HIPPO signaling resolves embryonic cell fate conflicts during establishment of pluripotency in vivo
Figures
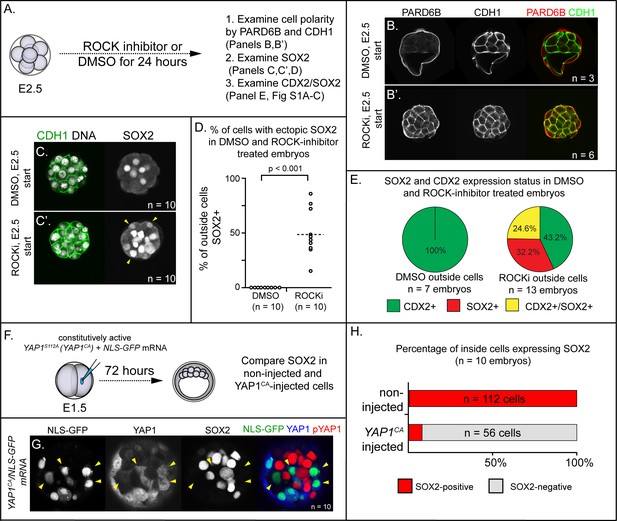
ROCK1/2 and nuclear YAP1 repress expression of SOX2.
(A) Experimental design: embryos were collected at E2.5 and treated with ROCK inhibitor Y-27632 (ROCKi) or DMSO (control) for 24 hr. (B–B’) Confocal images of apical (PARD6B) and basolateral (CDH1) membrane components in control and ROCKi-treated embryos. As expected, PARD6B and CDH1 are mislocalized to the entire cell membrane of all cells in ROCKi-treated embryos, demonstrating effective ROCK inhibition (n = number of embryos examined). (C–C’) In control embryos, SOX2 is detected only in inside cells, while in ROCKi-treated embryos, SOX2 is detected in inside and outside cells (arrowheads, outside cells; n = embryos). (D) Quantification of ectopic SOX2 detected in outside cells of control and ROCKi-treated embryos (p, student’s t-test, n = embryos). (E) SOX2 and CDX2 staining in outside cells of control and ROCKi-treated embryos. ROCK-inhibitor treatment leads to outside cells with mixed lineage marker expression (CDX2+/SOX2+). (F) Experimental design: embryos were collected at E1.5 and one of two blastomeres injected with mRNAs encoding YAP1CA and GFP. Embryos were cultured for 72 hr, fixed, and then analyzed by immunofluorescence and confocal microscopy. (G) SOX2 is detected non-injected inside cells. SOX2 is not detected in YAP1CA-overexpressing inside cells (arrowheads), n = embryos. (H) Across multiple embryos, all non-injected inside cells express SOX2, whereas the vast majority of YAP1CA-injected inside cells fail to express SOX2.
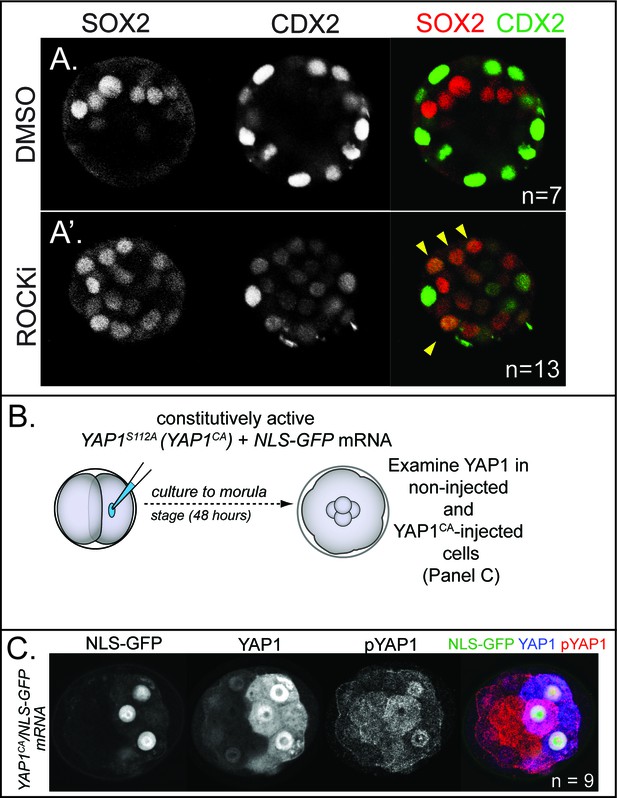
Effect of ROCK1/2 inhibition on Cdx2 expression and effect Yap1CA overexpression on YAP1 localization and phosphorylation.
