An unexpected INAD PDZ tandem-mediated plcβ binding in Drosophila photo receptors
Figures
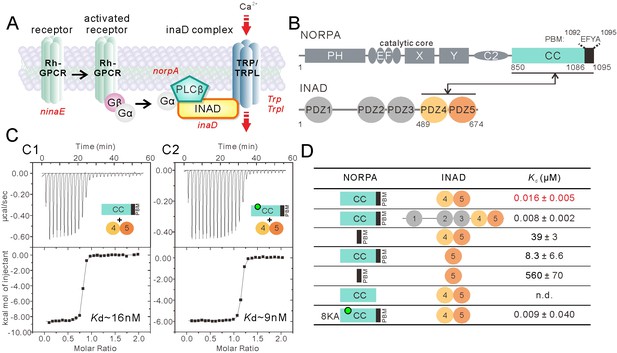
Super strong interaction between NORPA and INAD.
(A) Schematic cartoon diagram showing the pathway of Drosophila photo-transduction signaling. (B) Schematic diagram showing the domain organizations of NORPA and INAD. The interaction mediated by NORPA CC-PBM and INAD PDZ45 is illustrated. The color coding of the domains is kept throughout this paper. (C) Isothermal titration calorimetry (ITC)-based measurement of the binding between NORPA CC-PBM and INAD PDZ45 (C1), and between the 8KA mutation of NORPA CC-PBM and INAD PDZ45 (C2). The sites of the point mutations in the CC region of NORPA are indicated by a green dot. (D) Table summarizing the measured binding affinities between various forms of NORPA CC-PBM and INAD derived from ITC-based assays.
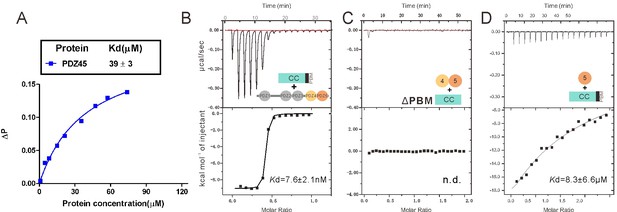
Characterization of the interaction between INAD PDZ45 and NORPA CC-PBM.
(A) Fluorescence polarization-based assay measuring the binding affinity between INAD PDZ45 and FITC-labeled NORPA PBM peptide (KTQGKTEFYA). (B) ITC-based measurement showing the binding affinity between the full-length INAD and NORPA CC-PBM. (C) ITC-based measurement showing that deletion of PBM from NORPA CC-PBM completely disrupted its binding to INAD PDZ45. (D) ITC-based measurement showing that isolated PDZ5 has a weak binding to NORPA CC-PBM.
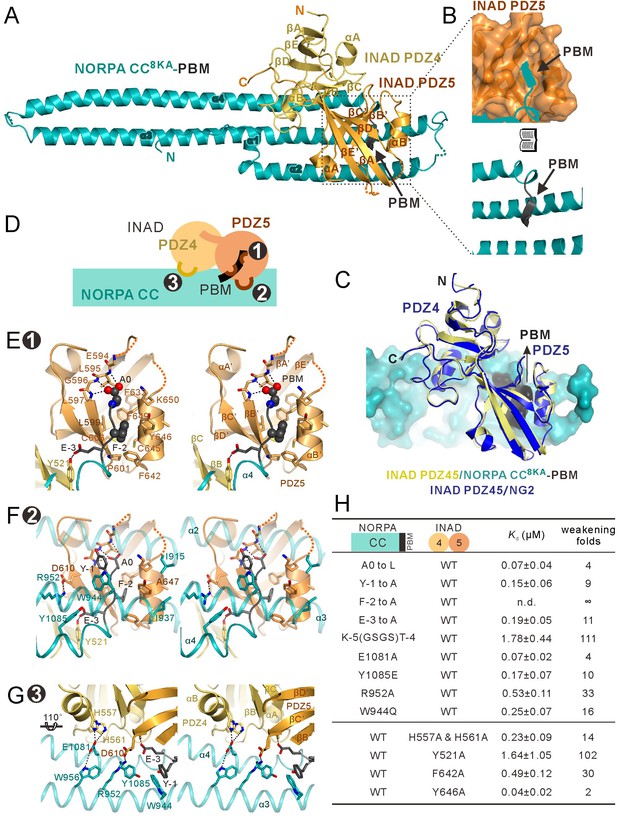
Structure of the NORPA CC8KA-PBM–INAD PDZ45 complex.
(A) Ribbon representation of the NORPA CC8KA-PBM–INAD PDZ45 complex structure. The disordered loops are drawn as dashed lines in the ribbon representation. (B) Open book view showing the binding interface between INAD PDZ5 and NORPA PBM. (C) Superimposition of INAD PDZ45–NG2 peptide structure (blue) over PDZ45 in complex with NORPA (yellow). (D) Schematic cartoon diagram summarizing the binding mode of the NORPA CC8KA-PBM–INAD PDZ45 complex with three characteristic binding sites detailed in panels E–G. (E–G) Stereoviews showing the interaction interfaces between INAD PDZ45 and NORPA CC-PBM. The side chains of the residues involved in the inter-domain interactions are drawn in the stick representation. The complex interface is divided into three parts, the PDZ5–PBM interaction site ((E) site 1), the PDZ5/CC packing site ((F) site 2), and the PDZ4/CC binding site ((G) site 3). (H) Table summarizing the measured binding affinities, showing that mutations of the critical residues in the NORPA CC-PBM–INAD PDZ45 interface weakened or even abolished the interaction.
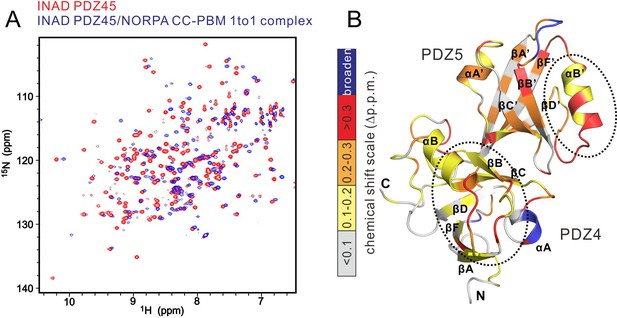
NMR-spectroscopy-based characterization of the INAD PDZ45–NORPA CC-PBM interaction.
(A) Overlay plot of the 1H-15N heteronuclear single quantum coherence spectroscopy (HSQC) spectra of INAD PDZ45 alone (in red) and in complex with an equal molar ratio of unlabeled NORPA CC-PBM (in blue), showing significant chemical shift changes of INAD PDZ45 induced by NORPA CC-PBM binding. (B) Calculation of backbone amide chemical shift differences as a function of the residue number of INAD PDZ45 between its apo-form and in complex with NORPA CC-PBM. The calculated shift changes were mapped onto the ribbon diagram of the INAD PDZ45 structure. In this representation, the combined 1H and 15N chemical shift changes are defined as: Δp.p.m. = [(ΔδHN)2 + (ΔδN × αN)2 ]1/2, where ΔδHN and ΔδN represent chemical shift differences of amide proton and nitrogen chemical shifts of each residue of INAD PDZ45. The scaling factor (αN) used to normalize the 1H and 15N chemical shifts is 0.17. The PDZ45 areas showing the most obvious chemical shifts as the result of NORPA interaction are highlighted by dash circles.
