Inadequate BiP availability defines endoplasmic reticulum stress
Figures
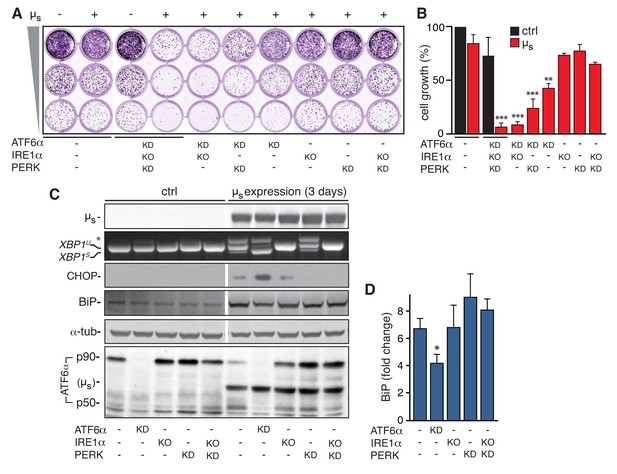
ATF6α is essential but IRE1α and PERK are dispensable for restoring ER homeostasis upon µs expression.
(A–D) In HeLa-µs cells, IRE1α was deleted (KO), and ATF6α and PERK were silenced (KD) either alone or in combination, or not (-), as indicated. (A) Cells were seeded upon 1:5 serial dilution into 24-well plates, and treated with 0.5 nM mifepristone (Mif) to induce expression of µs where indicated (+). After 7 days of growth, cells were fixed and stained with crystal violet. (B) Staining in (A) was quantitated as a measure for cell growth. Mean and s.e.m. are shown in a bar graph; n = 2. (C) Expression of µs was induced for 0 or 3 days. Immunoblotting of lysates from cells that were sufficiently viable upon the insult for analysis revealed levels of µs, BiP, CHOP, α-tubulin, and ATF6α processing (i.e. release of the p50 cleavage product from the p90 precursor); cross-reaction of the secondary antibody against anti-ATF6α with µs is denoted (µs). RT-PCR fragments corresponding to spliced (XBPS) and unspliced (XBPU) were separated on gel. A hybrid product that is formed during the PCR reaction is denoted by an asterisk. (D) BiP levels in (C) were quantitated and expressed as fold change upon µs expression compared to untreated cells. Mean and s.e.m. are shown in a bar graph; n=2-5. Statistical significance of differences in growth (B), or in expression levels (D), was tested by ANOVA (*p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001).
-
Figure 1—source data 1
- https://doi.org/10.7554/eLife.41168.003
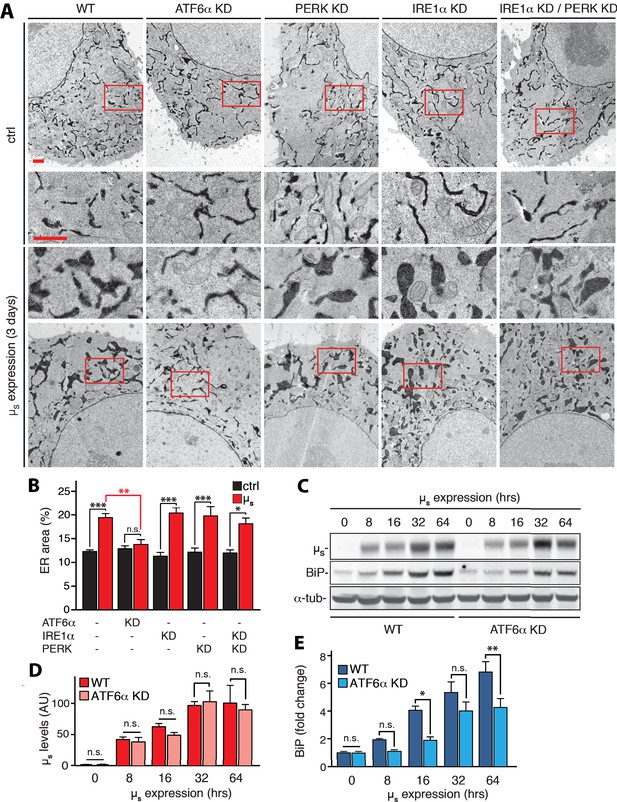
ATF6α is essential but IRE1α and PERK are dispensable for upregulation of ER chaperones and ER expansion in response to µs expression.
