TRIM28 promotes HIV-1 latency by SUMOylating CDK9 and inhibiting P-TEFb
Figures
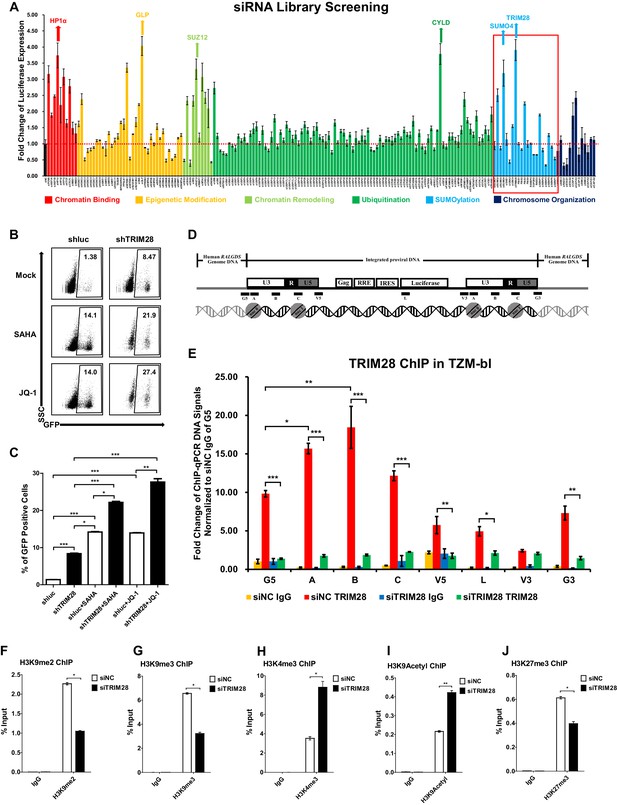
TRIM28 suppresses HIV-1 expression and contributes to HIV-1 latency.
(A) A siRNA library targeting 182 human genes was transfected into TZM-bl cell line, respectively. Three distinct siRNAs targeting each gene were transfected as a mixture. Forty-eight hours post-transfection, cells were harvested and the activity of luciferase from cell lysates was measured. Fold changes were calculated for each gene compared to negative control siRNA (siNC). (B–C) shRNA constructs were packaged into recombinant lentiviruses and infected J-Lat 10.6. The reactivation efficiency was measured by the GFP-positive percentage which was shown in the top right corner. SAHA and JQ-1 were used as positive controls. (D) Eight ChIP-qPCR primers targeting HIV-1 reporter provirus were designed. G5: Cellular DNA and viral 5’LTR junction; A: Nucleosome 0 assembly site; B: Nucleosome-free region; C: Nucleosome one assembly site; V5: Viral 5’LTR and gag leader sequence junction; L: Luciferase region; V3: Viral poly purine tract and 3’LTR junction; G3: Viral 3’LTR and cellular DNA junction. (E) ChIP assay with antibody against TRIM28 was performed in TZM-bl cell line. All the ChIP-qPCR DNA signals were normalized to siNC IgG of G5. (F–J) ChIP assays with antibodies against H3K9me2, H3K9me3, H3K4me3, H3K9Acetyl and H3K27me3 were performed in TZM-bl cell lines. Data represents mean ±SEM in triplicates. p-Values were calculated by Student’s t-test. *p<0.05, **p<0.01, ***p<0.001.
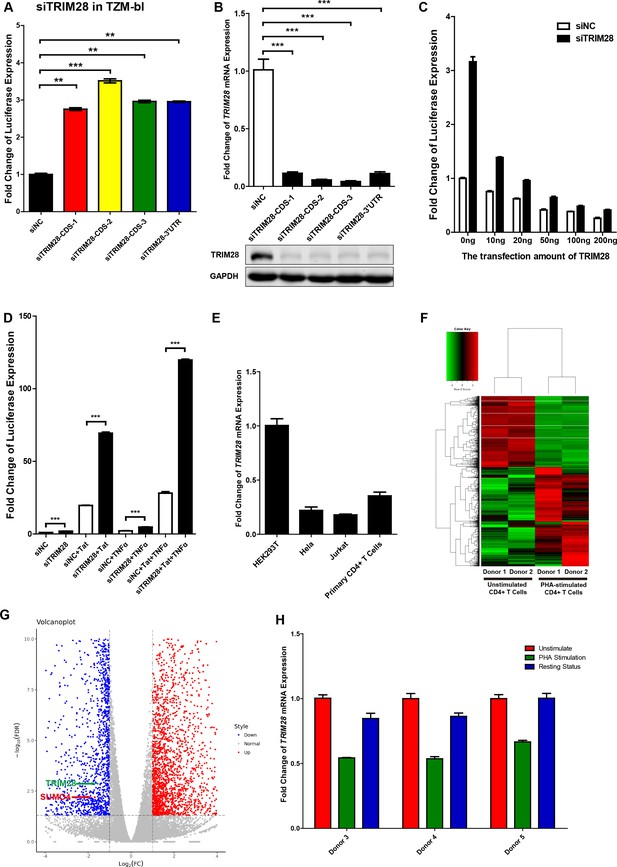
TRIM28 suppresses HIV-1 expression and is upregulated upon activation by PHA.
(A) TRIM28 in TZM-bl cells was knocked down by siRNAs targeting the coding sequence and 3’UTR of TRIM28 mRNA. The luciferase from clarified lysates was quantitated and normalized to siNC. Data represents mean ±SEM in triplicates. P values were calculated by Student’s t-test. **p<0.01, ***p<0.001. (B) The knockdown efficiency of different TRIM28 siRNAs was confirmed by qPCR and western blot. Data represents mean ±SEM in triplicates. p-Values were calculated by Student’s t-test. ***p<0.001. (C) Endogenous TRIM28 in TZM-bl cells was knocked down by siRNA targeting 3’UTR or treated with siNC. Different gradients of TRIM28 construct were co-transfected. The luciferase from lysate supernatants was quantitated and normalized to the siNC control which was not co-transfected with TRIM28. (D) Endogenous TRIM28 in TZM-bl cells was knocked down by siRNAs or treated with siNC. HIV-1 Tat construct and TNFα were separately co-treated with siRNAs or joint used with siRNAs. The luciferase from clarified lysates was quantitated and normalized to the siNC which has no additive. Data represents mean ± SEM in triplicates. p-Values were calculated by Student’s t-test. ***p<0.001. (E) The expression of TRIM28 in different cells was quantitated by qPCR and normalized to HEK293T group. β-actin mRNA was set as internal reference. (F–G) Freshly isolated CD4+ T cells from two healthy donors were stimulated with PHA for 2 days or left untreated. Total mRNAs from each group were extracted and proceeded to RNA-Seq. Differentially expressed genes, which were filtered with log2FC of 1 and PvalueFDR cutoff of 0.05, were plotted as heatmap (F) and volcanoplot (G). Arrow-pointed scatters indicated TRIM28 and SUMO4. (H) CD4+ T cells from three healthy donors were stimulated with PHA for 2 days or left untreated. One part of PHA-activated CD4+ T cells was washed for removing PHA and cultured in RPMI1640 which contained low IL-2 for 1 month. Then, resting CD4+ T cells were isolated from long-term cultured CD4+ T cells. Total RNAs from unstimulated (red), PHA-stimulated (green) and resting (blue) CD4+ T cells were extracted and proceeded to qPCR. TRIM28 from each group was quantitated and normalized to unstimulated group. β-actin mRNA was set as internal reference.
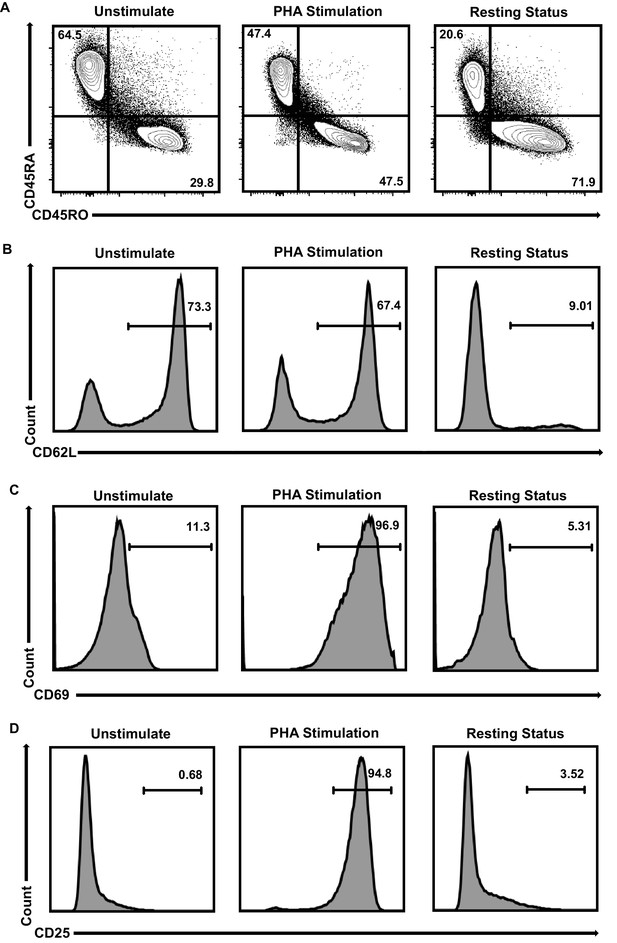
Primary CD4+T cells populations’ identities.
(A–D) CD4+ T cells were stimulated with PHA for 2 days or left untreated. One part of PHA-activated CD4+ T cells was washed for removing PHA and cultured in RPMI1640 which contained low IL-2 for 1 month. Then, resting CD4+ T cells were isolated from long-term cultured CD4+ T cells. Both of unstimulated (left panels), PHA-stimulated (middle panels) and resting (right panels) CD4+ T cells were incubated with antibodies against CD4, CD45RA, CD45RO, CD62L, CD69 and CD25, and analyzed by flow cytometry. Most of the unstimulated CD4+ T cells were naïve cells which were CD45RA+/CD45RO-/CD62L+/CD69-/CD25-. After PHA stimulation, CD4+ T cells started to change into CD45RA-/CD45RO+/CD62L- and express high amounts of activation markers CD69 and CD25 which were the identities of activated effector cells. Most of long-term cultured resting CD4+ T cells were CD45RA-/CD45RO+/CD62L-/CD69-/CD25- which were the identities of effector memory CD4+ T cells.