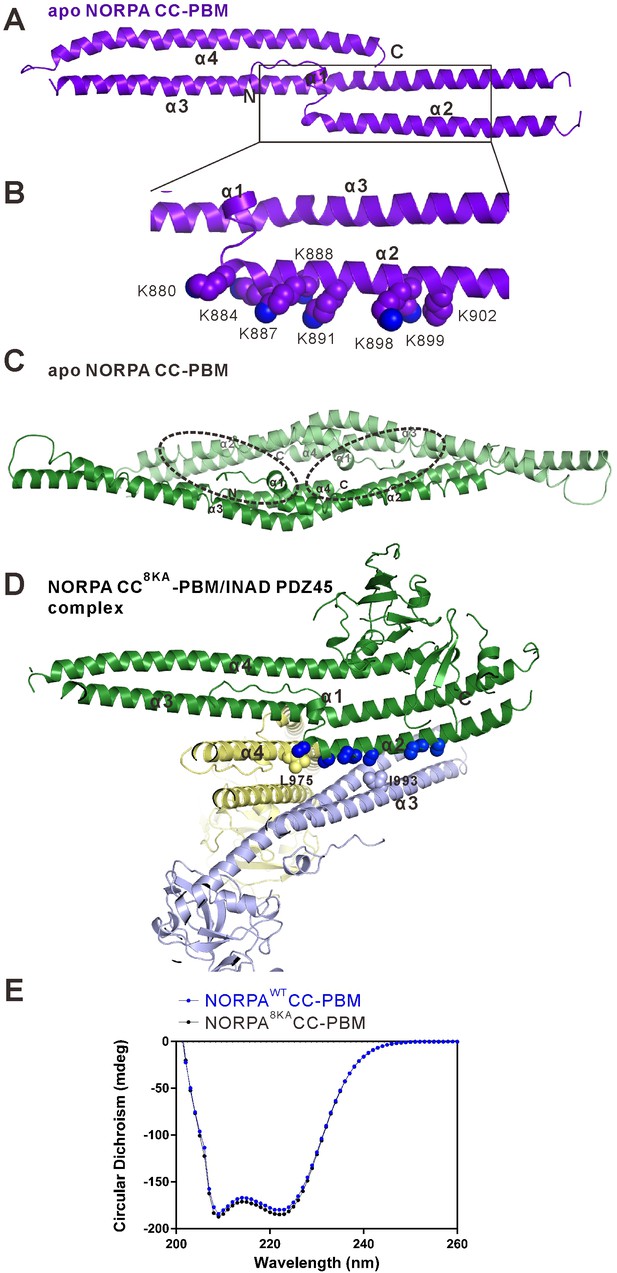
Crystal structure of the NORPA CC-PBM domain.
(A) Ribbon representation of the NORPA CC-PBM. The NORPA CC forms an elongated three helix bundle with an N-terminal short helix packed to the center of the helix bundle. The PBM is not defined in this structure. (B) Ribbon combined sphere representations showing a Lys-rich surface in the α2 helix of the NORPA CC domain. These eight Lys residues are aligned in the solvent-exposed face of α2, which presumably prevented crystal growth of the unmethylated NORPA CC-PBM. (C) Ribbon diagram showing the crystal packing of the methylated apo NORPA CC dimer in each asymmetric unit of the crystal. The PDZ45 binding surface (highlighted with dashed circles) overlaps with the crystal packing surface. (D) Ribbon combined with sphere diagram representations showing the crystal packing of the NORPA CC8KA-PBM–INAD PDZ45 complex. The Ala residues substituting Lys in the α2 helix are directly involved in the crystal packing of the complex. (E) Circular dichroism spectra of the NORPA CC-PBM (blue line) and NORPA CC8KA-PBM (black line).
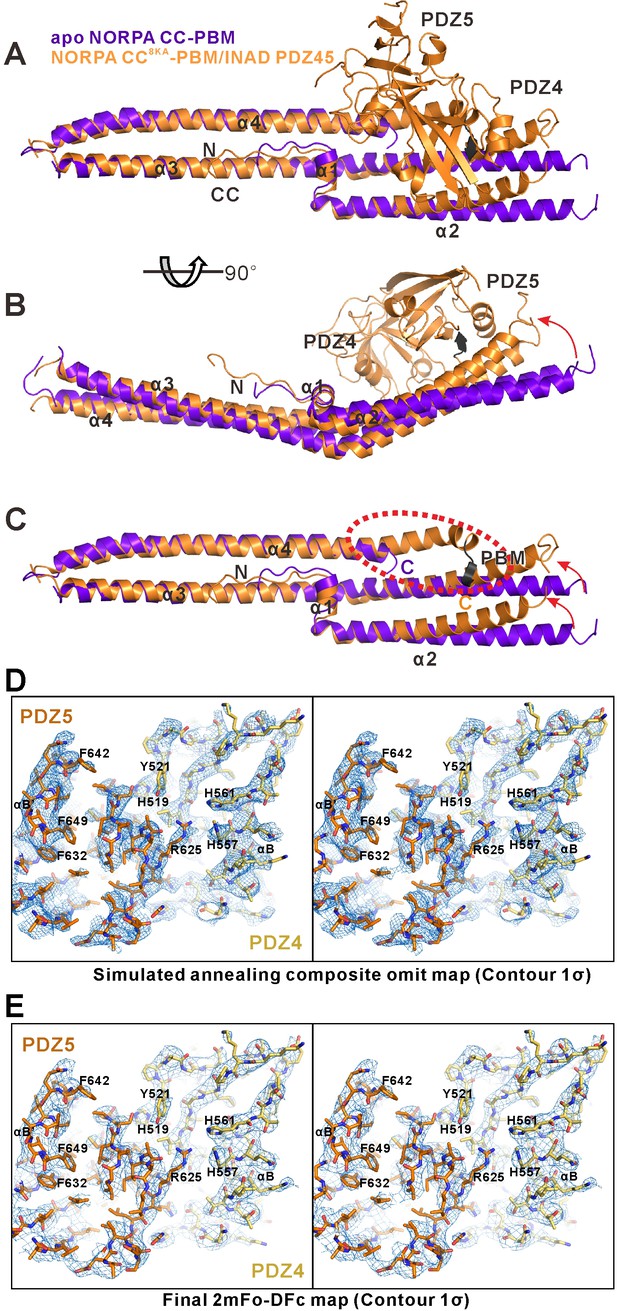
Alignment of the apo NORPA CC-PBM and the NORPA CC8KA-PBM–INAD PDZ45 complex structures showing the INAD PDZ45 binding-induced conformational changes of the NORPA CC-PBM.