(A) Confocal images of CDX2 and SOX2 in control embryos and embryos treated with ROCKi for 24 hr starting at E2.5.In control embryos, CDX2 is specific to outside cells and SOX2 is specific to inside cells (n = embryos). (A’) Treatment with ROCKi leads to ectopic SOX2 in outside cells which is often co-expressed with CDX2 (arrowheads, n = embryos). (B) YAP1CA was injected into one of two blastomeres at the 2 cell stage and evaluated 48 hr later. (C) In non-injected cells, YAP1 is exclusively nuclear in outside cells while pYAP1 is exclusively cytoplasmic in inside cells. By contrast, YAP1 is detected in the nucleus of Yap1CA-injected cells, regardless of their position, demonstrating that YAP1CA is constitutively nuclear. Additionally, analysis of pYAP1 in YAP1CA-injected cells shows that Yap1CA can still be phosphorylated on non-mutated residues, but this is not sufficient to alter YAP1 nuclear localization (n = embryos).
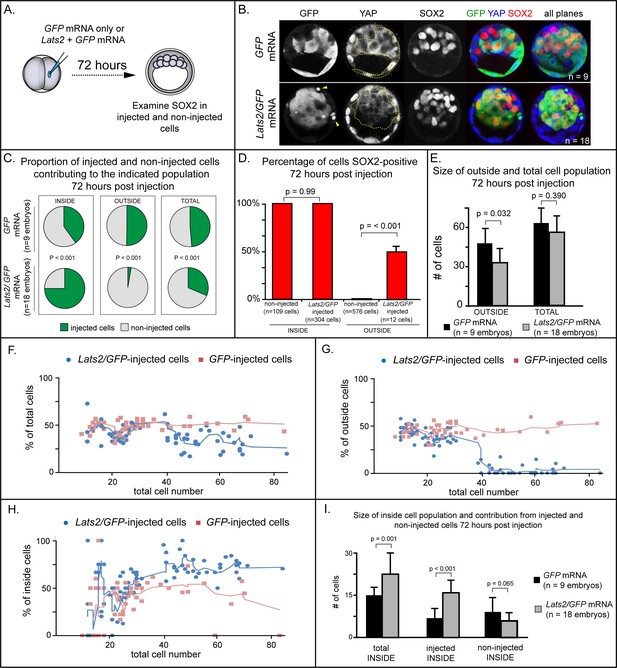
LATS2 kinase is sufficient to direct cells to inner cell mass fate.
(A) Embryos were collected at E1.5 and one of two blastomeres was injected with mRNAs encoding LATS2 and GFP or GFP alone. Embryos were cultured for 72 hr, fixed, and then analyzed by immunofluorescence and confocal microscopy. (B) Cells injected with GFP (dotted line) contributed to trophectoderm and inner cell mass, while cells injected with Lats2 and GFP (dotted line) contributed almost exclusively to the inner cell mass, leaving only cellular fragments in the trophectoderm (arrows), suggestive of cell death (n = embryos). (C) Proportion of inside, outside, and total cell populations across multiple embryos, which were comprised of non-injected cells, or cells injected with either GFP or GFP/Lats2 mRNAs. Cells injected with GFP/Lats2 were overrepresented within the inside cell population and underrepresented in the outside and total cell populations, relative to cells injected with GFP alone (P, chi-squared test). (D) Percentage of SOX2-positive cells within non-injected and GFP-injected or Lats2/GFP-injected populations observed inside and outside of the embryo. SOX2 was detected in all of the Lats2/GFP-injected inside cells, and in half of the rare, Lats2/GFP-injected outside cells (same number of embryos as in panel C) (p, student’s t-test). (E) Average number of outside and total cells per embryo. The average number of outside cells is reduced in embryos injected with Lats2/GFP, relative to GFP-injected (p, student’s t-test). (F) Proportion of GFP and Lats2/GFP-injected cells, relative to total cell number, over the course of development to the ~80-cell blastocyst (solid lines = average of indicated data point and four previous data points). (G) Data as shown in panel H, shown relative to outside cell number. (H) Data as shown in panel H, shown relative to inside cell number. (I) Contribution of injected and non-injected cells to the inside cell population, following injection with GFP or Lats2/GFP. Injection with Lats2/GFP increases the overall number of inside cells compared to injection with GFP only through increasing the number of injected cells contributing to the inside cell population, without affecting the number of non-injected cells contributing to the inside cell population (p, student’s t-test).