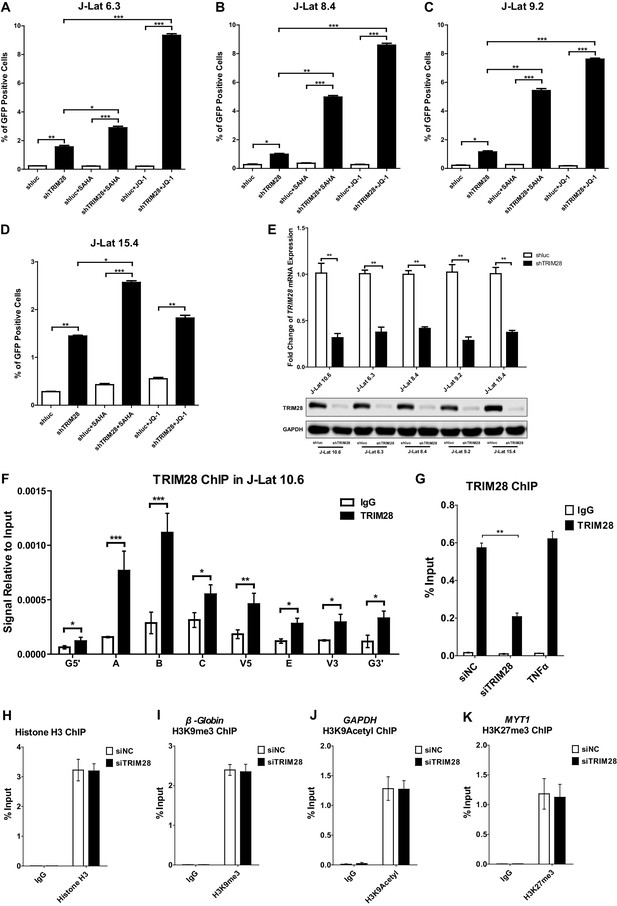
TRIM28 contributes to HIV-1 latency and is enriched on HIV-1 LTR.
(A–D) J-lat 6.3, 8.4, 9.2 and 15.4 cell lines were treated as in Figure 1B. The reactivation efficiency for each group was analyzed as in Figure 1C. Data represents mean ±SEM in triplicates. p-Values were calculated by Student’s t-test. *p<0.05, **p<0.01, ***p<0.001. (E) The knockdown efficiency of shTRIM28 in different cell lines was confirmed by qPCR and western blot. (F) ChIP assay with antibodies against TRIM28 and normal rabbit IgG was performed in J-Lat 10.6 cell line. All the ChIP-qPCR DNA signals were normalized to IgG of G5’. Data represents mean ±SEM in triplicates. p-Values were calculated by Student’s t-test. *p<0.05, **p<0.01, ***p<0.001. G5’ represented cellular DNA and viral 5’LTR junction; E represented envelop; G3’ represented viral 3’LTR and cellular DNA junction; A, B, C, V5 and V3 represented as in Figure 1D (Supplementary file 2). (G) ChIP assay with antibodies against TRIM28 and normal rabbit IgG was performed in TZM-bl cell lines which were treated with negative control, TRIM28 siRNAs and TNFα, respectively. Data represents mean ±SEM in triplicates. p-Values were calculated by Student’s t-test. **p<0.01. (H–K) Data represented positive controls of siTRIM28-related ChIP. ChIP assay with antibodies against Histone H3, H3K9me3, H3K9Acetyl, H3K27me3 and normal rabbit IgG was performed in TZM-bl cell lines which were treated with negative control and TRIM28 siRNAs, respectively. For Histone H3 ChIP, ChIP-qPCR DNA signals were normalized to Input of ‘B’ which represented the nucleosome free region of HIV-1 LTR (H). ChIP-qPCR DNA signals were normalized to input of the promoter of β-Globin for H3K9me3 ChIP (I). ChIP-qPCR DNA signals were normalized to input of the promoter of GAPDH for H3K9Acetyl ChIP (J). ChIP-qPCR DNA signals were normalized to Input of the promoter of MYT1 for H3K27me3 ChIP (K).
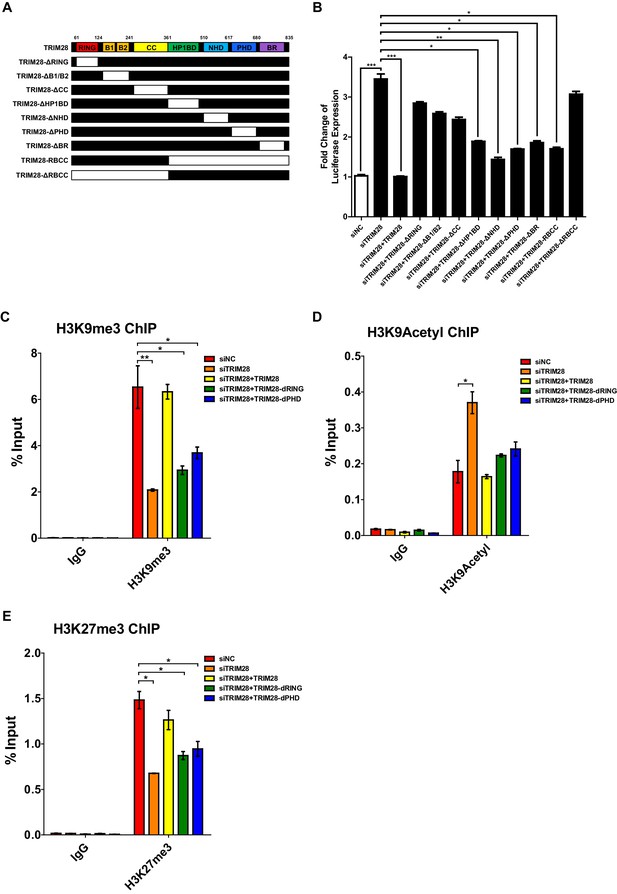
Both RING and PHD domains E3 ligase activities are important for repressive epigenetic modifications.
(A) Schematic of wild-type TRIM28 and nine TRIM28 mutants. (B) Endogenous TRIM28 was knocked down by siRNA targeting 3’UTR in TZM-bl cells and re-expressed with wild type and different TRIM28 mutants. The luciferase activity was measured. Data represents mean ±SEM in triplicates. p-Values were calculated by Student’s t-test. *p<0.05, **p<0.01, ***p<0.001. (C–E) Endogenous TRIM28 in TZM-bl cells was knocked down by siRNA targeting 3’UTR of TRIM28 mRNA. Another three groups whose endogenous TRIM28 was knocked down were overexpressed with wild type TRIM28 construct or TRIM28 mutants without RING or PHD domain, respectively. ChIP assays with antibodies against H3K9me3, H3K9Acetyl and H3K27me3 were performed for each group. Data represents mean ±SEM in triplicates. p-Values were calculated by Student’s t-test. *p<0.05, **p<0.01.
Positive controls for ChIP.
(A–D) Data represented positive controls of siTRIM28-related ChIP. Endogenous TRIM28 in TZM-bl cells was knocked down by siRNA targeting 3’UTR of TRIM28 mRNA. Another three groups whose endogenous TRIM28 was knocked down were overexpressed with wild-type TRIM28 construct or TRIM28 mutants without RING or PHD domain, respectively. ChIP assay with antibodies against Histone H3, H3K9me3, H3K9Acetyl, H3K27me3 and normal rabbit IgG was performed for each group. For Histone H3 ChIP, ChIP-qPCR DNA signals were normalized to Input of ‘B’ which represented the nucleosome-free region of HIV-1 LTR (A). ChIP-qPCR DNA signals were normalized to Input of the promoter of β-Globin for H3K9me3 ChIP (B). ChIP-qPCR DNA signals were normalized to Input of the promoter of GAPDH for H3K9Acetyl ChIP (C). ChIP-qPCR DNA signals were normalized to Input of the promoter of MYT1 for H3K27me3 ChIP (D).
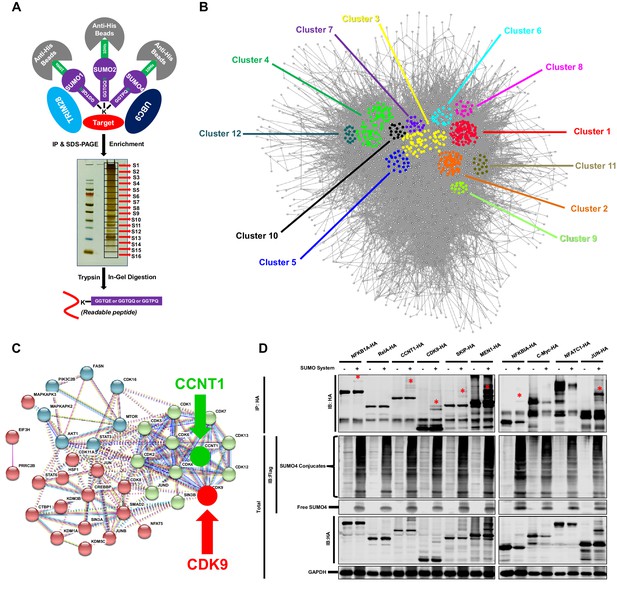
TRIM28 SUMOylates many transcription factors and transferases.