(A, B and C) Superimposition of the apo-form NORPA CC-PBM structure (purple) and the NORPA CC8KA-PBM in the NORPA CC8KA-PBM/INAD PDZ45 complex structure (orange), showing the similar overall helical bundle organization of the NORPA CC (C). The right part of the CC domain is slightly bent because of the packing of PDZ45 (highlighted with red arrows in panels B and C). The C-terminal PBM region of the NORPA CC (highlighted with a red dashed oval), which is disordered in the apo-form crystal structure, is stabilized in the complex structure through interaction with INAD PDZ45 (C). (D and E) Representative simulated annealing composite omit map (D) and the final 2mFo-DFc map (E) of the INAD PDZ45–NORPA CC-PBM complex. For clarity, the NORPA CC-PBM part is not shown and the INAD PDZ45 is shown as a stick model. Bulky residues are highlighted. Both maps are contoured at the 1σ level. The simulated annealing composite omit map was calculated using the Phenix software suite.
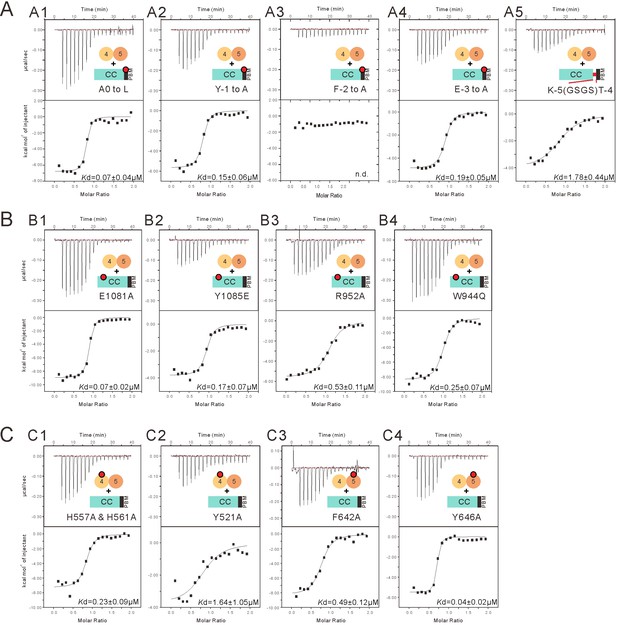
Impact of the mutations on the binding affinity between INAD PDZ45 and the NORPA CC-PBM.
(A) ITC-based measurements comparing the binding affinities between INAD PDZ45 and the NORPA CC-PBM with different mutations in its PBM. (A1) A0 was mutated to L. (A2) Y-1 was mutated to A. (A3) F-2 was mutated to A. (A4) E-3 was mutated to A. (A5) A flexible GSGS sequence was inserted between CC and PBM. (B) ITC-based measurements comparing the binding affinities between INAD PDZ45 and the NORPA CC-PBM with different mutations in the CC region as indicated. (C) ITC-based measurements comparing the binding affinities between different INAD PDZ45 mutants and the NORPA CC-PBM.
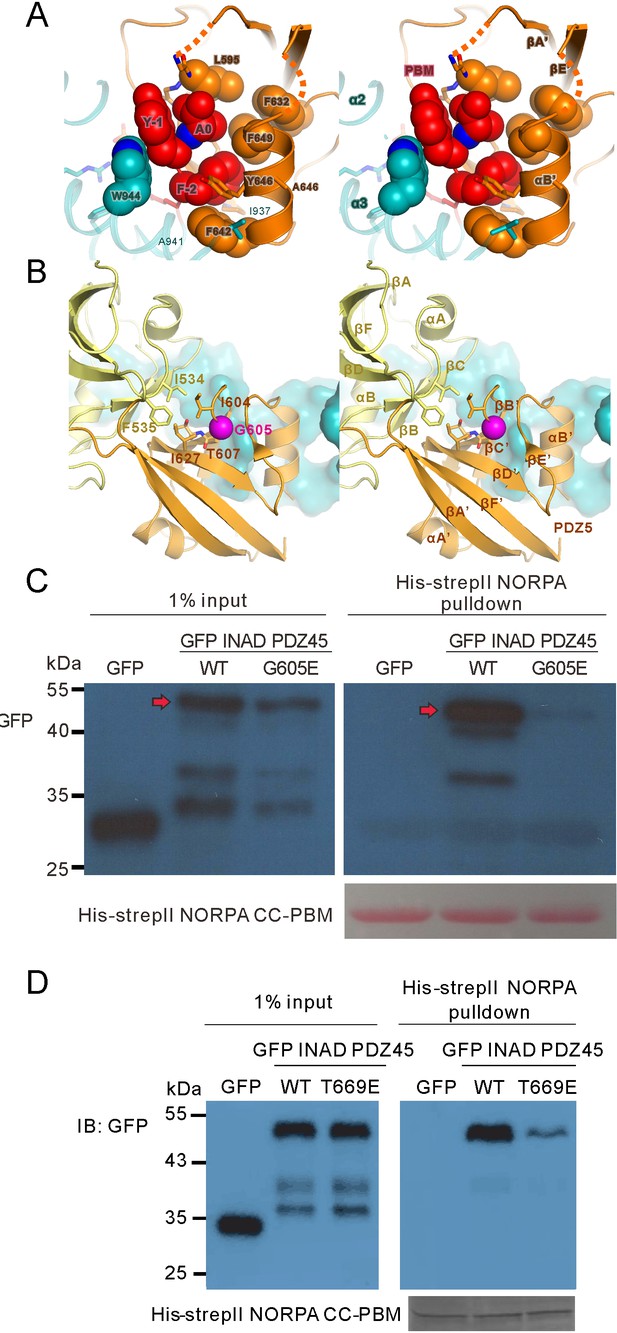
Biochemical and structural analysis of the INAD PDZ45–NORPA CC-PBM interaction.
(A) Stereoviews showing the binding of the NORPA PBM (in red) in the canonical PBM binding groove of INAD PDZ5 (in orange). W944 from CC (in cyan) interacts with Y-1 of the NORPA CC-PBM. (B) Stereoviews showing that Gly605 (in purple) is located in the hydrophobic interface coupling site between PDZ4 and PDZ5 of the INAD PDZ45 tandem. The G605E mutation impairs the inter-domain coupling of PDZ45. (C) Pull-down assay showing that the G605E mutation of INAD (corresponding to the inaD2 mutant) largely impairs the interaction between NORPA and INAD PDZ45. (D) Pull-down assay showing the T669E mutation of INAD PDZ45, which impaired the domain coupling of PDZ4 and PDZ5, largely weakening its interaction with NORPA.