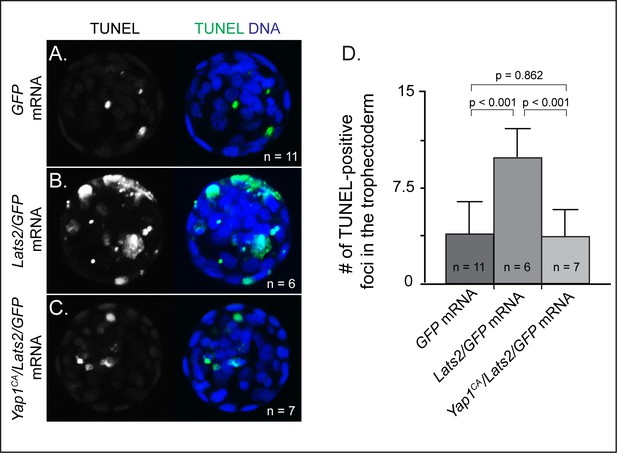
Lats2-overexpressing cells die on the surface of the embryo (A) Merge of all confocal sections from TUNEL assay performed on an embryo injected with GFP mRNA into one blastomere at the two-cell stage and then cultured until the blastocyst stage.
Note that GFP fluorescence does not survive the TUNEL assay (n = embryos). (B) Embryo injected in one of two cells with Lats2 and GFP mRNA and then cultured until the blastocyst stage shows elevated TUNEL-positive foci in the outside cells (n = embryos). (C) Embryo injected in one of two cells with Yap1CA, Lats2 and GFP mRNA and then cultured until the blastocyst stage shows reduced TUNEL-positive foci in the outside cells (n = embryos). (D) Quantification, across indicated sample sizes, of the average number of TUNEL-positive foci per embryo (t = student’s t-test, n = embryos).
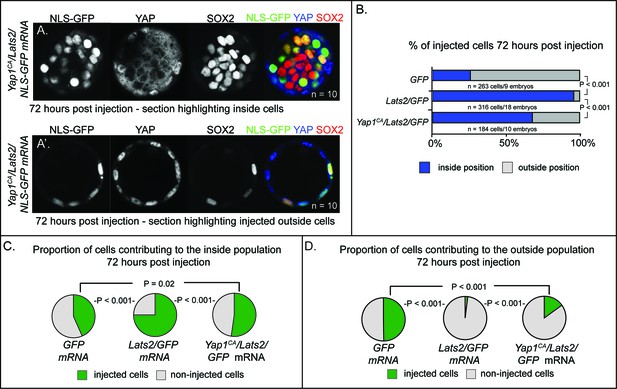
LATS2 drives cells to an inside position by inhibiting YAP1 activity (A–A’) Cooverexpression of Yap1CA and Lats2 partially rescues the ability of Lats2-overexpressing cells to contribute to trophectoderm and to repress Sox2.
Panel (A) shows a confocal section that includes the inside cell population of a Yap1CA/Lats2 injected embryo, showing inhibition by Yap1CA on Sox2 expression in some Lats2-overexpressing inside cells. Panel (A’) shows a confocal section of the same embryo and highlights the contribution of cells cooverexpressing Yap1CA and Lats2 to the trophectoderm (n = embryos). (B) Contribution of injected cells to inside and outside embryo compartments. Yap1CA-overexpression partially reverses the effect of Lats2-overexpression on cellular localization to the inside of the embryo (P, chi-squared test). (C) Proportion of non-injected cells and injected cells contributing to the inside population in embryos injected with the indicated mRNAs. Yap1CA-cooverexpression reduces the proportion of Lats2-overexpressing cells observed in the inside population (P, chi-squared test, n = embryos). (D) Proportion of non-injected cells and injected cells contributing to the outside population in embryos injected with the indicated mRNAs. Yap1CA-cooverexpression increases the proportion of Lats2-overexpressing cells observed in the outside population (P, chi-squared test, n = embryos).
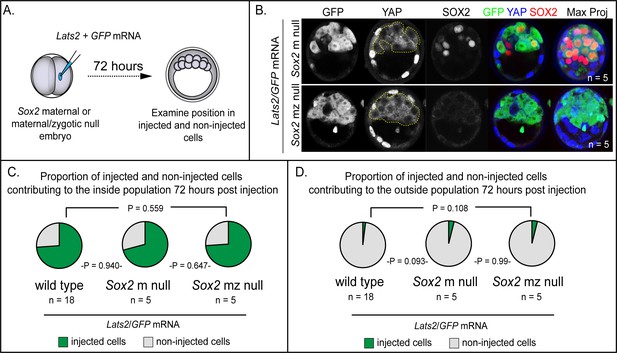
LATS2 directs inner cell mass fate independently of Sox2 (A) Lats2 and GFP or GFP alone were overexpressed in embryos lacking maternal or maternal and zygotic Sox2.