(A,B) HeLa-µs cells in which UPR transducers were ablated by silencing alone or in combination, or not (WT), as indicated, were induced with 0.5 nM Mif to express µs for 3 days or not. The cells harbor APEX-KDEL, a modified version of pea peroxidase that is targeted to the ER, and that catalyzes polymerization of 3,3’-diaminobenzidine tetrahydrochloride (DAB) upon treatment with H2O2 to obtain DAB precipitates (dark), revealing the extent of the ER in electron micrographs. Boxed areas are shown by 3-fold magnification; scale bars represent 1 µm (A). The extent of ER expansion was assessed as described (Bakunts et al., 2017), and the percentage of the area within the cytoplasm corresponding to ER was determined and depicted in bar graphs (B). Mean and s.e.m. are shown, n = 10–20. (C–E) Cells were induced to express µs for the indicated times. Levels of µs (D) and BiP (E) were quantitated from (C), and replicate experiments. (D) Levels in WT of µs at 64 hr were set at 100 that was scaled to levels of BiP in WT at 64 hr such as to reflect a ratio of µs to BiP of 2:3, that is an estimate for this ratio at day three based on earlier quantitations that we have described (Bakunts et al., 2017). Mean and s.e.m. are shown in bar graphs; n = 2–5. Statistical significance in the extent of ER areas in the electron micrographs between µs-expressing or non-expressing cells (black), or between µs-expressing WT or ATF6α ablated cells (red) (B), or in expression levels (D,E) was tested by ANOVA (n.s., not significant; *p≤0.05; **p≤0.01; ***p≤0.001).
-
Figure 2—source data 1
- https://doi.org/10.7554/eLife.41168.005
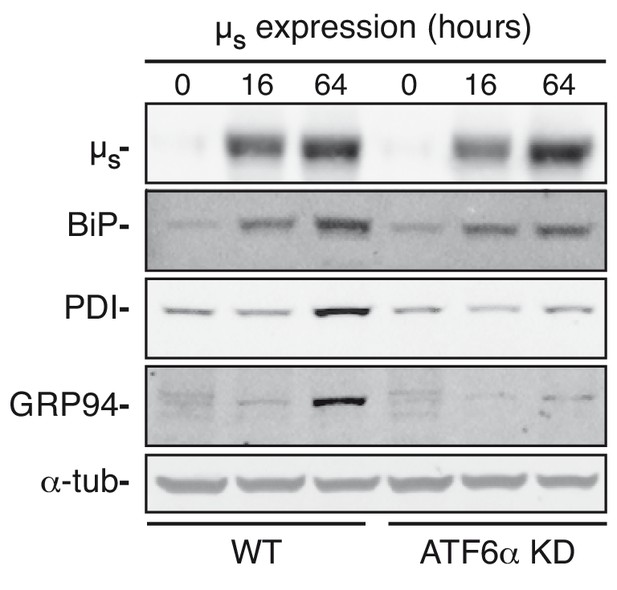
ATF6α ablation compromises ER chaperone upregulation upon µs expression.
HeLa-µs cells in which ATF6α was silenced or not (WT), as indicated, were induced with 0.5 nM Mif to express µs for the indicated times. Levels of µs, BiP, GRP94, PDI, and α-tubulin, were assessed by immunoblotting.
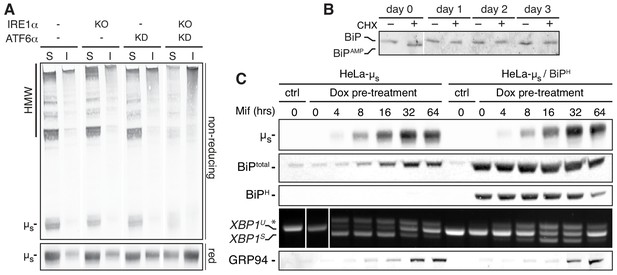
ER stress correlates with the extent of ER chaperones being engaged and becomes cytotoxic when their capacity is exceeded.