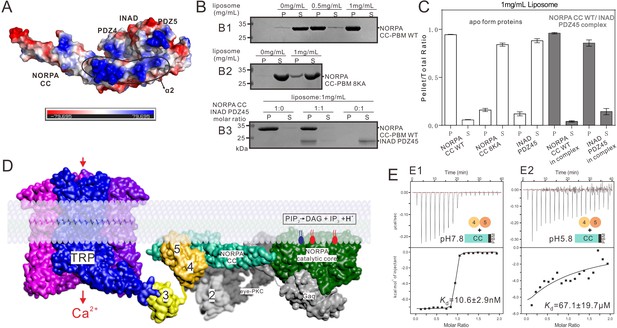
Summary model of the INAD PDZ45–NORPA CC-PBM interaction in Drosophila photon-transduction.
(A) Surface representation showing the electrostatic potential of the INAD PDZ45–NORPA CC-PBM complex. The ± 80 kT/e potential isocontours are shown as blue (positively charged) and red (negatively charged) surfaces, respectively. This electrostatic potential analysis was generated by Pymol (https://www.pymol.org). (B) Lipid sedimentation assay showing the binding properties of NORPA CC-PBM WT (B1), NORPA CC8KA-PBM mutant (B2), and the INAD PDZ45–NORPA CC-PBM complex (B3) to liposomes prepared from bovine brain lipid extracts. Fractions labeled 'S' and 'P' represent proteins that are present in the supernatants and pellets after centrifugation, denoting lipid-free and lipid-bound forms of the proteins, respectively. (C) Quantification of the sedimentation-based assay of the lipid binding capacities of the proteins shown in panel B. The results are from three independent batches of sedimentation assays and are represented as mean ± SD. (D) Surface combined cartoon representation showing a model of the INAD-organized signaling complex underneath the rhabdomere plasma membranes. In this model, the INAD PDZ345 tandem can position NORPA and the TRP channel in close proximity on the 2D membrane plane. (E) ITC-based measurement of the binding between the NORPA CC-PBM and INAD PDZ45 at pH 7.8 (E1) and at pH 5.8 (E2), showing acidification-induced weakening of the binding between NORPA and INAD.
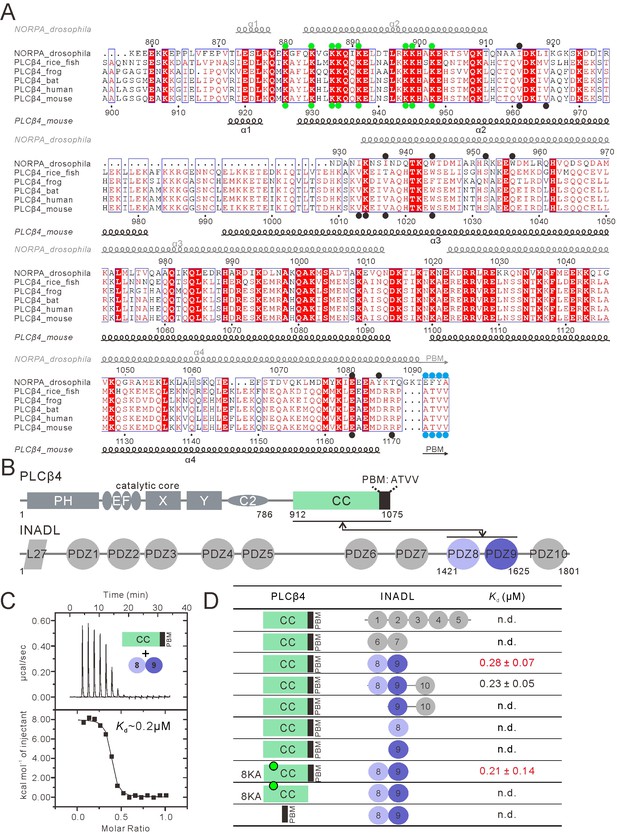
PLCβ4 and NORPA share a similar PDZ tandem binding mode.
(A) Multiple sequence alignment of NORPA with PLCβ4 proteins from various animals by ClusterW and ESpript (espript.ibcp.fr/ESPript/ESPript/). Strictly conserved residues are boxed in white on a red background, and highly conserved residues are boxed in red on a white background. Helical structures as well as the PBM β-strand are depicted. The PBMs of NORPA and PLCβ4 are highlighted by blue dots. The key residues of the NORPA CC (or the PLCβ4 CC) involved in the interaction with INAD PDZ45 (or INADL PDZ89) are highlighted by black dots. The Lys residues mutated to Ala in order to facilitate crystallizations are highlighted by green dots. (B) Schematic diagram showing the domain organizations of PLCβ4 and INADL. The interaction is mediated by the PLCβ4 CC-PBM and INADL PDZ89 as indicated. The color coding of the domains is kept throughout this paper. (C) ITC-based measurement of the binding between the PLCβ4 CC-PBM and INADL PDZ89. (D) Table summarizing the measured dissociation constants between various constructs of PLCβ4 and INADL derived from ITC-based assays.
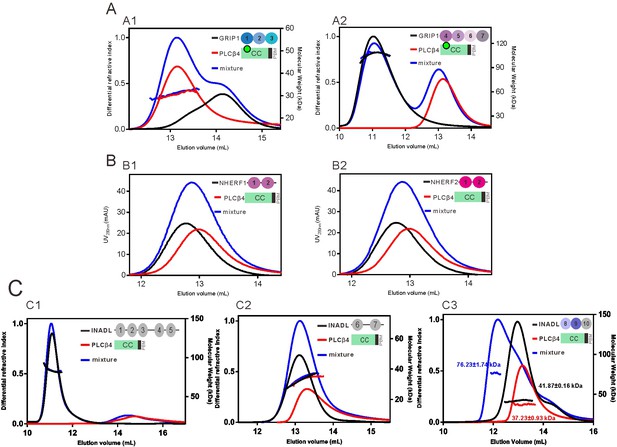
The PLCβ4 CC-PBM specifically interacts with INADL.
(A) The PLCβ4 CC8KA-PBM has no detectable interaction with GRIP1 PDZ1-3 (A1) or PDZ4-7 (A2). Analytical gel filtration chromatography analysis coupled with static light scattering analysis showing that mixing of PLCβ4 CC8KA-PBM with GRIP1 PDZ domains showed no obvious elution volume changes for either PLCβ4 CC8KA-PBM or GRIP1 PDZ domains. (B) Analytical gel filtration chromatography analysis showing that PLCβ4 CC-PBM WT has no detectable interaction with NHERF1 PDZ12 (B1) or with NHERF2 PDZ12 (B2). (C) PLCβ4 CC-PBM WT specifically interacts with INADL PDZ8-10. Analytical gel filtration chromatography analysis coupled with static light scattering analysis showed that the mixing of INADL PDZ8-10 with PLCβ4 CC-PBM WT induced an obvious elution volume shift of CC-PBM (C3), whereas mixing of either INADL PDZ1-5 or PDZ6-7 with CC-PBM PLCβ4 did not induce an elution peak shift (C1 and C2).
