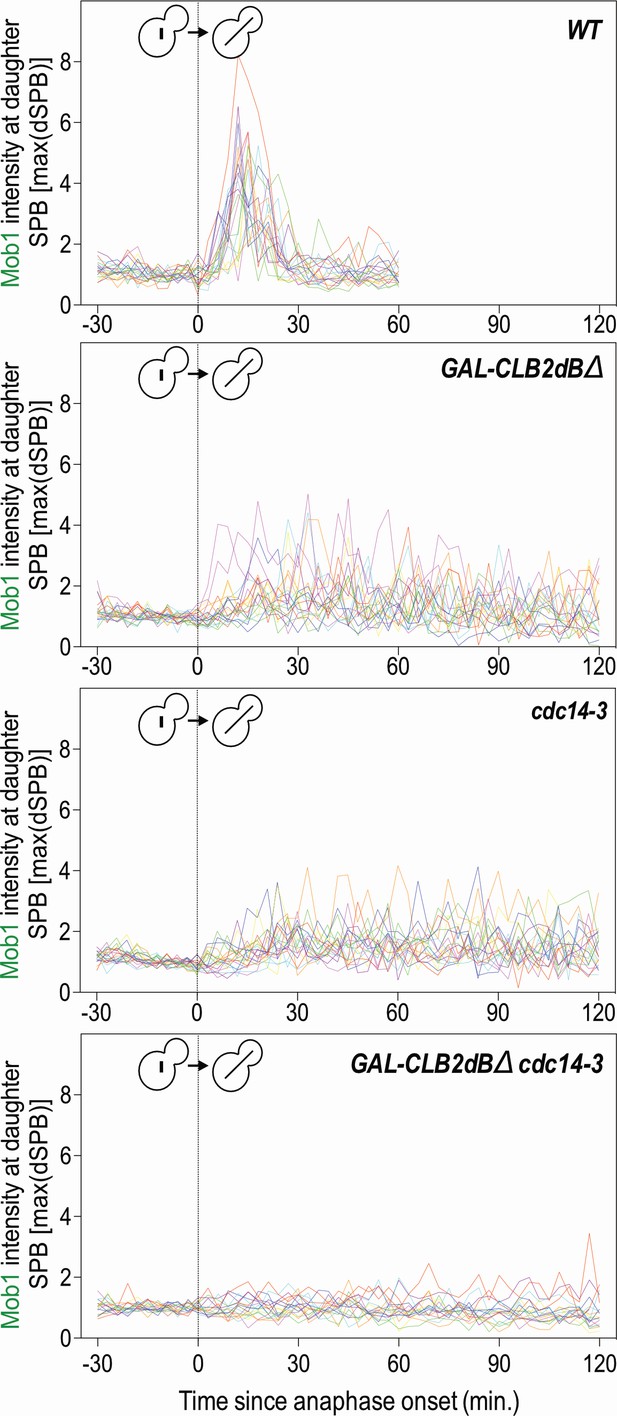
Clb2 degradation and Cdc14 released from the nucleolus by the FEAR network activate the MEN.
Related to Figure 4B–C. Traces of 20 individual cells from Figure 4B. GAL-CLB2dBΔ and cdc14-3 cells prevented Mob1 localization to the daughter SPB.
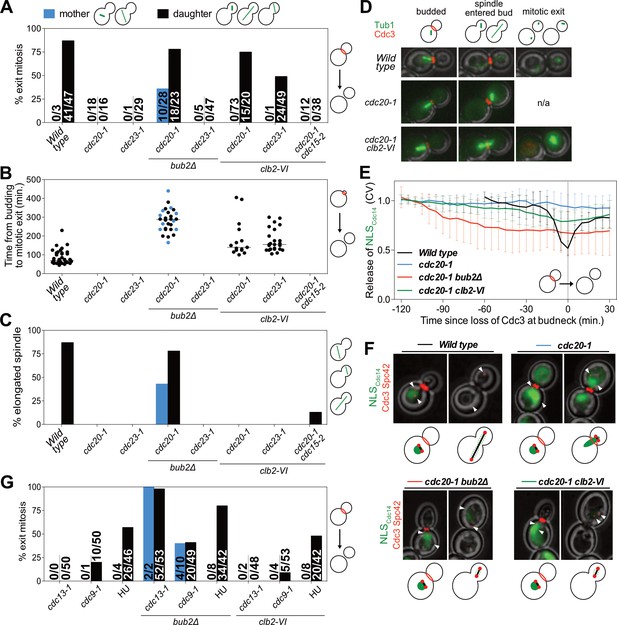
Mitotic CDKs prevent MEN activation in metaphase.