(A) HeLa-µs cells, in which IRE1α (KO) and/or ATF6α (KD) was ablated, or not (-), as indicated, were induced with 0.5 nM Mif to express µs for 24 hr. Samples were lysed in NP40 and equivalent amounts of soluble (S) and insoluble (I) fractions resolved under reducing (red) or non-reducing conditions, blotted and decorated with anti-µs. (B) HeLa-µs cells were induced with 0.5 nM Mif to express µs for the indicated times and treated with or without 100 μg/ml CHX for 3 hr before harvesting. Samples were analyzed by iso-electric focusing (IEF) to separate AMPylated (BiPAMP) from non-AMPylated BiP, which were detected by immunoblotting, as described (Preissler et al., 2015). To allow a better comparison between samples, considering the upregulation of BiP upon µs expression, approximately 15 µg of lysates were loaded for the 0 day samples, while only 2.5 µg were loaded for the other days. (C) HeLa-µs-derived cells, harboring Dox-inducible hamster BiP (HeLa-µs/BiPH), were treated for 2 days with 50 nM Dox to induce hamster BiP expression, while WT HeLa-µs cells were mock-treated with 50 nM Dox, before both cell lines were induced with 0.5 nM Mif to express µs for the indicated times. Immunoblotting of lysates revealed levels of µs, total BiP, hamster BiP, and GRP94. XBP1 mRNA splicing was assessed as in Figure 1C.
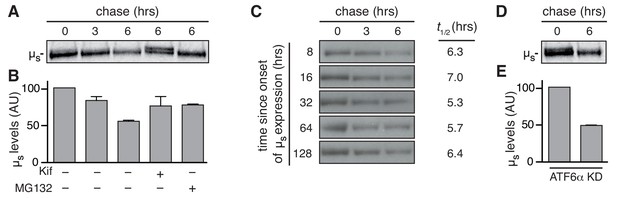
ERAD accounts for disposal of µs in a robust manner.
HeLa-µs cells, in which ATF6α was ablated (D,E), as indicated, or not (A-C) were pulse labeled for 10 min and chased with excess unlabeled cysteine and methionine for the indicated times after 24 hr (A,B,D,E) or at various times (C), as indicated, after induction of µs expression with 0.5 nM Mif, in the absence (A,C,D) or presence (A) of 10 µM MG132 or 30 µM Kif, as indicated (+). Signals were quantitated and the signal after 0 hr chase was set at 100; mean and s.e.m. are shown in bar graphs (B,E); linear fitting of the quantitations of (C) were used to calculate the t½ of µs at various time points after induction of its expression; see column on the right of panel.
-
Figure 4—source data 1
- https://doi.org/10.7554/eLife.41168.012
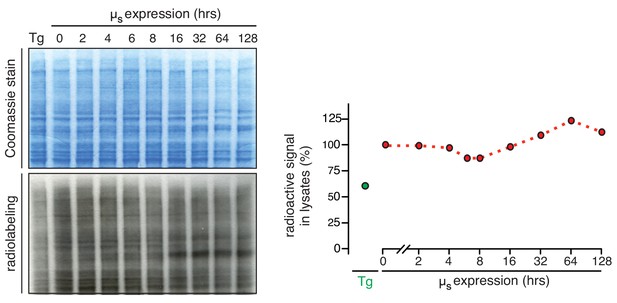
Translation is transiently attenuated upon µs overexpression to a marginal extent.
HeLa-µs cells were induced with 0.5 nM Mif to express µs for the indicated times, or treated with 4 µM Tg for 45 min before they were pulse labeled for 10 min with 35S labeled methionine and cysteine for 10 min at the indicated times after induction. Lysates were separated by gel electrophoresis and total proteins were visualized by Coomassie, while radio-labeled proteins were detected and quantified by phosphor imaging. Labeling efficiency was compared to that in cells before induction, which was set at 100%.