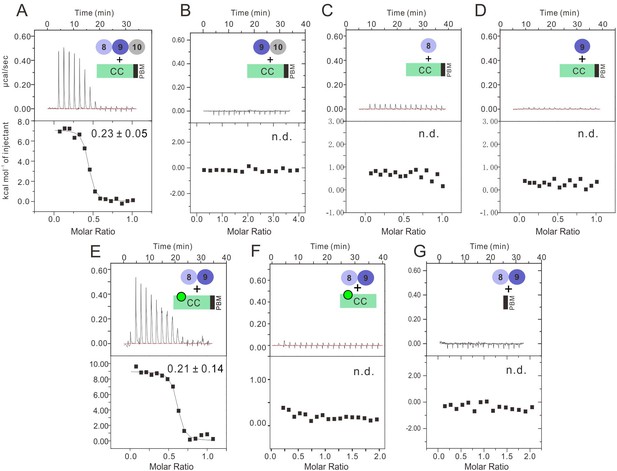
Characterization of the binding of INADL PDZ89 and the PLCβ4 CC-PBM.
ITC-based measurement showing the binding affinities of INADL PDZ domains with PLCβ4 CC-PBM. (A) INADL PDZ8-10 and PLCβ4 CC-PBM. (B) INADL PDZ9-10 and PLCβ4 CC-PBM. (C) INADL PDZ8 and NORPA CC-PBM. (D) INADL PDZ9 and PLCβ4 CC-PBM. (E) INADL PDZ89 and PLCβ4 CC8KA-PBM mutant. (F) INADL PDZ8-9 and PLCβ4 CC8KA. (G) INADL PDZ8-9 and PLCβ4 PBM.
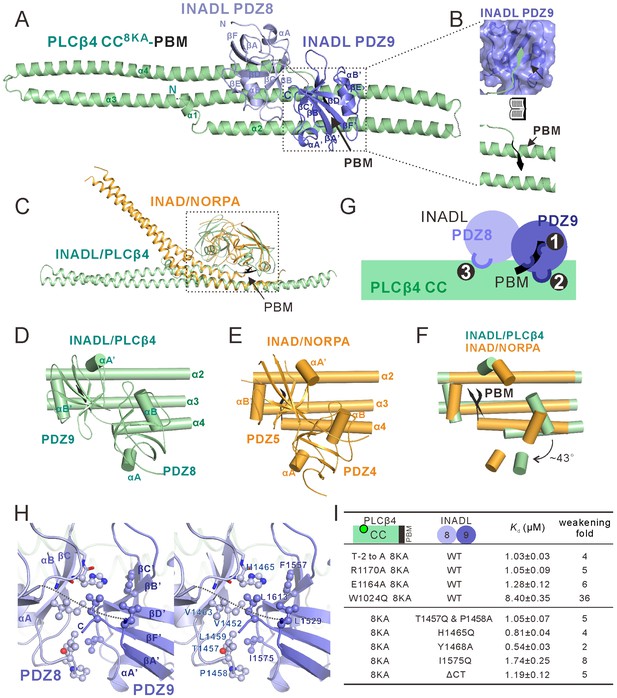
Structure of the PLCβ4 CC8KA-PBM–INADL PDZ89 complex.
(A) Ribbon representation of the PLCβ4 CC8KA-PBM–INADL PDZ89 complex structure. The disordered loops are drawn as dashed lines in the ribbon representation. (B) Open-book view showing the interaction interface between INADL PDZ9 and the PBM of PLCβ4. (C) Superimposition of the NORPA CC8KA-PBM–INAD PDZ45 complex structure (yellow) with the PLCβ4 CC8KA-PBM–INADL PDZ89 complex structure (green), showing the similar binding mode of the two complexes. (D–F) Comparison of the PDZ tandem orientations in the PLCβ4 CC8KA-PBM–INADL PDZ89 (D) and the NORPA CC8KA-PBM–INAD PDZ45 (E) complexes. (F) The superposition of the CC8KA-PBM portion of NORPA and PLCβ4 from the two complexes. Only the αA and αB helices of the PDZ domains are shown to illustrate the orientation differences of the PDZ domains in the two complexes. (G) Schematic cartoon diagram summarizing the interaction mode of the PLCβ4 CC8KA-PBM–INADL PDZ89 complex with three characteristic binding sites (Sites 1, 2, and 3; detailed in Figure 5—figure supplement 1B and C). (H) Stereoview showing the interaction in the interfaces between INADL PDZ8 and PDZ9. In this drawing, the side chains of the residues involved in the inter-domain interactions are drawn in the stick and ball representations. (I) A table summarizing the measured dissociation constants shows that mutations of the critical residues in the PDZ8 and PDZ9 interface can weaken the PLCβ4 CC8KA-PBM–INADL PDZ89 interaction.
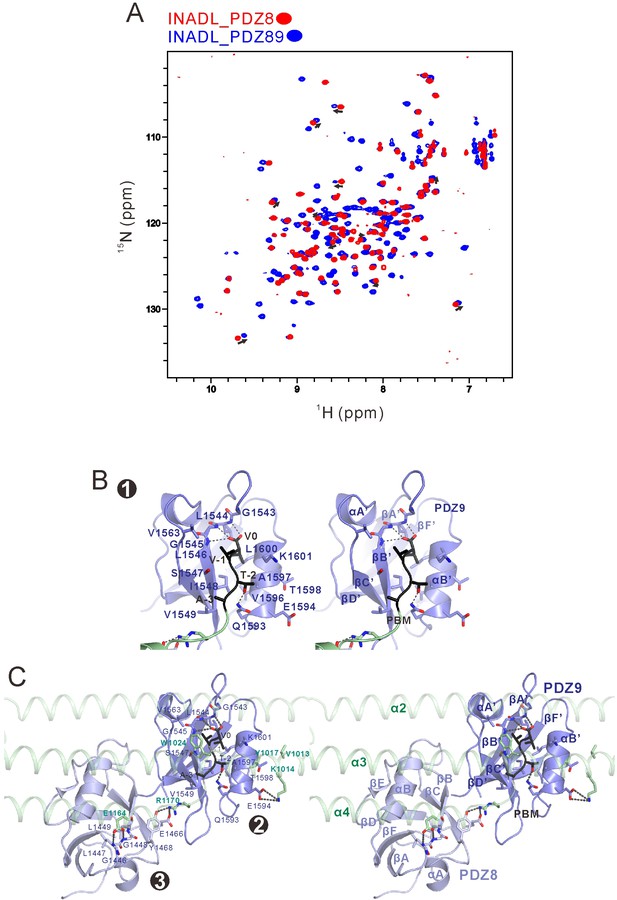
Detailed analysis of the interaction between INADL PDZ89 and the PLCβ4 CC-PBM.