(A) Schematic of global site-specific SUMO-MS. His-tagged SUMO mutants were co-overexpressed with UBC9 and TRIM28. The SUMOylated proteins were enriched by His-tag beads and separated by SDS-PAGE. Gel fragments were excised and subjected to separate in-gel digestions. The digested peptides were desalted and analyzed by nanoscale LC-MS/MS. (B) SUMOylated proteins were analyzed with STRING. The network were further analyzed by MCODE. Twelve highly interconnected functional subclusters were extracted and shown in different colors. (C) Transferases and transcription factors were clustered by k-means clustering and visualized with STRING analysis. (D) Ten HA-tagged various transcriptional factors were overexpressed with Flag-tagged SUMO proteins, UBC9 and TRIM28. The targeted proteins were immunoprecipitated (IP) by anti-HA-tag beads followed by immunoblotting (IB) with anti-HA and –Flag antibodies. Asterisk (*) indicated the SUMOylated bands.
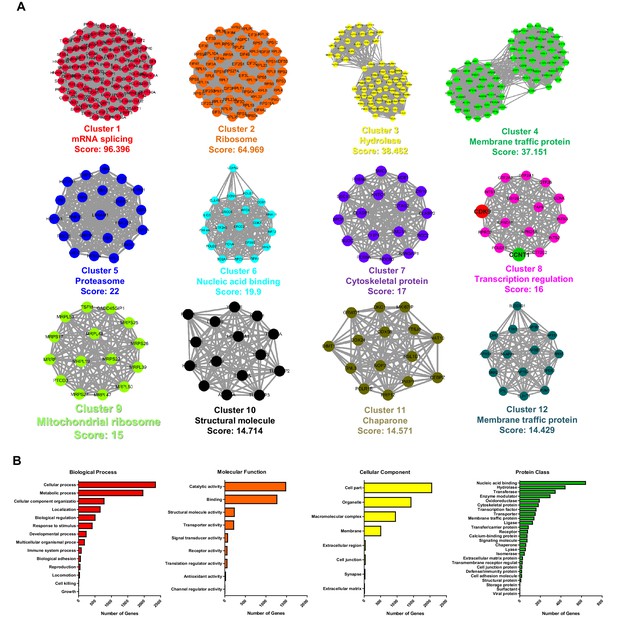
STRING, MCODE and GO analysis of proteins SUMOylated by TRIM28.
(A) Twelve subclusters, which were extracted by MCODE analysis, were separately plotted. The STRING and MCODE analyses were performed with the following settings: a significance threshold below 10−7, interaction confidence of 0.7, a degree cutoff of 3, a node score cutoff of 0.1, a maximum depth of 2, a K-Core of 5, and haircut. Cluster name and corresponded interconnectivity score were shown below each cluster. (B) Biological process analysis, molecular function analysis, cellular component analysis and protein class analysis were conducted for the identified SUMOylated proteins.
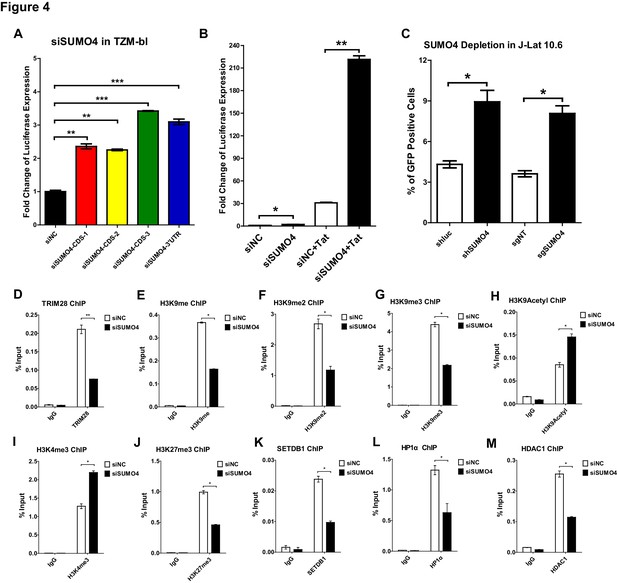
SUMO4 suppresses HIV-1 expression and contributes to HIV-1 latency.
(A) SUMO4 in TZM-bl cells was knocked down by siRNAs targeting the coding sequence and 3’UTR of SUMO4 mRNA. The luciferase from clarified lysates was quantitated and normalized to siNC. Data represents mean ± SEM in triplicates. p-Values were calculated by Student’s t-test. **p<0.01, ***p<0.001. (B) SUMO4 in TZM-bl cells was knocked down by siRNAs or treated with siNC. HIV-1 Tat construct was co-treated with siRNAs. The luciferase from clarified lysates was quantitated and normalized to the siNC which had no additive. Data represents mean ±SEM in triplicates. p-Values were calculated by Student’s t-test. *p<0.05, **p<0.01. (C) shRNA or sgRNA constructs targeting luciferase (shluc), non-target (sgNT) and SUMO4 (shSUMO4 and sgSUMO4) were packaged into recombinant lentiviruses and infected J-Lat 10.6. The reactivation efficiency was measured by the GFP positive percentage. Data represents mean ±SEM in triplicates. p-Values were calculated by Student’s t-test. *p<0.05. (D–M) SUMO4 in TZM-bl cells was knocked down by siRNA targeting SUMO4 mRNA. ChIP assays with antibodies against TRIM28, H3K9me, H3K9me2, H3K9me3, H3K9Acetyl, H3K4me3, H3K27me3, SETDB1, HP1α and HDAC1 were performed for each group. Data represents mean ±SEM in triplicates. p-Values were calculated by Student’s t-test. *p<0.05, **p<0.01.
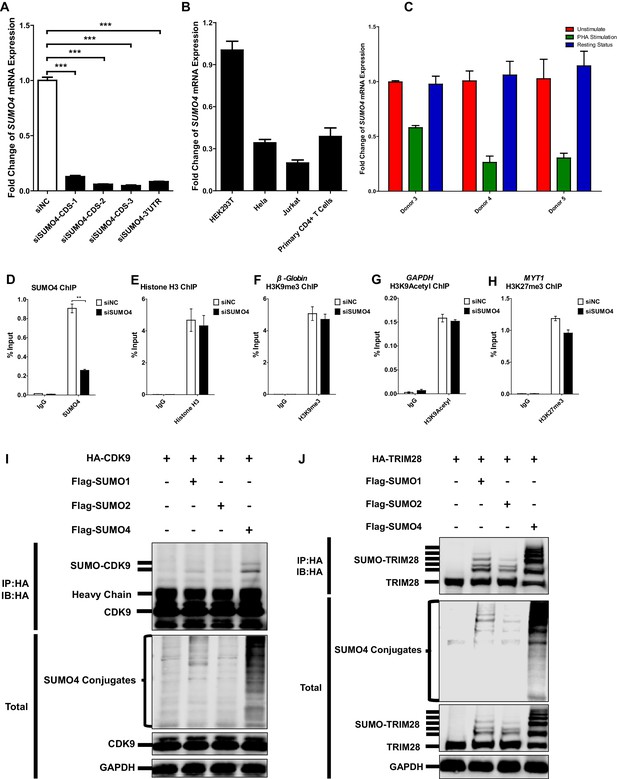
SUMO4 is uregulated upon activation by PHA and is the major paralog used by CDK9 and TRIM28.
(A) The knockdown efficiency of different SUMO4 siRNAs was confirmed by qPCR. Data represents mean ±SEM in triplicates. p-Vvalues were calculated by Student’s t-test. ***p<0.001. (B) The expression of SUMO4 in different cells was quantitated by qPCR and normalized to HEK293T group. β-actin mRNA was set as internal reference. (C) CD4+ T cells from three healthy donors were stimulated with PHA for 2 days or left untreated. One part of PHA-activated CD4+ T cells was washed for removing PHA and cultured in RPMI1640 which contained low IL-2 for 1 month. Then, resting CD4+ T cells were isolated from long-term cultured CD4+ T cells. Total RNAs from unstimulated (red), PHA-stimulated (green) and resting (blue) CD4+ T cells were extracted and proceeded to qPCR. SUMO4 from each group was quantitated and normalized to unstimulated group. β-actin mRNA was set as internal reference. (D–H) Data represented positive controls of siSUMO4-related ChIP. SUMO4 in TZM-bl cells was knocked down by siRNA targeting SUMO4 mRNA or treated with siNC. ChIP assays with antibodies against SUMO4, Histone H3, H3K9me3, H3K9Acetyl and H3K27me3 were performed for each group. For SUMO4 and Histone H3 ChIP, ChIP-qPCR DNA signals were normalized to Input of ‘B’ which represented the nucleosome-free region of HIV-1 LTR (D–E). ChIP-qPCR DNA signals were normalized to Input of the promoter of β-Globin for H3K9me3 ChIP (F). ChIP-qPCR DNA signals were normalized to Input of the promoter of GAPDH for H3K9Acetyl ChIP (G). ChIP-qPCR DNA signals were normalized to Input of the promoter of MYT1 for H3K27me3 ChIP (H). Data represents mean ±SEM in triplicates. p-Values were calculated by Student’s t-test. **p<0.01. (I) HA-tagged CDK9 was co-overexpressed with Flag-tagged SUMO1, Flag-tagged SUMO2 and Flag-tagged SUMO4, respectively. CDK9 was IP with anti-HA beads, followed by IB with antibodies against HA-tag, Flag-tag and GAPDH in total samples (lower panel), and IB with antibody against HA-tag in IP samples (upper panel). (J) HA-tagged TRIM28 was co-overexpressed with Flag-tagged SUMO1, Flag-tagged SUMO2 and Flag-tagged SUMO4, respectively. TRIM28 was IP with anti-HA beads, followed by IB with antibodies against HA-tag, Flag-tag and GAPDH in total samples (lower panel), and IB with antibody against HA-tag in IP samples (upper panel).