-
Figure 4—figure supplement 1—source data 1
- https://doi.org/10.7554/eLife.41168.010
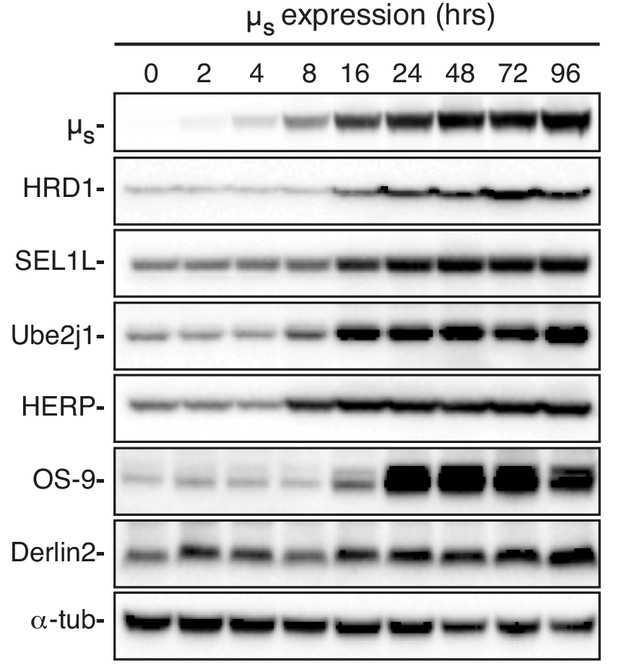
Levels of various ERAD components are induced in response to µs expression.
HeLa-µs cells were induced with 0.5 nM Mif to express µs for the indicated times. Levels of µs, HRD1, SEL1L, Ube2j1, HERP, Derlin-2, OS-9, and α–Tubulin were assessed by immunoblotting.
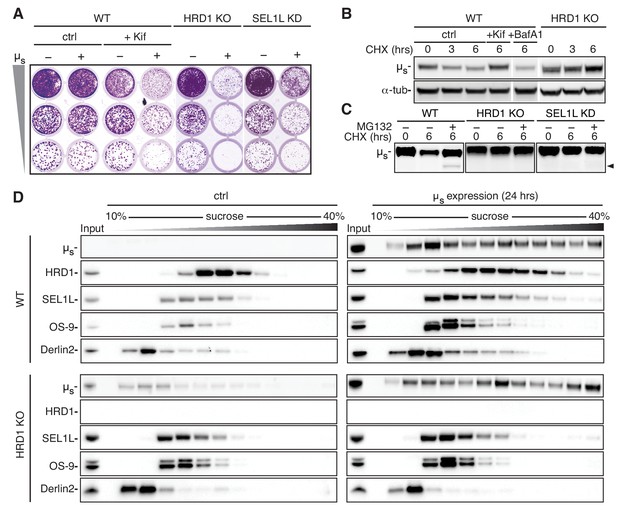
ERAD of µs is mediated through the HRD1 complex.
(A) Growth assay as in Figure 1A of HeLa-µs cells, in which HRD1 was deleted (KO), SEL1L was silenced (KD), or not (WT). Cells were treated with 0.5 nM mifepristone (Mif) to induce expression of µs (+), or not (-), and WT cells were treated with Kif or not (ctrl), as indicated. (B,C) Immunoblots of µs harvested from WT, HRD1 KO (B,C), or SEL1L KD (C) HeLa-µs cells that were induced with 0.5 nM Mif to express µs for 4 hr and then treated for the indicated times with 100 μg/ml CHX either alone (B,C), in combination with 20 mM Kif, 100 nM BafA1, or not (ctrl) (B), or 10 µg/ml MG132 (C), as indicated. The arrowhead indicates the deglycosylated form of µs. (D) HeLa-µs WT or HRD1 KO cells were induced with Mif (0.5 nM) for 24 hr to express µs or not (ctrl) , as indicated. Samples were lysed in 1% lauryl maltose neopentyl glycol (LMNG) and sedimented over a 10–40% sucrose gradient. Levels of µs, HRD1, SEL1L, Derlin-2, and OS-9 were detected by immunoblotting. Note that in HRD1 KO cells leaky expression of µs becomes apparent due to the lack of ERAD. At low expression levels, however, µs does not form high molecular weight aggregates, indicative of the adequacy of the chaperoning machinery.