(A) Overlay plot of the 1H-15N HSQC spectra of INADL PDZ8 (in red) and PDZ89 (in blue), showing that a subset of peaks from PDZ89 nicely overlap with the peaks of isolated PDZ8. This analysis indicates that there is minimal conformational coupling between PDZ8 and PDZ9 in the isolated PDZ89 tandem. (B) Stereoviews showing the binding of PLCβ4 PBM (in black) with the canonical PBM binding groove of INADL PDZ9 (in blue). (C) Stereoviews showing the interaction interfaces between INADL PDZ89 and PLCβ4 CC-PBM.
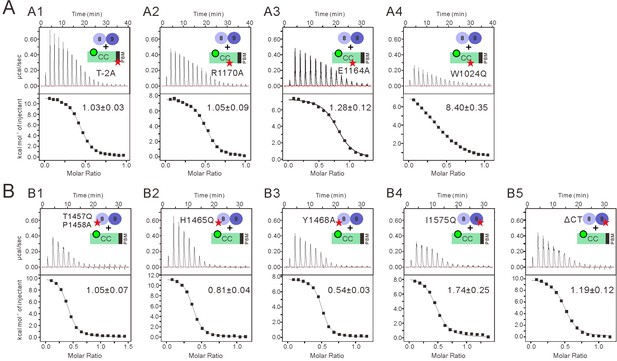
Impact of the mutations on the binding affinity between INADL PDZ89 and the PLCβ4 CC-PBM.
(A) ITC-based measurements comparing the binding affinities between the WT INADL PDZ89 and various forms of PLCβ4 CC8KA-PBM. (A1) T-2 was substituted with Ala. (A2) R1170 was substituted with Ala. (A3) E1164 substituted with Ala. (A4) W1024 was substituted with Gln. (B) ITC-based measurements comparing the binding affinities between variant mutants of INADL PDZ89 and the PLCβ4 CC8KA-PBM. (B1) T1457Q and P1458A-PDZ89. (B2) H1465Q-PDZ89. (B3) Y1468A-PDZ89. (B4) I1575Q-PDZ89. (B5) Truncation of the C-terminal 10-residue extension following PDZ9 in the PDZ89 tandem.
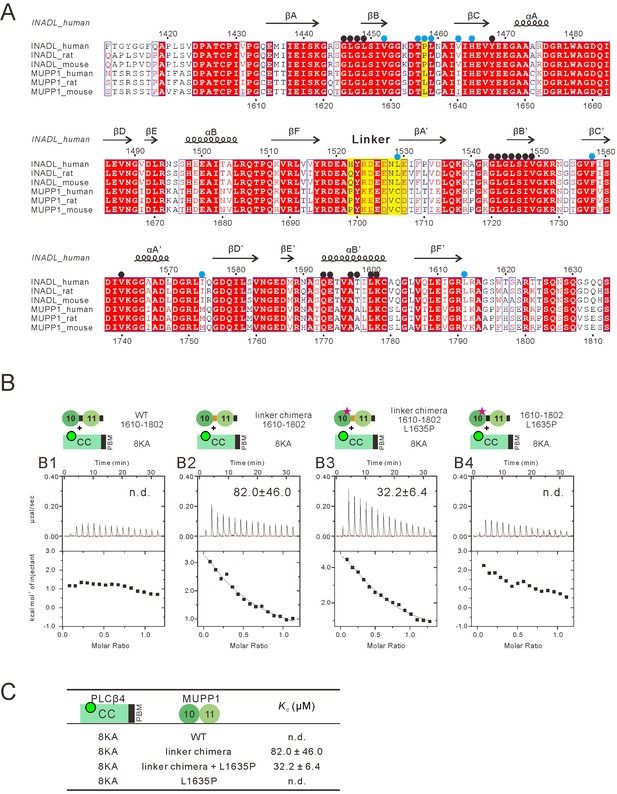
INADL PDZ89 instead of MUPP1 PDZ10-11 specifically interacts with PLCβ4.
(A) Sequence alignment showing the multiple-PDZ-containing proteins MUPP1 PDZ10-11 shares high sequence identity with INADL PDZ89. Residues of INADL PDZ89 that are involved in binding interface for PLCβ4 are highlighted with black dots, residues involved in the domain coupling between INADL PDZ8 and PDZ9 are highlighted with blue dots. The sites used to create the MUPP1-INADL PDZ10-11 chimera are highlighted with yellow shading. (B) ITC-based measurements comparing the binding affinities of various chimeras of MUPP1 PDZ10-11 to the PLCβ4 CC8KA-PBM. (C) Table summarizing the measured dissociation constants for the bindig of the MUPP1 PDZ10-11 chimeras with the PLCβ4 CC8KA-PBM.
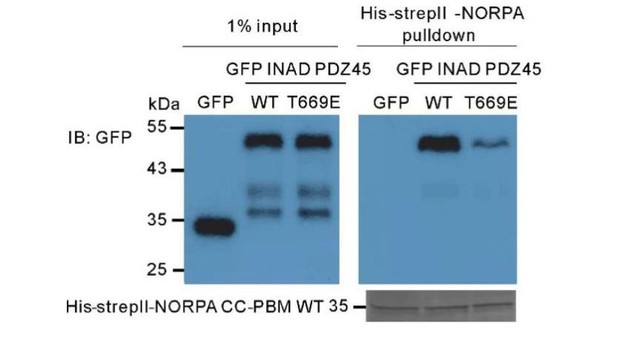
Pull-down assay showing that the T669E mutant of INAD PDZ45 impairs its binding to NORPA CC-PBM.
In this assay, GFP-fused T669E-INAD PDZ45 or WT INAD PDZ45 was expressed in HEK293T cells, and pulled down by purified His-StrepII-tagged NORPA CC-PBM as detailed in the Materials and methods section.
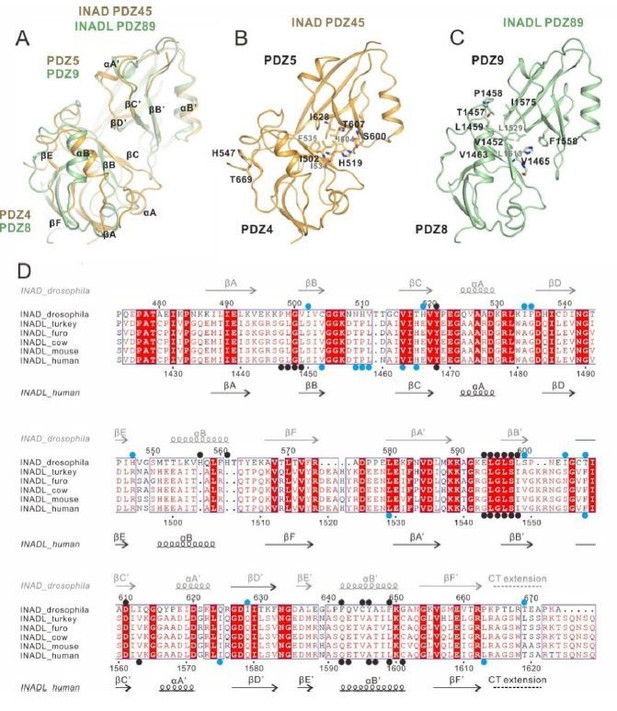
Comparison of the inter-PDZ domain packing of the INAD PDZ45 and INADL PDZ89 tandems in the two complex structures.