(B) Lats2/GFP-overexpressing cells (dotted line) contribute almost exclusively to the inner cell mass in the presence or absence of Sox2 (n = embryos). (C) Proportion of non-injected cells and cells injected with Lats2/GFP mRNAs contributing to inner cell mass in the indicated genetic backgrounds. No significant differences were observed based on embryo genotype, indicating that Sox2 is dispensable for inside positioning by Lats2-overexpression (P, chi-squared test; n = embryos). (D) Proportion of non-injected cells and cells injected with the indicated mRNAs contributing to trophectoderm in the indicated genetic backgrounds. No significant differences were observed based on embryo genotype (P, chi-squared test; n = embryos).
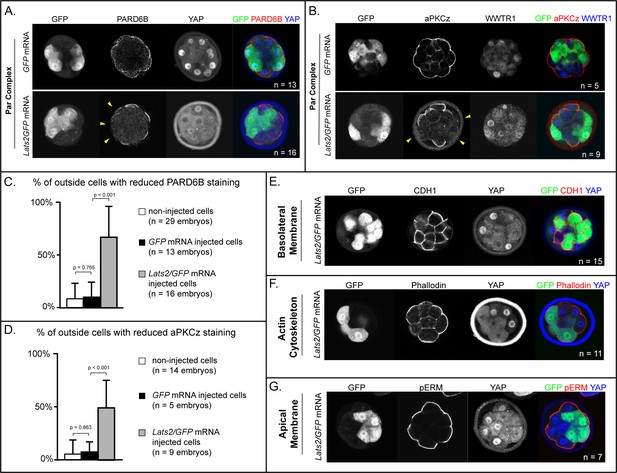
LATS2 antagonizes formation of the apical domain (A) In embryos at 16–32 cell stages, PARD6B is detectable in GFP-overexpressing and in non-injected cells, but not in Lats2-overexpressing cells (arrowheads, n = embryos).
(B) At 16–32 cell stages, aPKCz is detectable in GFP-overexpressing and in non-injected cells, but not in Lats2-overexpressing cells (arrowheads, n = embryos). (C) Quantification of embryos shown in panel A (p, student’s t-test). (D) Quantification of embryos shown in panel B (p, student’s t-test). (E) At 16–32 cell stages, CDH1 is localized to the basolateral membrane in both Lats2-overexpressing and non-injected cells (n = embryos). (F) At 16–32 cell stages, Phalloidin staining demonstrates that filamentous Actin is apically enriched in Lats2-overexpressing and non-injected cells (n = embryos). (G) At 16–32 cell stages, pERM is localized to the apical membrane in both Lats2-overexpressing and non-injected cells (n = embryos).
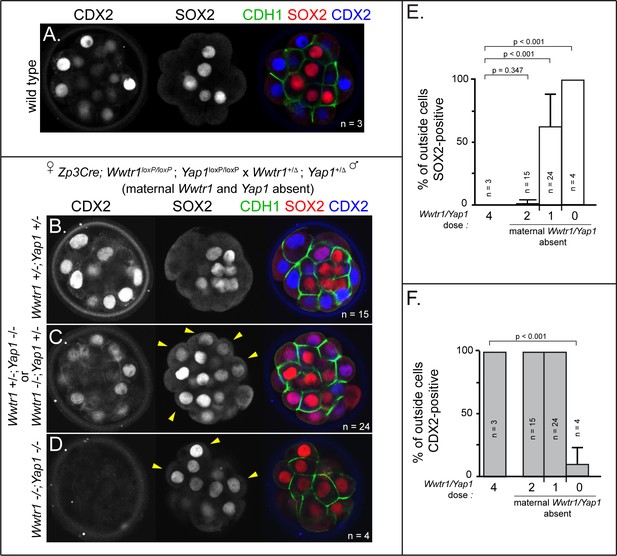
Wwtr1 and Yap1 are required to repress SOX2 expression in outside cells.