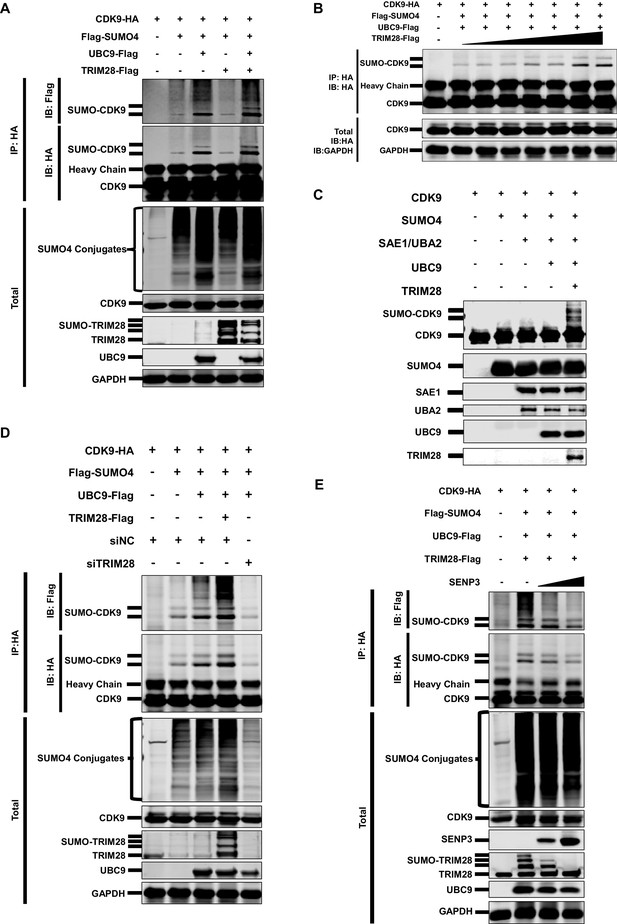
CDK9 is SUMOylated by TRIM28.
(A) HA-tagged CDK9 was co-overexpressed with Flag-tagged SUMO4, UBC9 or TRIM28. CDK9 was IP with anti-HA-tag beads, followed by IB with anti-HA and –Flag antibodies. TRIM28, UBC9 and GAPDH in total samples were IB with specific antibodies targeting each proteins. (B) HA-tagged CDK9 was co-overexpressed with Flag-tagged SUMO4, Flag-tagged UBC9 and different amount of Flag-tagged TRIM28. Target proteins were IB as in (A). (C) In vitro purified CDK9, SUMO4, SAE1, UBA2, UBC9 and TRIM28 were co-cultured in SUMO conjugation reaction buffer. Proteins including SUMOylated CDK9 were IB with antibodies against each targets. (D) HA-tagged CDK9 was co-overexpressed with Flag-tagged SUMO4, Flag-tagged UBC9 or Flag-tagged TRIM28, and siNC. In the last group, CDK9 was co-overexpressed with SUMO4, UBC9 and siRNA against TRIM28. Target proteins were IB as in (A). (E) HA-tagged CDK9 was co-overexpressed with Flag-tagged SUMO4, Flag-tagged UBC9, Flag-tagged TRIM28 or two gradients of SENP3. Target proteins were IB as in (A).
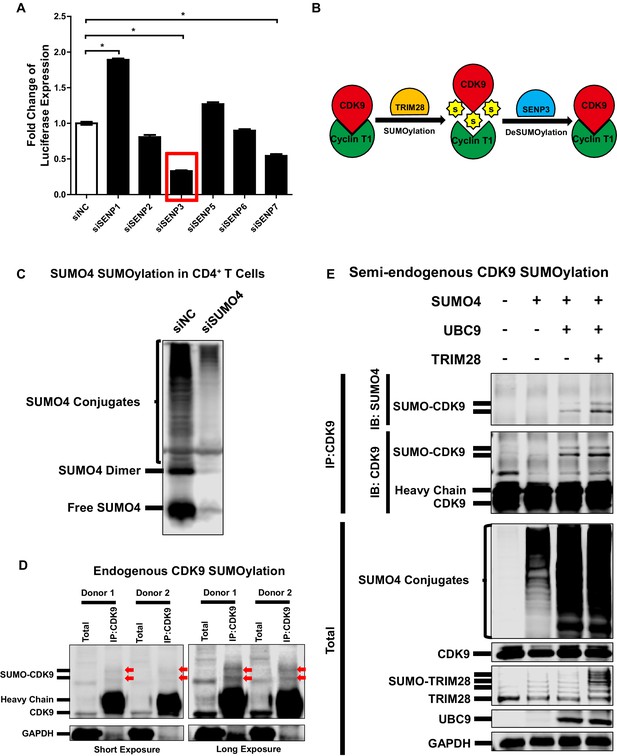
CDK9 is deSUMOylated by SENP3 and CDK9 SUMOylation occurs in primary CD4+T cells.
(A) siRNAs targeting six SENPs were transfected into TZM-bl cells. The luciferase from the clarified lysates of each group was quantitated and normalized to siNC. Data represents mean ±SEM in triplicates. p-Values were calculated by Student’s t-test. *p<0.05. (B) Model proposed based on Figure 5 and Figure 5—figure supplement 1. CDK9 which was subunit of P-TEFb complex was SUMOylated by TRIM28. The SUMO peptides were removed by SENP3-mediated deSUMOylation. (C) CD4+ T cells isolated from a healthy donor were transfected with siRNAs targeting negative control and SUMO4, respectively. Forty-eight hours later, the total lysates were immunoblotted with antibody against SUMO4. The lower band indicated free SUMO4. The middle band indicated SUMO4 dimer. The upper bands indicated SUMO4-SUMOylated cellular targets. (D) CD4+ T cells isolated from two healthy donors were lysed. The endogenous CDK9 was IP with antibody against CDK9. Both total samples and IP samples were IB with antibodies against CDK9 and GAPDH. Arrows indicated SUMOylated CDK9. (E) CD4+ T cells isolated from a heathy donor were transfected with SUMO4, UBC9 and TRIM28 constructs, or left untreated. Forty-eight hours later, endogenous CDK9 of the cell lysates was IP with antibody against CDK9. GAPDH, CDK9, UBC9, TRIM28 and SUMO4 were IB with corresponding antibodies in total samples. CDK9 and SUMO4 in IP samples were IB with antibodies against CDK9 and SUMO4 respectively.
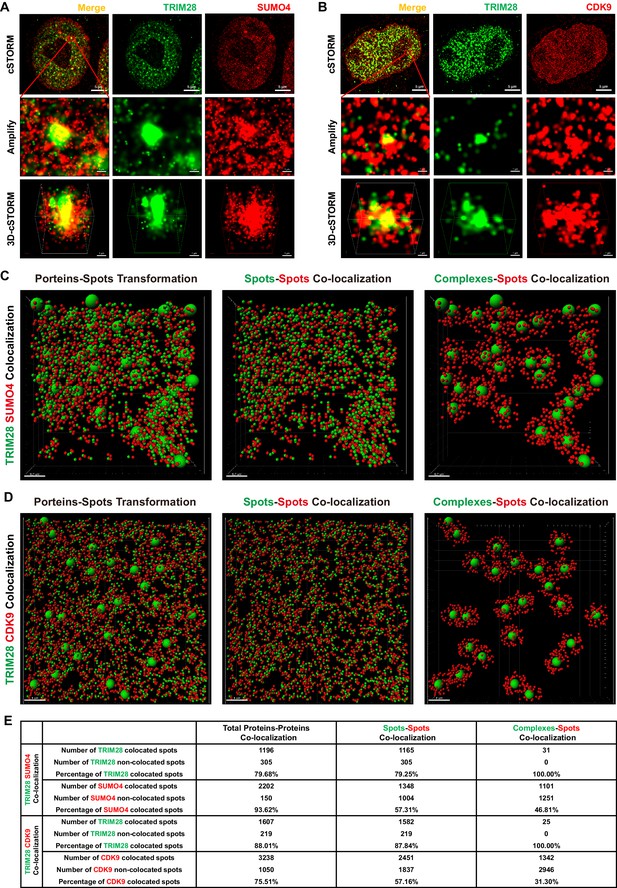
TRIM28 co-localizes with SUMO4 and CDK9.
(A) cSTORM image of endogenous TRIM28 and SUMO4 in HEK293T cells. The first row: the original whole nucleus; the second row: one of the amplified region of the nucleus; the third row: the 3D-cSTORM image of the amplified region. Merged views of TRIM28 and SUMO4 were shown on the left column. Endogenous TRIM28 was shown in the middle column and colored green. Endogenous SUMO4 was shown in the right column and colored red. Of note, DAPI and Hoechst were not allowed to dye DNA according to cSTORM protocol. (B) cSTORM image of endogenous TRIM28 and CDK9 in HEK293T cells. Each row was shown as in (A). First column: merged view of TRIM28 and CDK9, yellow indicating co-localization; second column: endogenous TRIM28 which was colored green; third column: endogenous CDK9 which was colored red. (C–D) cSTORM-imaged protein molecules and complexes were transformed into small or large spots based on their diameter. The left panel of each figure showed the original transformation. The middle panel showed spots-spots co-localization in compliance with the criterion of maximal distance of 10 nm. The right panel showed complexes-spots co-localization in compliance with the criterion of maximal distance of 100 nm. Green spots indicated TRIM28 molecules. Red spots indicated SUMO4 or CDK9 molecules. (E) Quantitation of co-localization of TRIM28 with SUMO4 or CDK9. Both of total proteins-proteins, spots-spots and complexes-spots co-localizations were measured.
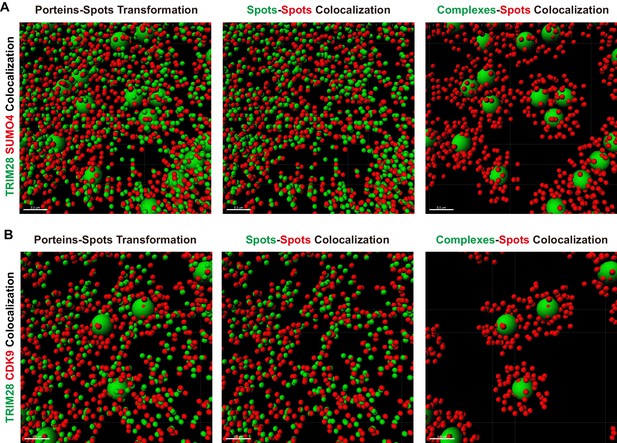
Amplified views of transformed co-localization.