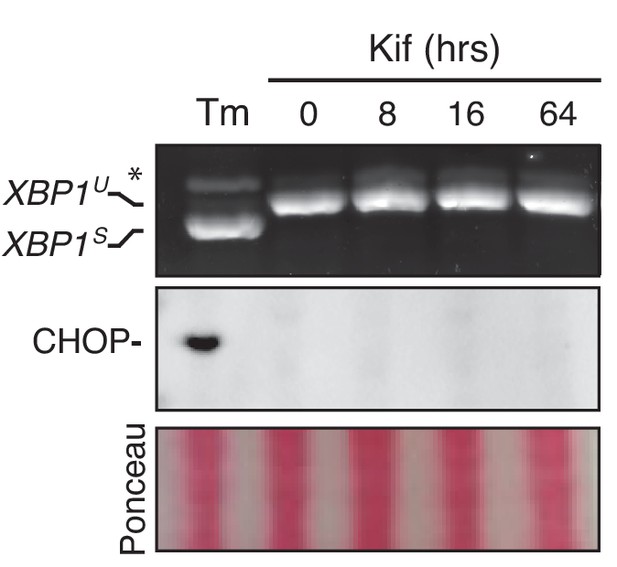
Inhibition of ERAD does not trigger the UPR under basal conditions.
HeLa-µs cells were treated with 30 µM Kif for the indicated times, or treated with 5 µg/ml Tm for 4 hr as a positive reference. XBP1 mRNA splicing and CHOP levels were assessed as in Figure 1C. Ponceau staining of the blot served as a loading control.
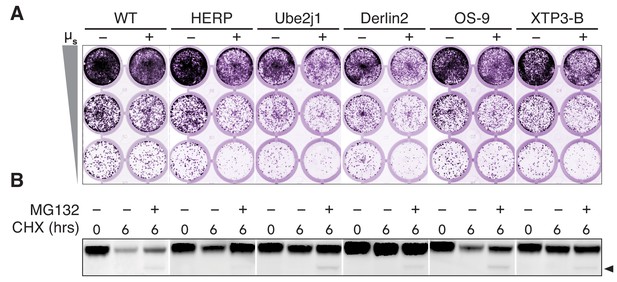
HERP, Ube2j1, and Derlin-2 are key for ERAD of µs.
(A) Growth assays as in Figure 1A of WT HeLa-µs cells, or in which Ube2j1, HERP, Derlin-2, OS-9, or XTP3-B were deleted by CRISPR/Cas9 in a non-clonal fashion (KO). (B) Immunoblot of µs harvested from these same cells that were induced with 0.5 nM Mif to express µs for 4 hrs and then treated (or not) for 6 hrs with 100 μg/ml CHX either alone or in combination with 10 µg/ml MG132, as indicated. The arrowhead marks deglycosylated µs.
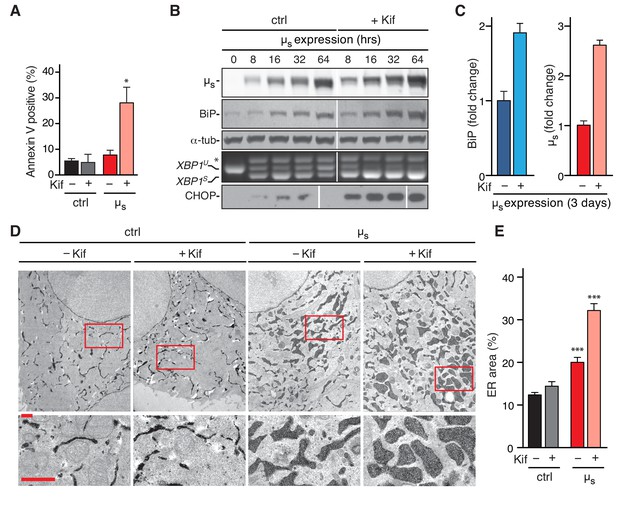
Abrogation of µs disposal through ERAD leads to BiP being permanently eclipsed, ER homeostatic failure, and apoptosis.