(A) CDX2 and SOX2 in wild type embryos at E3.25 (16–32 cell stages). CDX2 staining is more intense in outside cells than inside cells and SOX2 staining is specific to inside cells (n = embryos). (B) Embryos lacking maternal Wwtr1 and Yap1 with and heterozygous for Wwtr1 and Yap1 (which we consider to have 2 doses of WWTR1/YAP1) exhibit normal CDX2 and SOX2 expression (n = embryos). (C) Embryos lacking maternal Wwtr1 and Yap1 and heterozygous for either Wwtr1 or Yap1 (1 dose of WWTR1/YAP1) exhibit a high degree of ectopic SOX2 in outside cells (arrowheads), but continue to express CDX2, although the levels appear reduced (n = embryos). (D) Embryos lacking maternal and zygotic Wwtr1 and Yap1 (0 doses of WWTR1/YAP1) have the most severe phenotype, with a high degree of ectopic SOX2 in outside cells (arrowheads) and little or no detectable CDX2 (n = embryos). (E) Quantification of the percentage of outside cells in which ectopic SOX2 is detected in the presence of decreasing dose of Wwtr1 and Yap1 (t = student’s t-test, n = embryos). (F) Quantification of the percentage of outside cells in which CDX2 is detected in the presence of decreasing dose of Wwtr1 and Yap1 (t = student’s t-test, n = embryos).
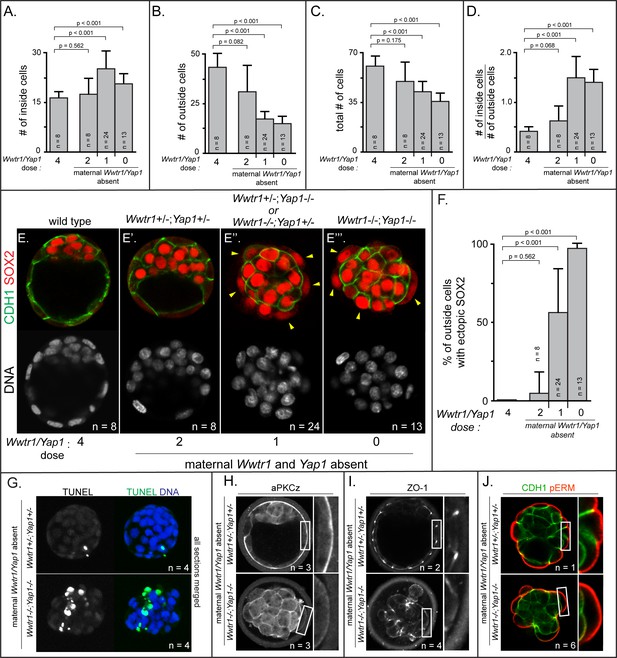
Positioning and epithelialization defects in embryos with Wwtr1 and Yap1 null alleles (A) Quantification of the average number of inside cells per embryo with decreasing dose of Wwtr1 and Yap1.
The number of inside cells increases as the dose of wild type Wwtr1 and Yap1 alleles is reduced (p, student’s t-test, n = embryos). (B) Quantification of the average number of outside cells per embryo with decreasing dose of Wwtr1 and Yap1. The number of outside cells decreases as the dose of wild type Wwtr1 and Yap1 alleles is reduced (p, student’s t-test, n = embryos). (C) Quantification of the average number of total cells per embryos with decreasing dose of wild type zygotic Wwtr1 and Yap1. The number of total cells decreases as the dose of wild type Wwtr1 and Yap1 is reduced (p, student’s t-test, n = embryos). (D) Quantification of the average ratio of inside to outside cells per embryo with decreasing dose of Wwtr1 and Yap1. The ratio of inside to outside cells increases as the dose of wild type Wwtr1 and Yap1 is reduced (p, student’s t-test, n = embryos). (E) Wild type embryos at E3.75 exhibit inner cell mass-specific expression of SOX2 (n = embryos). (E’) E3.75 embryos lacking maternal Wwtr1 and Yap1 and heterozygous for zygotic Wwtr1 and Yap1 cavitate and repress Sox2 in outside cells, leading to inner cell mass-specific expression of SOX2 similar to wild type embryos (n = embryos). (E’’) Embryos lacking maternal Wwtr1 and Yap1 but with only one wild type allele of Wwtr1 or Yap1 fail to cavitate and repress Sox2 in outside cells, leading to ectopic SOX2 in outside cells (arrowheads, n = embryos). (E’’’) Embryos lacking maternal and zygotic Wwtr1 and Yap1 fail to cavitate and repress Sox2 in outside cells, leading to ectopic SOX2 in outside cells (arrowheads, n = embryos). (F) Quantification of ectopic SOX2 detected in embryos such as those shown in panels