(A–D, G) The indicated strains containing mCherry-Cdc3 and GFP-Tub1 were grown at 37°C and imaged every 5 min for 5 (WT) or 8 (remaining strains) hours. (A) Spindle position was monitored and cells were apportioned by whether the spindle had entered the daughter cell (daughter) or remained in the mother cell (mother). Mitotic exit was measured by loss of Cdc3 from the bud neck. (B) Time from bud emergence until mitotic exit for the cells shown in (A). (C) Percent of cells from (A) that elongated their spindles (>3 μm). (D) Sample images for (A–C). Cdc3 is shown in red, microtubules in green. (E, F) Wild type (A40293), cdc20-1 (A40295), cdc20-1 bub2Δ (A40296), and cdc20-1 clb2-VI (A40292) cells containing mCherry-Cdc3, Spc42-mCherry, and NLSCdc14-GFP were grown at 37°C and imaged every 5 min for 4 (WT), or 8 (remaining strains) hours. Curves in (E) show mean and stdev. (n = 20 cells; see Figure 5—figure supplement 2 for individual traces). Data were normalized to the average intensity measured during the initial 20 min of the analysis. Data were centered so that loss of Cdc3 from the bud neck represents t = 0. cdc20-1 cells do not lose Cdc3 from the bud neck. We therefore analyzed cells for 2.5 hr after the spindle first moved into the bud. NLSCdc14-GFP localization was measured as coefficient of variation (CV = stdev/mean) within the cell. Sample images are shown in (F). Cdc3 is shown in red, microtubules in green. (G) Spindle position was monitored and mitotic exit was measured by loss of Cdc3 from the bud neck. 5 mg/ml hydroxyurea (HU) was used in indicated samples.
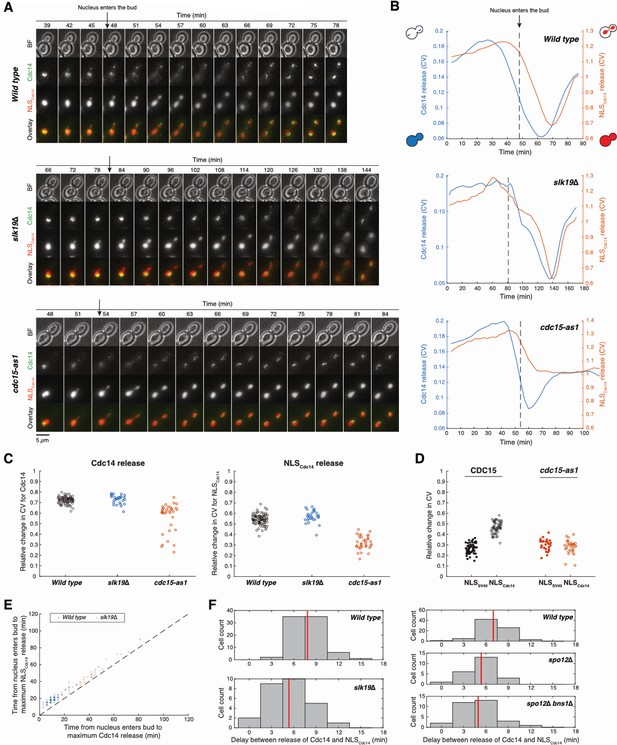
NLSCdc14 is a live-cell MEN activity reporter.