(A–B) cSTORM-imaged protein molecules and complexes were transformed and displayed as in Figure 6C–D.Data represented amplified views of each transformed co-localization images. Green spots indicated TRIM28 molecules. Red spots indicated SUMO4 or CDK9 molecules.
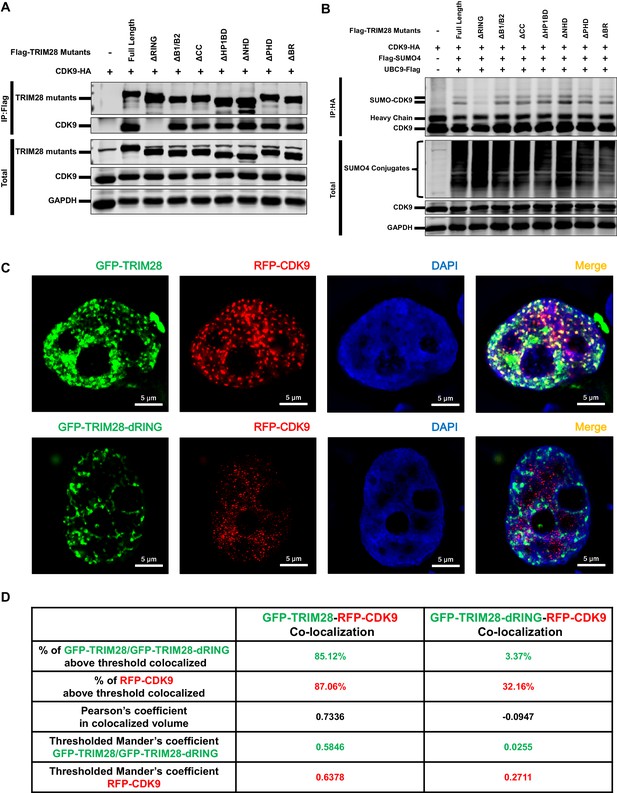
The RING domain of TRIM28 plays a key role in binding to and SUMOylating CDK9.
(A) HA-tagged CDK9 was co-overexpressed with Flag-tagged full length TRIM28 or domain-truncated TRIM28 mutants. Flag-tagged proteins were IP, followed by IB with antibodies against HA-tag, Flag-tag and GAPDH. (B) HA-tagged CDK9 was co-overexpressed with Flag-tagged SUMO4, Flag-tagged UBC9, Flag-tagged full length TRIM28 or Flag-tagged domain-truncated TRIM28 mutants. CDK9 was IP with anti-HA-tag beads, followed by IB with antibodies against HA-tag, Flag-tag and GAPDH. (C) GFP-tagged TRIM28 or TRIM28-dRING mutant was co-overexpressed with RFP-tagged CDK9 in HEK293T cells. The samples were fixed and dyed according to the immunofluorescence procedure, then visualized in Nikon A1 N-SIM. DAPI was used to dye DNA which was colored into blue. (D) Quantitation of co-localization of TRIM28 or TRIM28-dRING with CDK9. The percentage of co-localization was indicated by percentage of target protein voxels above threshold co-localized voxels. Both Pearson’s coefficient and thresholded Mander’s coefficient were used to evaluate co-localization. For Pearson’s coefficient, a value of 1 represents perfect co-localization, 0 no co-localization, and −1 perfect inverse co-localization. For thresholded Mander’s coefficient, a value of 1 represents perfect co-localization and 0 no co-localization.
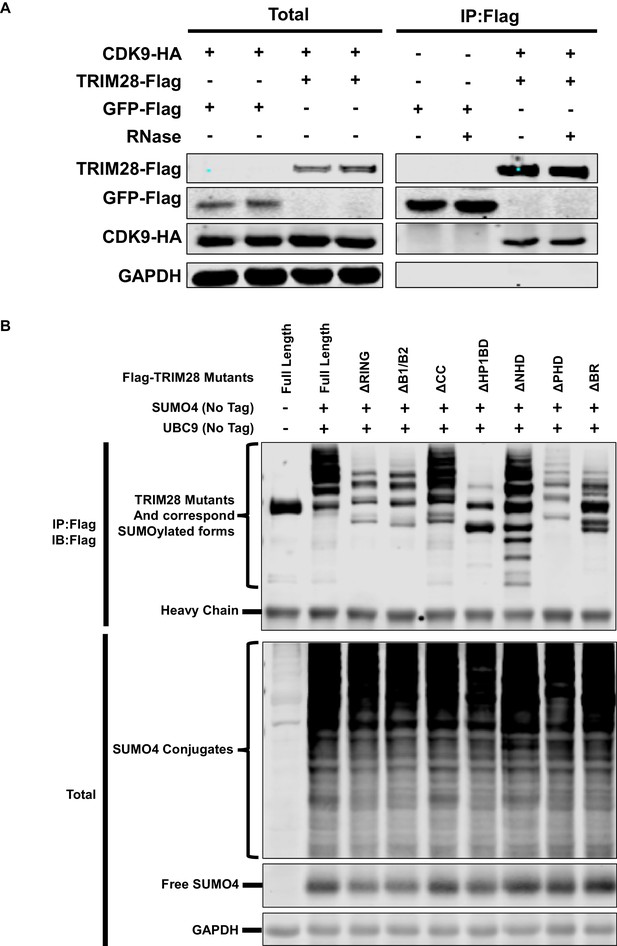
TRIM28 enriches CDK9 in the presence of RNase and SUMOylation status of TRIM28 mutants.
(A) Flag-tagged GFP and Flag-tagged TRIM28 were co-overexpressed with HA-tagged CDK9, respectively. Flag-tagged proteins were IP with anti-Flag beads. Flag-tagged proteins from another two similar groups were IP in the presence of RNase. Both total samples and IP samples were IB with antibodies against GAPDH, HA-tag and Flag-tag. (B) Flag-tagged full length TRIM28 and Flag-tagged TRIM28 mutants were co-overexpressed with SUMO4 and UBC9 respectively. Flag-tagged proteins were IP with anti-Flag beads followed by IB with antibodies against Flag-tag. Both total samples (lower panel) and IP samples (upper panel) were IB for each group.
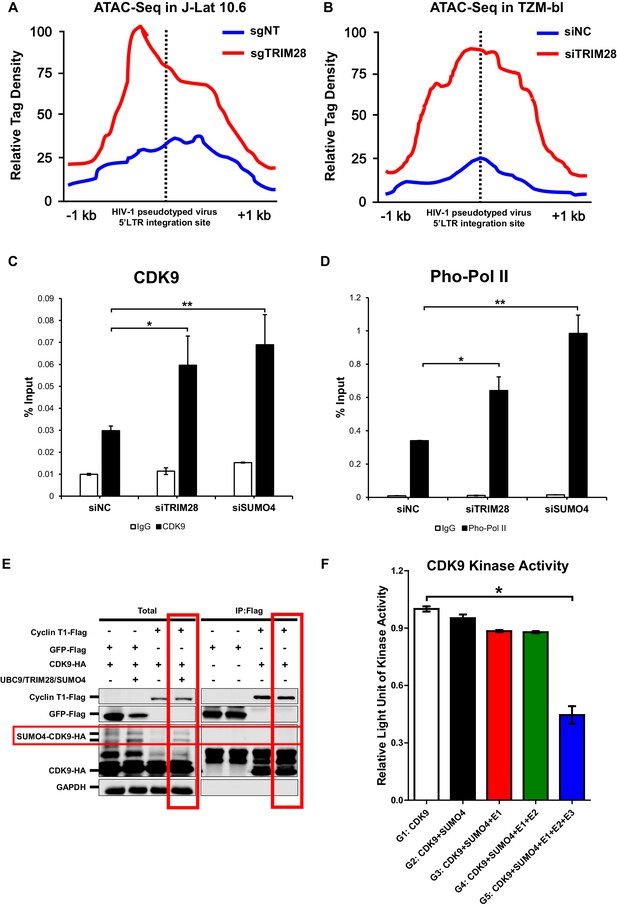
CDK9 function is reduced when SUMOylated by TRIM28.
(A–B) TRIM28-defective (sgTRIM28) J-Lat 10.6 cell line was generated by CRISPR-CAS9 technique. ATAC-Seq was conducted with sgNT and sgTRIM28 J-Lat 10.6 cell lines, as well as siNC and siTRIM28 TZM-bl cell lines. The tag reads of the HIV-1 pseudotyped virus/minigenome 5’LTR integration sites were counted and normalized to the total mapped reads, and represented as relative tag density. The highest tag density was set as 100. Figures showed 2 kb range centered the 5’LTR integration sites. (C–D) ChIP assays with antibodies against CDK9 and Ser2 Pho-Pol II were performed in TZM-bl cell lines which were treated with siNC, siSUMO4 and siTRIM28, respectively. (E) Cyclin T1 or GFP was co-overexpressed with CDK9 in the absence or presence of SUMO4, UBC9 and TRIM28. Cyclin T1 and GFP were IP followed by IB. (F) Fold change of kinase activity when CDK9 was SUMOylated. Data represents mean ±SEM in triplicates. p-Values were calculated by Student’s t-test. *p<0.05, **p<0.01.