(A,D,E) HeLa-µs cells, harboring APEX-KDEL (D,E) or not (A) were induced with (µs) or without (ctrl) 0.5 nM Mif for 3 days in the presence or absence of 30 µM Kif. (A) Percentages of Annexin V positive cells were assessed by cytometric analysis. Mean and s.e.m. are shown in a bar graph, n = 2–4. (B,C) HeLa-µs cells were induced to express µs for various times as indicated (B) or for 3 days in the absence or presence of 30 µM Kif. (C) Levels of µs, BiP, and α-tubulin as well as activation of the IRE1α and PERK branches of the UPR were assessed as in (Bakunts et al., 2017). (B) Levels of BiP and µs were assessed by quantitative immunoblotting as described (Bakunts et al., 2017), and depicted in bar graphs as in Figure 2D,E, such that the µs levels in the absence of Kif were scaled to BiP levels at a ratio of 2:3. Levels in the presence of Kif are expressed as a fold change compared to levels in the absence of Kif; mean and s.e.m. are shown; n = 2. (D) In cells harboring APEX-KDEL the extent of ER expansion was assessed as in Figure 2A. Boxed areas are shown by 3-fold magnification; scale bars represent 1 µm. The percentage of the dark area within the cytoplasm corresponding to ER was determined and depicted in bar graphs (E), mean and s.e.m. are shown, n = 10. Statistical significance of differences in Annexin V staining (A), or the extent of ER occupying cytosolic area in the electron micrographs (E) were tested by ANOVA (*p≤0.05; ***p≤0.001).
-
Figure 6—source data 1
- https://doi.org/10.7554/eLife.41168.017
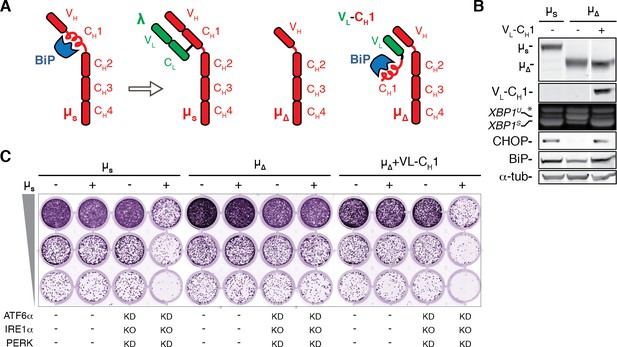
The BiP-sequestering CH1 domain of µs is necessary and sufficient to cause UPR activation and proteotoxic ER stress in absence of the UPR.
(A) Schematic representation of BiP associating with the CH1 domain of µs until it is displaced by the light chain (λ). Deletion of the CH1 domain (µ∆) abolishes BiP association, but through pairing of the VH and VL domains, the CH1 domain can associate in trans by virtue of a synthetic chimeric VL-CH1 construct. (B) HeLa cells were induced for 24 hr with 0.5 nM Mif to express the transgenes µs, µs∆CH1 (µ∆) alone or in conjunction with VL-CH1, as indicated. Immunoblotting of lysates revealed levels of µs, µ∆, VL-CH1, BiP, CHOP, and α-tubulin, as in Figure 1A. (C) Growth assay as in Figure 1A of HeLa cells inducibly expressing µs, µs∆CH1 (µ∆) in conjunction with VL-CH1 or not, and in which the UPR was ablated (i.e. IRE1α was deleted (KO), and ATF6α and PERK were silenced in combination), or not, as indicated.
Additional files
-
Supplementary file 1
List of cell lines and reagents.
- https://doi.org/10.7554/eLife.41168.019
-
Transparent reporting form
- https://doi.org/10.7554/eLife.41168.020