E-E’’’. The percentage of outside cells with ectopic SOX2 increases as the dose of wild type Wwtr1 and Yap1 alleles is reduced (p, student’s t-test, n = embryos). (G) TUNEL analysis of embryos lacking maternal Wwtr1 and Yap1 heterozygous for zygotic Wwtr1 and Yap1 or lacking maternal and zygotic Wwtr1 and Yap1. Extensive TUNEL staining is observed in embryos lacking maternal and zygotic Wwtr1 and Yap1 indicative of cell death. Max projections of all confocal sections from a single embryo are shown (n = embryos). (H) aPKCz staining in embryos lacking maternal Wwtr1 and Yap1, either heterozygous for zygotic Wwtr1 and Yap1 or with no zygotic Wwtr1 and Yap1. aPKC is not localized to the apical membrane of embryos with no zygotic Wwtr1 and Yap1 (n = embryos). (I) ZO-1 staining in embryos lacking maternal Wwtr1 and Yap1, either heterozygous for zygotic Wwtr1 and Yap1 or with no zygotic Wwtr1 and Yap1. ZO-1 is disorganized in embryos with no zygotic Wwtr1 and Yap1, suggesting that formation of a mature epithelium depends on Wwtr1 and Yap1 (n = embryos). (J) pERM and CDH1 staining in embryos lacking maternal Wwtr1 and Yap1, either heterozygous for zygotic Wwtr1 and Yap1 or with no zygotic Wwtr1 and Yap1. pERM is localized to apical membranes and CDH1 to basolateral membranes regardless of the dose of wild type Wwtr1 and Yap1 alleles (n = embryos).
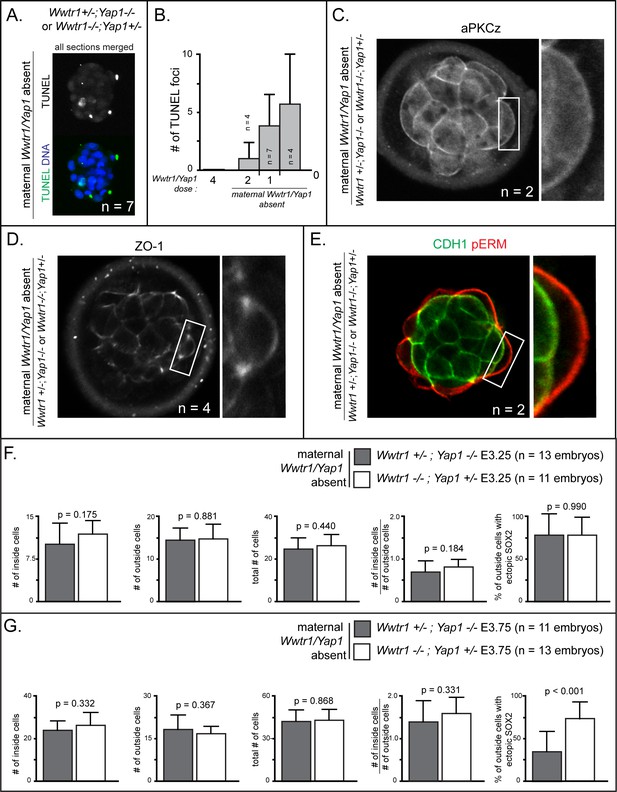
Increased cell death and epithelialization defects in embryos lacking maternal Wwtr1 and Yap1 with a single wild type allele of Wwtr1 or Yap1.
(A) TUNEL staining in embryos lacking maternal Wwtr1 and Yap1 with a single wild type allele of Wwtr1 or Yap1. Max projection of all confocal sections taken from a single embryo is shown (n = embryos) (B) Quantification of the average number of TUNEL foci per embryo in embryos with decreasing doses of Wwtr1 and Yap1 (n = embryos) (C) aPKCz staining in embryos lacking maternal Wwtr1 and Yap1 with a single wild type allele of Wwtr1 or Yap1. aPKC is not localized to the apical membrane of embryos lacking maternal Wwtr1 and Yap1 with a single wild type allele of Wwtr1 or Yap1 (n = embryos). (D) ZO-1 staining in embryos lacking maternal Wwtr1 and Yap1 with a single wild type allele of Wwtr1 or Yap1. aPKC is not localized to the apical membrane of embryos lacking maternal Wwtr1 and Yap1 with a single wild type allele of Wwtr1 or Yap1 (n = embryos). (E) pERM and CDH1 staining in embryos lacking maternal Wwtr1 and Yap1 with a single wild type allele of Wwtr1 or Yap1. pERM is correctly localized to apical membranes and CDH1 correctly localized to basolateral membranes in all embryos (n = embryos). (F) Average numbers per embryo of each stated category in embryos of indicated genotypes at E3.25. No differences were detected between the two genotypes at this stage (p, student’s t-test; n = embryos). (G) Average numbers per embryo of each stated category in embryos of indicated genotypes at E3.75. The only significant difference observed was in the degree of ectopic SOX2 detected in outside cells, a phenotype that was more severe in embryos lacking Wwtr1 (p, Student’s t-test; n = embryos).