(A) Superposition of INAD PDZ45 (in yellow) and INADL PDZ89 (in green). (B) Ribbon combined stick representation showing the domain packing interface of INAD PDZ45. (C) Ribbon combined stick representation showing the domain packing interface of INADL PDZ89. (D) Multiple sequence alignment of INAD PDZ45 and INADL PDZ89 from Drosophila to human by ClusterW and ESpript (espript.ibcp.fr/ESPript/ESPript/). The key residues involved in the PDZ domain coupling are highlighted in blue dots. The key residues of PDZ domains involved in the NORPA/PLCβ4 interaction interface are highlighted by black dots.
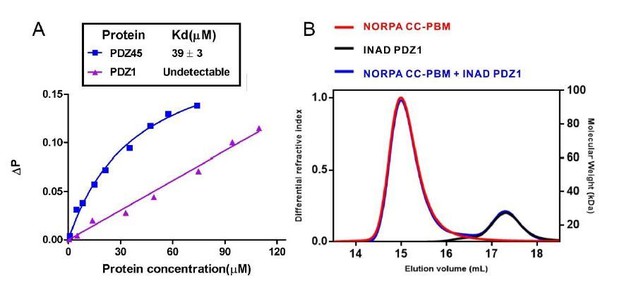
Binding analysis of INAD PDZ1 to NORPA PBM and NORPA CC-PBM.
(A) Fluorescence polarization assay showed that no interaction could be detected between NORPA PBM peptide (TQGKTEFYA) and INAD PDZ1. As a control, FITC-labeled NORPA PBM peptide bound to INAD PDZ45 with a Kd ~ 40 μM. (B) FPLC analysis showed INAD PDZ1 does not interact with NORPA CC-PBM.
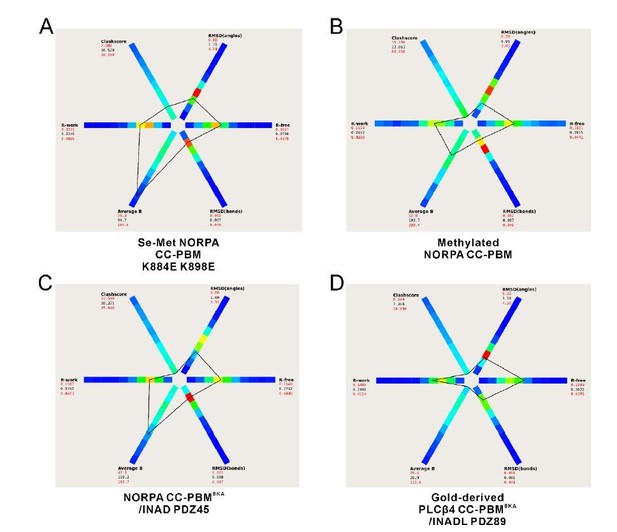
The statistics of our four structures compared to other structures at similar resolutions generated by the program POLYGON in the PHENIX software suite (Urzhumtseva et al., 2009).
https://doi.org/10.7554/eLife.41848.024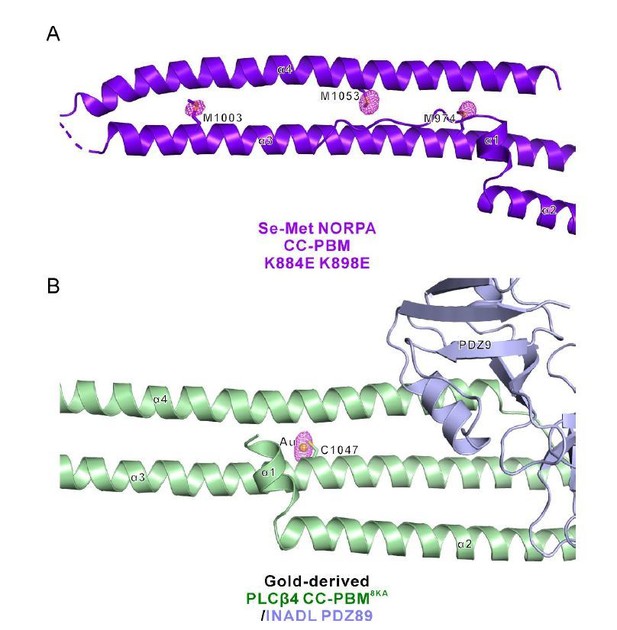
Phased anomalous difference electron density map.
(A) Three representative Se sites (M974, M1003, and M1053) of the Se-Met NORPA CC-PBM K884E K898E structure are shown and the electron density is contoured at a 3.0σ level. (B) One representative gold site of the Gold-derived PLCβ4 CC-PBM8KA/INADL PDZ89 structure is shown and the electron density is contoured at a 4.0σ level. The gold atom is conjugated to the sulfur of C1047 from PLCβ4 CC-PBM.
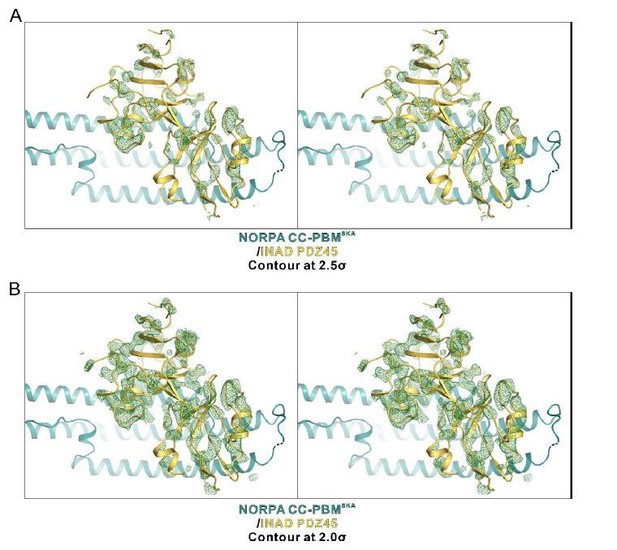
An omit map showing the PDZ45 module.