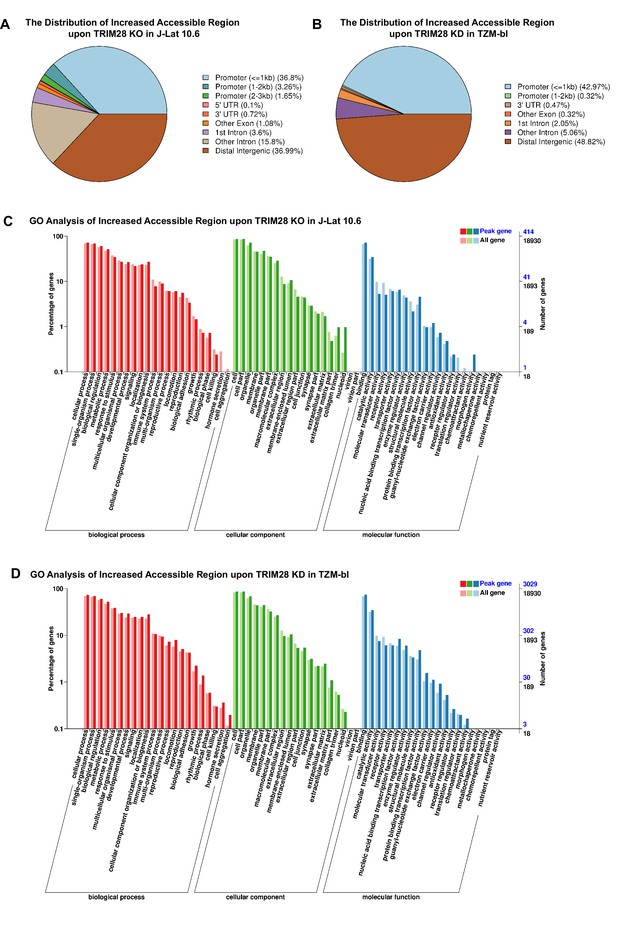
The distribution and GO analysis of increased accessible regions upon TRIM28 depletion.
(A–B) The distribution of increased accessible regions upon TRIM28 knockout in J-Lat 10.6 (A) and TRIM28 knockdown in TZM-bl (B). (C–D) GO analyses which included biological process analysis, cellular component analysis and molecular function analysis were used to classify genes with increased accessible regions upon TRIM28 knockout in J-Lat 10.6 (C) and TRIM28 knockdown in TZM-bl (D).
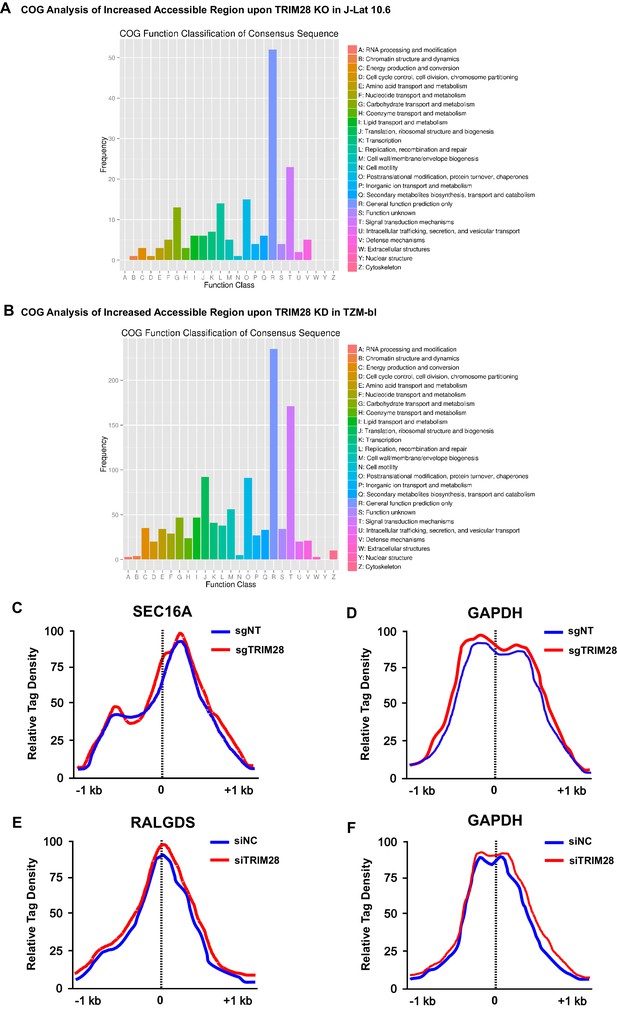
The COG analysis of increased accessible regions and the chromatin accessibility variations on target genes.
(A–B) Clusters of Orthologous Groups of proteins (COGs) analysis of increased accessible regions upon TRIM28 knockout in J-Lat 10.6 (A) and TRIM28 knockdown in TZM-bl (B). (C) The chromatin accessibilities of the promoter of SEC16A within which the integrated pseudotyped HIV-1 located were annotated in both wild type and TRIM28 knockout J-Lat 10.6 cell lines. (D) The chromatin accessibilities of the promoter of housekeeping gene GAPDH were annotated in both wild type and TRIM28 knockout J-Lat 10.6 cell lines. (E) The chromatin accessibilities of the promoter of RALGDS within which the HIV-1 reporter provirus located were annotated in both wild type and TRIM28 knockdown TZM-bl cell lines. (F) The chromatin accessibilities of the promoter of housekeeping gene GAPDH were annotated in both wild type and TRIM28 knockdown TZM-bl cell lines.
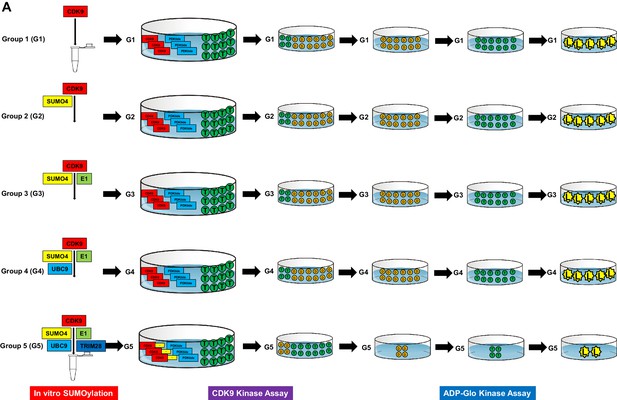
Schematic of in vitro SUMOylation assay and CDK9 kinase assay.
(A) In vitro expressed and purified CDK9 was incubated with SUMO system components (SUMO4, E1, UBC9 and TRIM28) or left untreated. Five groups were set. Group 1 (G1): CDK9 only; Group 2 (G2): CDK9 and SUMO4; Group 3 (G3): CDK9, SUMO4 and E1 (SAE1 and UBA2); Group 4 (G4): CDK9, SUMO4, E1 and E2 (UBC9); Group 5 (G5): CDK9, SUMO4, E1, E2 and E3 (TRIM28). After in vitro SUMOylation, CDK9 substrate PDKtides and ATP were added and incubated for 120 min at room temperature. The ADP which was consumed during CDK9 kinase assay was converted to ATP and quantitated by luciferase assay.
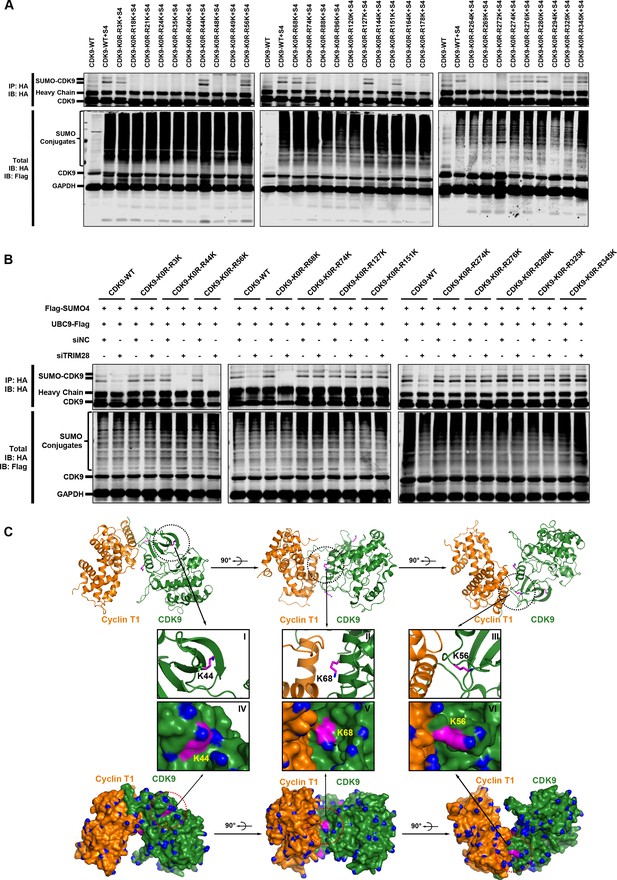
The Lys44, Lys56 and Lys68 residues of CDK9 are SUMOylated with SUMO4.
(A) Different HA-tagged CDK9 reversing mutation constructs or wild type CDK9 were co-overexpressed with SUMO4, UBC9 and TRIM28, respectively. CDK9 and CDK9 mutants were IP with anti-HA-tag beads followed by IB. S4: SUMO4. (B) HA-tagged wild type CDK9 and 12 identified SUMOylation site reversing mutation constructs were co-overexpressed with Flag-tagged SUMO4 and Flag-tagged UBC9. The endogenous TRIM28 was knocked down with siRNAs. CDK9 and CDK9 mutants were IP with anti-HA-tag beads followed by IB. Asterisks represented the constructs whose SUMOylation bands disappeared upon TRIM28 knockdown. (C) Three angles of co-crystal structure of Cyclin T1 and CDK9 (PDB ID: 4EC8). Three SUMOylation sites Lys44, Lys56 and Lys68 were shown in ball-and-stick models. The two upper panels showed the ribbon models, while two lower panels showed the surface models. The inner six framed figures which numbered from I to VI represented the amplification views of Lys44, Lys56 and Lys68 sites.