Related to Figure 5E and Figure 6D. (A) Representative images of Cdc14 (Cdc14-eGFP) and NLSCdc14 (NLSCdc14-TagRFP) release throughout the cell cycle in wild type (A39895), slk19Δ (A40177) and cells harboring the analog sensitive cdc15-as1 (A40095) treated with 1-NA-PP1. Arrows indicate the time-point when the nucleus enters the bud. (B) Quantification of Cdc14 release from the nucleolus and NLSCdc14 release into the cytoplasm of cells shown in (A). Release was quantified as the coefficient of variation (CV) of the signal intensity within the cell. Dashed lines indicate the time point when the nucleus enters the bud. (C) The extent of release of Cdc14 from the nucleolus (left) and NLSCdc14 translocation into the cytoplasm (right) in FEAR network (slk19Δ) and MEN (cdc15-as1) mutants. Relative changes in release is calculated as (CVmax-CVmin)/CVmax. NLSCdc14 release depended only on the MEN and not the FEAR network. (D) Comparison of relative changes in CV [(CVmax-CVmin)/CVmax] for a canonical nuclear localization signal NLSSV40 (2xmCherry-NLSSV40) and NLSCdc14 (NLSCdc14-GFP) during cell cycle in Wild type (CDC15, A40339) and MEN mutant (cdc15-as1, A40371) cells. The observed low level of relative changes in CV for NLSSV40 in both cases and NLSCdc14 in cdc15-as1 is due to nuclear division. (E) Time of Cdc14 and NLSCdc14 release relative to the time the nucleus enters the bud for Wild type and FEAR mutant (slk19Δ). The data points are set to a transparency value of 0.2 and thus the shade correlates with number of observations. The dashed line represents the identity line. Both releases were delayed in the FEAR network mutant. (F) Time delays between maximum release of Cdc14 and NLSCdc14 for Wild type and FEAR mutants (slk19Δ, spo12Δ, spo12Δbns1Δ). Red lines represent mean values. The reduced delay (increased correlation) between the two release events in FEAR network mutants is consistent with the interpretation that without the FEAR network both processes are the result of MEN activity.
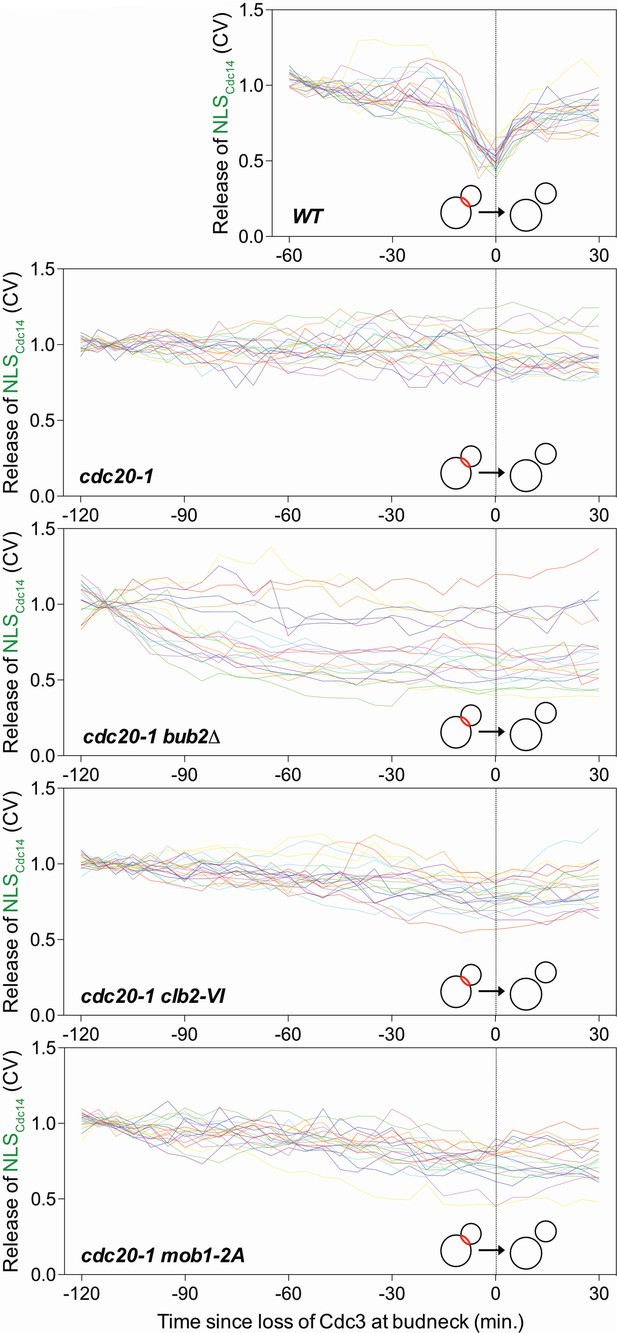
Clb2 degradation and Mob1 dephosphorylation promote MEN activation.