The Fo-Fc density map is generated by deleting the INAD PDZ45 part from the final model and contoured at 2.5σ (A) and 2.0σ (B), respectively.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (E. coli) | BL21(DE3) | Novagen | Cat #69450 | |
| Strain, strain background (Escherichia coli) | B834(DE3) | Novagen | Cat #69041 | |
| Strain, strain background (E. coli) | Rosseta (DE3) | Novagen | Cat #70954 | |
| Cell line (human) | HEK293T | ATCC | Cat #CRL-3216; RRID:CVCL_0063 | |
| Transfected construct (plasmid) | GFP-INAD Drosophila PDZ45 WT | This paper | N/A | |
| Transfected construct (plasmid) | GFP-INAD Drosophila PDZ45 G605E | This paper | N/A | |
| Transfected construct (plasmid) | GFP-INAD Drosophila PDZ45 T669E | This paper | N/A | |
| Recombinant protein | Drosophila INAD FL WT: aa M1-A674 | This paper | NCBI: NM_166566.1 | |
| Recombinantprotein | Drosophila INAD PDZ45 WT: aa K473-A674 | This paper | NCBI: NM_166566.1 | |
| Recombinantprotein | Drosophila INAD PDZ5 WT: aa L580-P665 | This paper | NCBI: NM_166566.1 | |
| Recombinantprotein | Drosophila NORPA CC-PBM WT: aa E863-A1095 | This paper | NCBI: NM_080330.4 | |
| Recombinant protein | Drosophila NORPA CC WT: aa E863-T1092 | This paper | NCBI: NM_080330.4 | |
| Recombinantprotein | Drosophila NORPA CC8KA-PBM: aa E863-A1095; K880A and K884A and K887A and K888A and K891A and K898A and K899A K902A | This paper | NCBI: NM_080330.4 | |
| Recombinantprotein | Drosophila NORPA CC-PBM K-5(GSGS)T-4: aa D852-K1090, GSGS, T1091-A1095 | This paper | NCBI: NM_080330.4 | |
| Recombinantprotein | Human INADL PDZ1-5 WT: aa M126-D776 | This paper | NCBI: NM_176877.3 | |
| Recombinantprotein | Human INADL PDZ6-7 WT: aa G1054-P1332 | This paper | NCBI: NM_176877.3 | |
| Recombinantprotein | Human INADL PDZ89 WT: aa L1421-T1625 | This paper | NCBI: NM_176877.3 | |
| Recombinantprotein | Human INADL PDZ8-10 WT: aa S1422-D1801 | This paper | NCBI: NM_176877.3 | |
| Recombinantprotein | Human INADL PDZ9-10 WT: aa E1530-D1801 | This paper | NCBI: NM_176877.3 | |
| Recombinantprotein | Human INADL PDZ8 WT: aa S1422-N1528; | This paper | NCBI: NM_176877.3 | |
| Recombinantprotein | Human INADL PDZ9 WT: aa E1530-R1659 | This paper | NCBI: NM_176877.3 | |
| Recombinantprotein | Human INADL PDZ89 ∆CT: aa L1421-R1615 | This paper | NCBI: NM_176877.3 | |
| Recombinantprotein | Mouse PLCβ4 CC-PBM WT: aa E912-V1175 | This paper | NCBI: NM_013829.2 | |
| Recombinantprotein | Mouse PLCβ4 CC8KA –PBM: aa: E912-V1175; K926A and K930A and K933A and K934A and K937A and K944A and K945A and K948A | This paper | NCBI: NM_013829.2 | |
| Recombinant protein | Mouse PLCβ4 CC8KA: aa E912-A11728KA | This paper | NCBI: NM_013829.2 | |
| Recombinant protein | INADL-PLCβ4 fusion: human INADL aa L1421-T1625, GGGGSGGGGSGGEGS, mouse PLCβ48KAaa E912-V11758KA | This paper | NCBI: NM_176877.3; NM_013829.2 | |
| Recombinant protein | Mouse GRIP1 PDZ1-3 WT: aa M1-A334 | This paper | NCBI: NM_130891.2 | |
| Recombinant protein | Rat GRIP1 PDZ4-7 WT: aa Q463-N1069 | This paper | NCBI: NM_032069.1 | |
| Recombinant protein | Human NHERF1 FL WT: aa M1-L358 | This paper | NCBI: NM_004252.4 | |
| Recombinant protein | Human NHERF2 FL WT: aa M1-F337 | This paper | NCBI: NM_001130012.2 | |
| Recombinant protein | Mouse MUPP1 PDZ10-11 WT: aa G1610-P1802 | This paper | NCBI: NM_001305284.1 | |
| Synthesized peptide | Mouse PLCβ4 PBM 1166EMDRRPATVV1175 | Synthesized by Chinapeptides Co. Ltd. | N/A | |
| Synthesized peptide | Drosophila NORPA PBM 1086KTQGKTEFYA1095 | Synthesized by Chinapeptides Co. Ltd. | N/A | |
| Antibody | Anti-GFP (B-2) (mouse mAb) | Santa Cruz Biotechnology | Cat# sc-9996; RRID:AB_627695 | Dilution factor: 1:1000 |
| Antibody | Anti-Mouse IgG (Goat polyAb) | Sigma | Cat# A4416; RRID:AB_258167 | Dilution factor: 1:20000 |
| Commercial assay or kit | Clone Express II, One-Step Cloning Kit | Vazyme Biotech Co., Ltd | Cat #C112 | |
| Commercial assay or kit | ViaFect transfection reagent | Promega Corporation | Cat #E4981 | |
| Software, algorithm | Origin7.0 | OriginLab | http://www.originlab.com/; RRID: SCR_002815 | ITC titration data analysis |
| Software, algorithm | GraphPad Prism | GraphPad Software Inc. | http://www.graphpad.com/scientific-software/prism; RRID: SCR_002798 | FITC titration data analysis |
| Software, algorithm | HKL2000 | HKL Research Inc. | http://www.hkl-xray.com/ | Diffraction data processing and scaling |
| Software, algorithm | CCP4 | PMID: 21460441 | http://www.ccp4.ac.uk/; RRID: SCR_007255 | Crystal structure determination |
| Software, algorithm | PHENIX | PMID: 20124702 | http://www.phenix-online.org/; RRID: SCR_014224 | Model building and refinement |
| Software, algorithm | PyMOL | DeLano Scientific LLC | http://www.pymol.org/; RRID: SCR_000305 | Structure figure plot |
| Software, algorithm | ASTRA6.1 | Wyatt Technology Corporation | http://www.wyatt.com/products/software/astra.html | Light-scattering data analysis |
| Software, algorithm | NMRPipe | NIH | https://spin.niddk.nih.gov/NMRPipe/ref/index.html | NMR data processing |
| Software, algorithm | Sparky | UCSF Sparky | https://www.cgl.ucsf.edu/home/sparky/ | NMR data analysis |
Additional files
-
Supplementary file 1
Statistics of data collection and model refinement for NORPA CC-PBM structures, INAD–NORPA complex structure and INADL–PLCβ4 complex structure.
- https://doi.org/10.7554/eLife.41848.018
-
Transparent reporting form
- https://doi.org/10.7554/eLife.41848.019