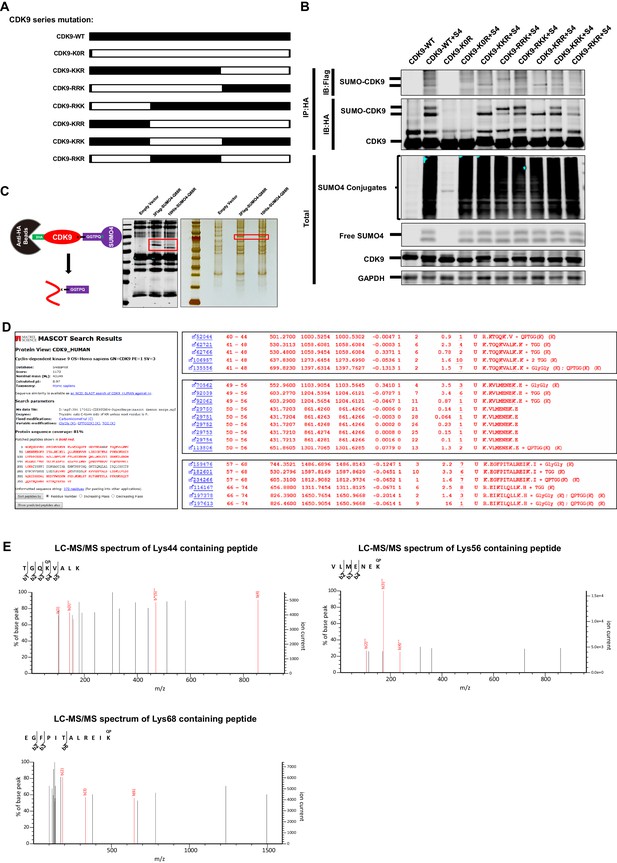
The Lys44, Lys56 and Lys68 residues of CDK9 are SUMOylated with SUMO4.
(A) Schematic of different CDK9 mutants. CDK9-K0R indicated that all lysines had been mutated to arginines. CDK9-KKR indicated that the third part of CDK9 had mutated all lysines to arginines. The left five clones were mutated similarly. (B) HA-tagged wild type CDK9 and different CDK9 mutants were co-overexpressed with Flag-tagged SUMO4, Flag-tagged UBC9 and Flag-tagged TRIM28. HA-tagged proteins were IP with anti-HA beads followed by IB with antibodies against HA-tag and Flag-tag. Both total samples (lower panel) and IP samples (upper panel) were IB for each group. (C) Schematic of target-specific SUMO-MS. HA-tagged CDK9 was co-overexpressed with Flag-tagged SUMO4-Q88R or His-tagged SUMO4-Q88R respectively. Anti-HA-tag beads were used to IP CDK9 and SUMO-CDK9 (left panel). The SUMOylation efficiency was determined by IB (middle panel). HA-tagged targets were separated by SDS-PAGE and developed by silver staining (right panel). Red frames indicated that SUMO-CDK9. SUMO-CDK9 was cut out and conducted in-gel digestion. The digested peptides were desalted and proceeded to nanoscale LC-MS/MS. (D) Results of the target-specific SUMO-MS of CDK9. Protein sequence coverage was 81% covering nearly all lysines. Three SUMOylation sites which were identified through SUMOylation assay were shown on the right panel. (E) Second-order mass spectra of CDK9 SUMOylation sites Lys44, Lys56 and Lys68.
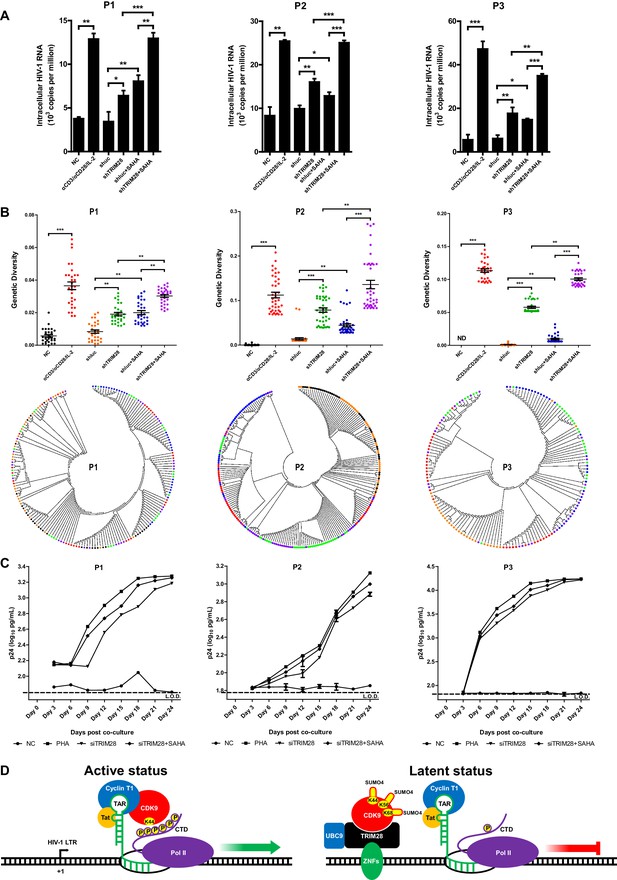
TRIM28 depletion reactivates latent HIV-1 in cells from HIV-1-infected individuals.
(A) shRNAs targeting luciferase and TRIM28 were packaged into lentiviruses and infected CD4+ T cells from HIV-1-infected individuals. Unstimulated CD4 +T cells were used as negative control (NC). Stimulation with αCD3/αCD28/IL-2 was used as positive control. Intracellular HIV-1 RNA was isolated and quantitated by qPCR. Experiments were conducted in three HIV-1-infected individuals. (B) The experiment setting was as in (A). Envelope V1 to V3 region from intracellular HIV-1 RNAs was reverse-transcribed and PCR-amplified. The PCR products were TA-ligated in pMD-18 T vector. At least 60 single clones were picked from each group and sequenced. The sequences from each group were aligned and the genetic diversity index was calculated and analyzed by Mann-Whitney U-test. The upper panel showed the statistical analysis results. The lower panel indicated the bootstrap consensus trees which were generated based on HIV-1 sequences. *p<0.05, **p<0.01, ***p<0.001. (C) Resting CD4+ T cells from HIV-1-infected individuals were isolated and nucleofected with siRNAs targeting negative control or TRIM28. Seventy-two hours later, PHA-stimulated uninfected CD4+ T cells were added into each group and co-cultured for another 27 days. The supernatants were collected and half-changed every 3 days. P24 antigens in supernatants were measured with ELISA and plotted in log10 scale. Dashed lines indicated the limit of detection (L.O.D.) of 50 pg/ml. Triplicates were represented by mean ±SEM. (D) Schematic of TRIM28-mediated HIV-1 latency.
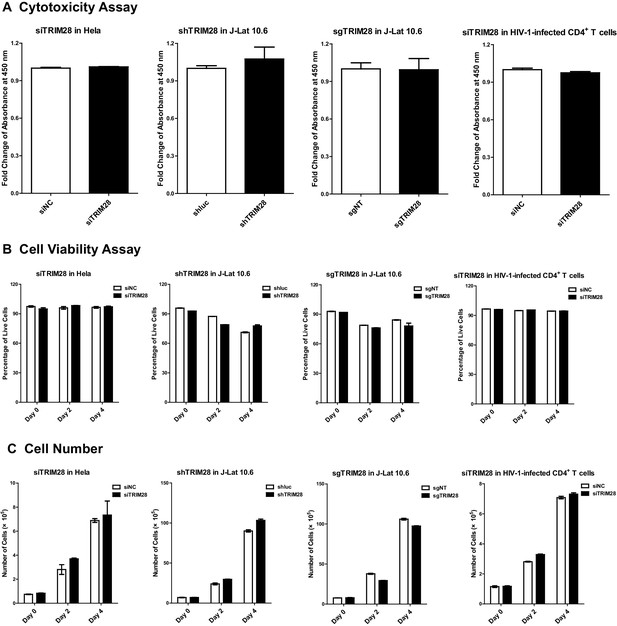
Cytotoxicity assay, cell viability assay and cell number counting used to evaluate the toxicity of targeting TRIM28.
(A) TRIM28 in Hela cells and HIV-1-infected CD4+ T cells was knocked down by siRNA targeting TRIM28. ShRNA and sgRNA lentiviruses targeting TRIM28 were used to knock down TRIM28 and knock out TRIM28 in J-Lat 10.6 respectively. Cell Counting Kit-8 (CCK-8) reagents were incubated with wild type and TRIM28-deficient cells for 3 hr followed by measuring the absorbance at 450 nm using a microplate reader. Fold changes of absorbance in each group were normalized to wild-type groups. (B) The experiment setup was conducted as in (A). The percentages of viable cells were quantitated every 2 days by measuring the percentages of amine-reactive fluorescent dye non-permeant cells. (C) The experiment setup was conducted as in (A). Cell numbers were recorded every 2 days for both wild-type and TRIM28-deficient cells.
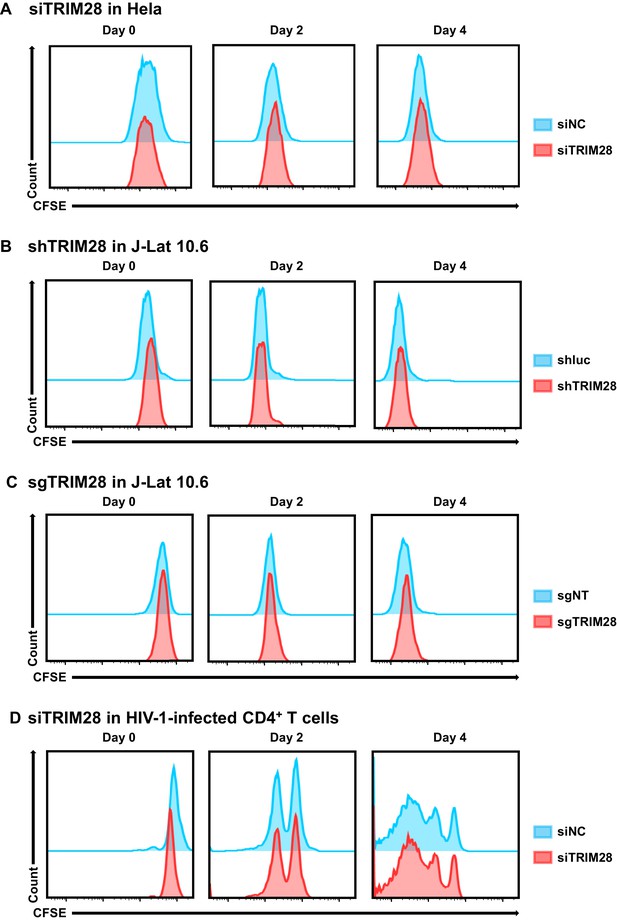
Cell proliferation assay used to evaluate the toxicity of targeting TRIM28.