Related to Figure 5E,F and Figure 6D. Traces of 20 individual cells from Figure 5E and Figure 6D. In wild type cells diffusion of NLSCdc14 into the cytoplasm coincides with Cdc3 loss from the bud neck, indicating MEN activation. cdc20-1 cells never lost Cdc3 from the bud neck and did not release the NLSCdc14 into the cytoplasm. cdc20-1 bub2Δ, cdc20-1 clb2-VI, and cdc20-1 mob1-2A cells which lost Cdc3 from the bud neck also released NLSCdc14.
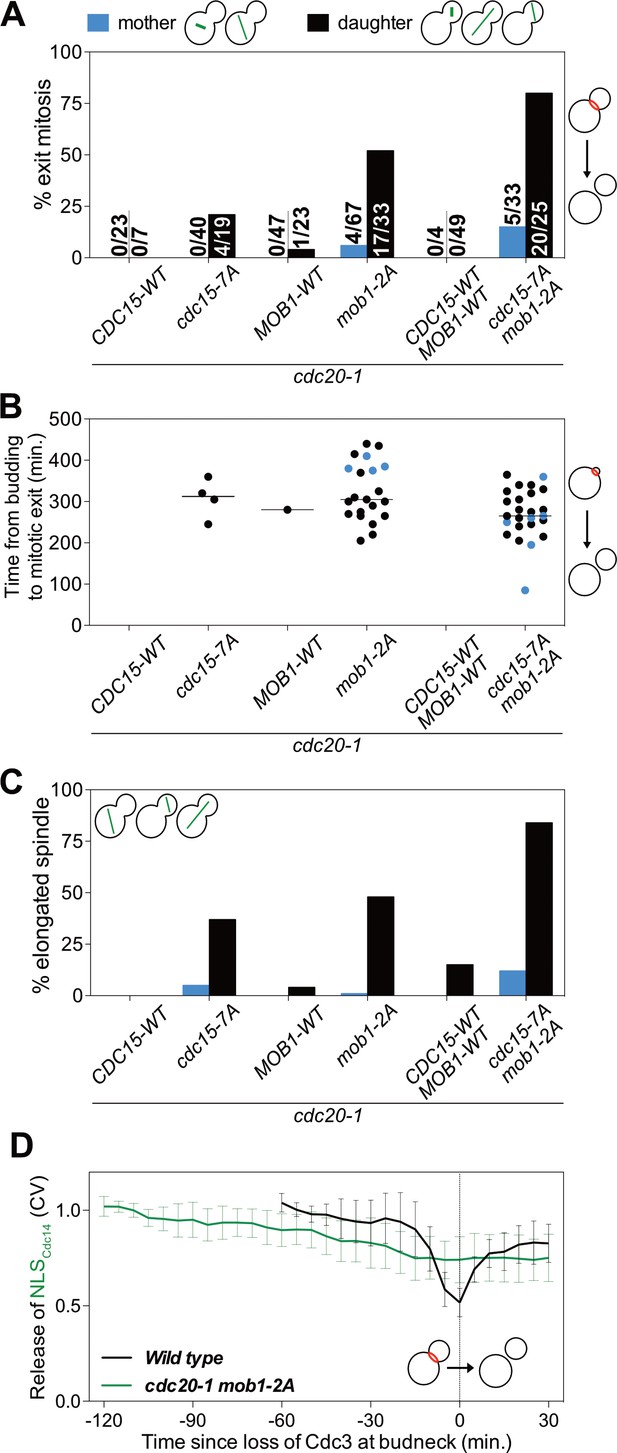
CDK activity inhibits the MEN by phosphorylating Cdc15 and Mob1.