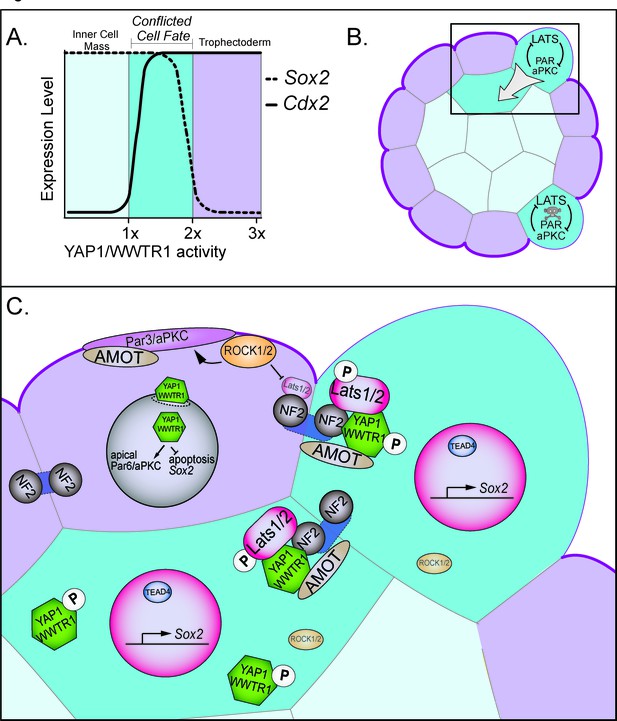
Resolution of cell fate conflicts in the preimplantation mouse embryo.
(A) The expression of Sox2 and Cdx2 is differentially sensitive to YAP1/WWTR1 activity, leading to co-expression of both lineage markers in cells when YAP1/WWTR1 activity levels are intermediate. (B) During division from the 16 to the 32-cell stage, cells that inherit the apical membrane repress HIPPO signaling and maintain an outside position. However, cells that inherit a smaller portion of the apical membrane would initially elevate their HIPPO signaling. We propose that elevated HIPPO then feeds back onto polarity by further antagonizing PAR-aPKC complex formation, leading to a snowball effect on repression of Sox2 expression, and thus ensuring that SOX2 is never detected in outside cells because these cells are rapidly internalized or apoptosed. (C) A closeup of the boxed region in panel B. In most outside cells, low LATS2 activity enables high levels of YAP1/WWTR1 activity, which repress Sox2 and apoptosis and promote Cdx2 expression and apical localization of aPKC and PARD6B, which in turn repress the HIPPO pathway. In rare outside cells, LATS2 activity becomes elevated, leading to lower activity of YAP1/WWTR1, which then leads these cells to become internalized or to undergo apoptosis.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Mus musculus) | CD-1 | Charles River Laboratories | RRID:IMSR_CRL:22 | |
| Strain, strain background (M. musculus) | Sox2tm1.1Lan | Smith et al. (2009) PMID:19666824 | RRID:IMSR_JAX:013093 | mixed background, Sox2 null refers to recombined allele |
| Strain, strain background (M. musculus) | Wwtr1 conditional allele; Wwtr1tm1.1Eno; Wwtr1loxp | Xin et al., 2013 Xin et al., 2013 PMID:23918388 | MGI:5544289 | mixed background, ‘Wwtr1-’ or ‘Wwtr1’ refers to recombined allele |
| Strain, strain background (M. musculus) | Yap1 conditional allele; Yap1tm1.1Eno; Yap1loxp | Xin et al., 2011 Xin et al., 2011 PMID:22028467 | MGI:5446483 | mixed background, , ‘Yap1-’ or ‘Yap1’ refers to recombined allele |
| Strain, strain background (M. musculus) | Tg(Zp3-cre) 93Knw; Zp3Cre | de Vries et al., 2000 de Vries et al., 2000 de Vries et al., 2000 PMID:10686600 | RRID:MGI:3835503 | mixed background |
| Strain, strain background (M. musculus) | 129-Alpl tm(cre)Nagy | Lomelí et al., 2000 Lomelí et al., 2000 Lomelí et al., 2000 PMID:10686602 | RRID:IMSR_ JAX:008569 | mixed background |
| Antibody | mouse anti-CDX2 | Biogenex | BioGenex Cat# AM392; RRID:AB_2650531 | (1:200) |
| Antibody | goat anti -SOX2 | Neuromics | Neuromics Cat# GT15098; RRID:AB_2195800 | (1:200) |
| Antibody | rabbit anti-PARD6B | Novus Biologicals | Novus Cat# NBP1-87337; RRID:AB_11034389 | (1:100) |
| Antibody | rabbit anti-PARD6B | Santa Cruz Biotechnology | Santa Cruz Biotechnology Cat# sc-67393; RRID:AB_2267889 | (1:100) |
| Antibody | mouse anti-PKCζ | Santa Cruz Biotechnology | Santa Cruz Biotechnology Cat# sc-17781; RRID:AB_628148 | (1:100) |
| Antibody | mouse anti-YAP1 | Santa Cruz Biotechnology | Santa Cruz Biotechnology Cat# sc-101199; RRID:AB_1131430 | (1:200) |
| Antibody | mouse anti-pYAP1 | Cell Signaling Technology | Cell Signaling Technology Cat# 4911; RRID:AB_2218913 | (1:800) |
| Antibody | chicken anti-GFP | Aves Labs | Aves Labs Cat# GFP-1020; RRID:AB_10000240 | (1:2000) |
| Antibody | rat anti- CDH1 | Sigma- Aldrich | Sigma-Aldrich Cat# U3254; RRID:AB_477600 | (1:500) |
| Antibody | mouse anti-ZO1 | Thermo Fisher Scienctific | Thermo Fisher Scientific Cat# 33–9100; RRID:AB_2533147 | (1:1000) |
| Recombinant DNA reagent | Lats2 mRNA; LATS2 | Nishioka et al., 2009 Nishioka et al., 2009 Nishioka et al., 2009 PMID:19289085 | pcDNA3.1 -pA83-Lats2; RIKEN: RDB12200 | In Vitro Transcription template for Lats2 mRNA |
| Recombinant DNA reagent | Yap1CA mRNA; YAP1CA | Nishioka et al., 2009 Nishioka et al., 2009 Nishioka et al., 2009 PMID:19289085 | pcDNA3.1-pA83 -HA-Yap-S112A; RIKEN: RDB12194 | In Vitro Transcription template for Yap1CA mRNA |
| Recombinant DNA reagent | nls-GFP mRNA; nls-GFP | Ariotti et al., 2015 Ariotti et al., 2015 Ariotti et al., 2015 PMID:26585296 | Addgene: Plasmid #67652 | In Vitro Transcription template for nls- GFP mRNA |
| Recombinant DNA reagent | pCS2-EGFP; EGFP mRNA; GFP mRNA; GFP | Chazaud et al., 2006 PMID: 16678776 | In Vitro Transcription template for GFP mRNA | |
| Commercial assay or kit | mMessage mMachine Sp6 Transcription Kit | Thermo Fisher Scienctific | Thermo Fisher Scientific Cat# AM1340 | |
| Commercial assay or kit | mMessage mMachine T7 Transcription Kit | Thermo Fisher Scienctific | Thermo Fisher Scientific Cat# AM1344 | |
| Commercial assay or kit | MEGAClear Transcription Clean-Up Kit | Thermo Fisher Scienctific | Thermo Fisher Scientific Cat# AM1908 | |
| Commercial assay or kit | In-Situ Cell Death Detection Kit, Fluorescein; TUNEL assay | Sigma-Aldrich | Sigma-Aldrich Cat# 11684795910 | |
| Commercial assay or kit | Extract-N -Amp Kit | Sigma- Aldrich | Sigma- Aldrich Cat # XNAT2 | |
| Chemical compound, drug | Y-27632; ROCK-inhibitor | Millipore | Millipore Cat# SCM075 | |
| Software, algorithm | Adobe Photoshop | Adobe | RRID:SCR_014199 | |
| Software, algorithm | Fiji | http://fiji.sc | RRID:SCR_002285 |
Additional files
-
Supplementary file 1
Summary of embryos recovered from Wwtr1;Yap1 germline null females.
- https://doi.org/10.7554/eLife.42298.014
-
Supplementary file 2
Mean and standard deviation of cell counts for every experimental treatment.
- https://doi.org/10.7554/eLife.42298.015
-
Supplementary file 3
Allele-specific primers used for determining embryo and mouse genotypes.
- https://doi.org/10.7554/eLife.42298.016
-
Transparent reporting form
- https://doi.org/10.7554/eLife.42298.017