(A–C) The indicated strains containing mCherry-Cdc3 and GFP-Tub1 were grown at 37°C and imaged every 5 min for 8 hr. (A) Spindle position was monitored and cells apportioned by whether the spindle entered the daughter cell (daughter) or remained in the mother cell (mother). Mitotic exit was measured by loss of Cdc3 from the bud neck. (B) Time from bud emergence until mitotic exit for the cells shown in (A). (C) Percent of cells from (A) that elongated their spindles (>3 μm). (D) Wild type (A40293), or cdc20 mob1-2A (A40292) cells containing mCherry-Cdc3, Spc42-mCherry, and NLSCdc14-GFP were grown at 37°C and imaged every 5 min for 4 (WT), or 8 (cdc20-1 mob1-2A) hours. Curves show mean and stdev. (n = 20 cells; see Figure 5—figure supplement 2 for individual traces). Data were normalized to the average intensity measured during the initial 20 min of the analysis. Data were centered so that loss of Cdc3 from the bud neck represents t = 0. Release of NLSCdc14-GFP was measured as coefficient of variation (CV = stdev/mean) of NLSCdc14-GFP intensity within the cell.
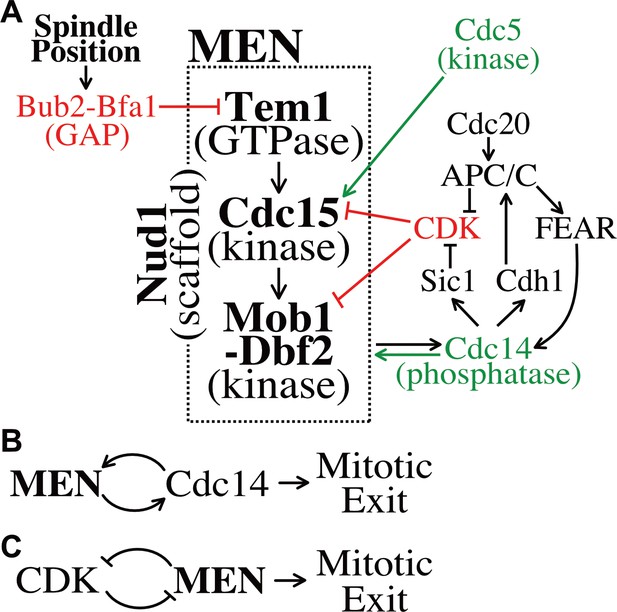
Temporal and spatial signals distribute regulation across multiple MEN components.
(A) The Mitotic Exit Network and its regulators. Direct positive regulators are shown in green, direct negative regulators in red. See text for details. (B) The MEN and Cdc14 act in a positive feedback loop to make mitotic exit ultrasensitive. (C) CDK and the MEN regulate each other through a double-negative feedback loop isolating mitotic exit ultrasensitivity to anaphase.
Additional files
-
Supplementary file 1
Yeast strains.
Yeast strains used in this study.
- https://doi.org/10.7554/eLife.41139.015
-
Supplementary file 2
Plasmids.
Plasmids used in this study.
- https://doi.org/10.7554/eLife.41139.016
-
Transparent reporting form
- https://doi.org/10.7554/eLife.41139.017