(A–D) The experiment setup was conducted as in Figure 10—figure supplement 1. On Day 0, cells from each group were stained with CFSE. The percentage and mean fluorescence intensity (MFI) of CFSE-positive cells were analyzed by flow cytometry every 2 days.
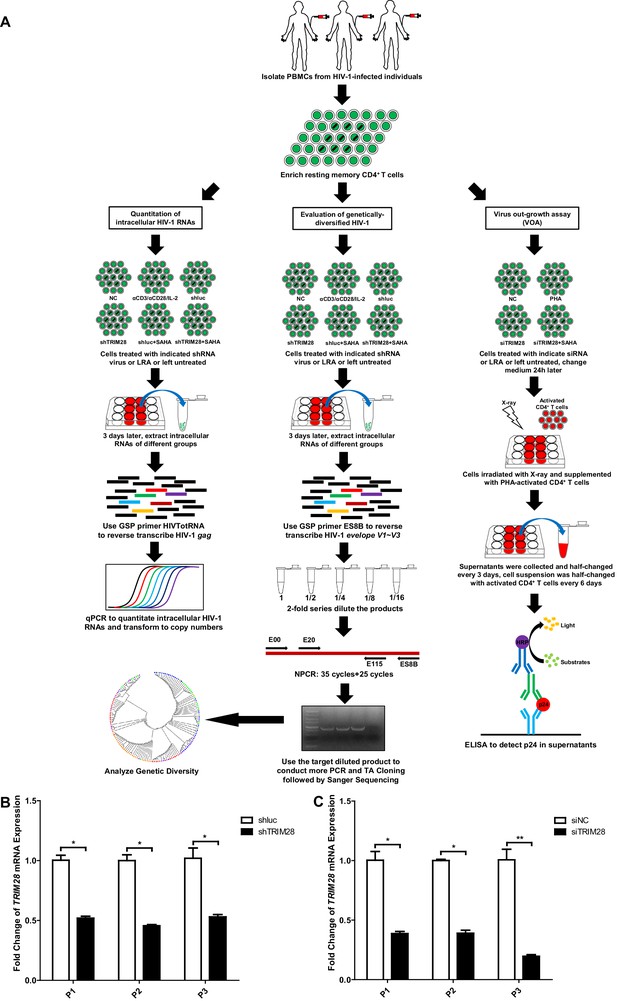
TRIM28 depletion reactivates latent HIV-1 in cells from HIV-1-infected individuals.
(A) Schematic of experiments on primary CD4 +T cells from HIV-1-infected individuals. (B) The knockdown efficiency of shTRIM28 in HIV-1-infected CD4+ T cells. (C) The knockdown efficiency of nucleofection of siRNAs targeting TRIM28. Data represents mean ±SEM in triplicates. p-Values were calculated by Student’s t-test. *p<0.05, **p<0.01.
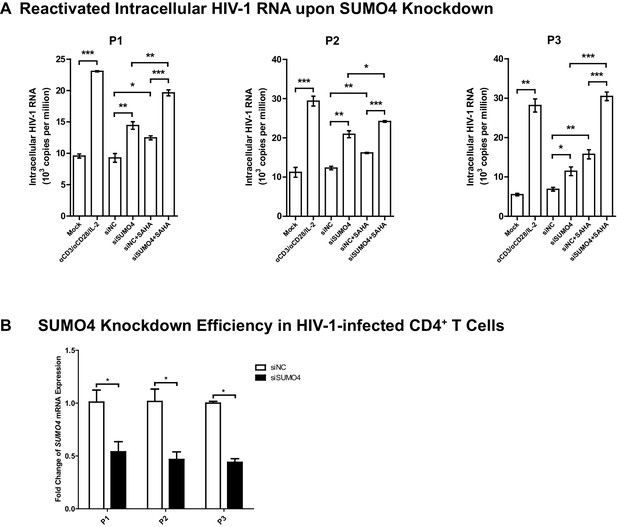
SUMO4 depletion reactivates latent HIV-1 in cells from HIV-1-infected individuals.
(A) SiRNAs targeting NC and SUMO4 were nucleofected into CD4+ T cells from HIV-1-infected individuals. Unstimulated CD4+ T cells were used as Mock. Stimulation with αCD3/αCD28/IL-2 was used as positive control. Intracellular HIV-1 RNA was isolated and quantitated by qPCR. Experiments were conducted in three HIV-1-infected individuals. Data represents mean ±SEM in triplicates. p-Values were calculated by Student’s t-test. *p<0.05, **p<0.01, ***p<0.001. (B) The knockdown efficiency of nucleofection of siRNAs targeting SUMO4. Data represents mean ±SEM in triplicates. p-Values were calculated by Student’s t-test. *p<0.05.
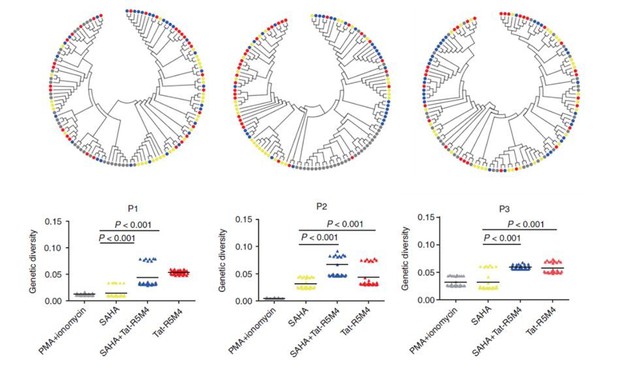
Attenuated Tat Protein Tat-R5M4 can reactivate more genetically-diversified HIV-1.
(Geng et al., 2016, Molecular Therapy, PMID: 27434587).
Videos
3D-cSTORM movie of the 3D co-localization of TRIM28 with SUMO4.
Green spots indicate TRIM28. Red spots indicate SUMO4.
3D-cSTORM movie of the 3D co-localization of TRIM28 with CDK9.
Green spots indicate TRIM28. Red spots indicate CDK9.
Transformed 3D-cSTORM movie of the 3D co-localization of TRIM28 with SUMO4.
Green spots indicate TRIM28. Red spots indicate SUMO4.
Transformed 3D-cSTORM movie of the 3D co-localization of TRIM28 with CDK9.
Green spots indicate TRIM28. Red spots indicate CDK9.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Escherichia coli) | E.coli DH5α: F-, φ 80dlacZ ΔM15, Δ(lacZYA -argF )U169, deoR , recA1 , endA1 , hsdR17 (rK-, mK+), phoA, supE44 , λ-, thi −1, gyrA96 , relA1 | Takara | Cat#9057 | |
| Strain, strain background (Escherichia coli) | E. coli HB101: F-, hsdS20(rB-, mB-), recA13, ara-14, proA2, lacY1, galK2, rpsL20 (str), xyl-5, mtl-1,supE44, leuB6, thi-1. | Takara | Cat#9051 | |
| Strain, strain background (Escherichia coli) | E.coli BL21: F-, ompT, hsdSB (rB-mB-), gal, dcm | Takara | Cat#9126 | |
| Strain, strain background (Escherichia coli) | E.coli Stbl3: F-, mcrB, mrr, hsdS20 (rB-, mB-), recA13, supE44, ara-14, galK2, lacY1, proA2, rpsL20 (StrR), xyl-5, λ- leu, mtl-1 | ThermoFisher | Cat#C7381201 | |
| Cell line (Homo sapiens) | HEK293T | ATCC | CRL-3216; RRID: CVCL_0063 | female |
| Cell line (Homo sapiens) | HeLa | ATCC | CCL-2; RRID: CVCL_0030 | female |
| Cell line (Homo sapiens) | TZM-bl | NIH AIDS Reagent Program | Cat#8129 | female |
| Cell line (Homo sapiens) | J-Lat 6.3 | PMID: 12682019 | NIH AIDS Reagent Program Cat#9846 | Dr. Eric Verdin (The Buck Institute for Research on Aging, Novato, CA, USA) |
| Cell line (Homo sapiens) | J-Lat 8.4 | PMID: 12682019 | NIH AIDS Reagent Program Cat#9847 | Dr. Eric Verdin (The Buck Institute for Research on Aging, Novato, CA, USA) |
| Cell line (Homo sapiens) | J-Lat 9.2 | PMID: 12682019 | NIH AIDS Reagent Program Cat#9848 | Dr. Eric Verdin (The Buck Institute for Research on Aging, Novato, CA, USA) |
| Cell line (Homo sapiens) | J-Lat 10.6 | PMID: 12682019 | NIH AIDS Reagent Program Cat#9849 | Dr. Eric Verdin (The Buck Institute for Research on Aging, Novato, CA, USA) |
| Cell line (Homo sapiens) | J-Lat 15.4 | PMID: 12682019 | NIH AIDS Reagent Program Cat#9850 | Dr. Eric Verdin (The Buck Institute for Research on Aging, Novato, CA, USA) |
| Biological sample (Homo sapiens) | Blood samples from healthy individuals | Guangzhou Blood Center, Guangzhou | http://www.gzbc.org/ | |
| Biological sample (Homo sapiens) | Blood samples from HIV-1- infected individuals | Department of Infectious Diseases, Guangzhou 8th People’s Hospital, Guangzhou | http://gz8h.com.cn/ | |
| Antibody | Mouse Monoclonal anti-TRIM28 Antibody | Proteintech | Cat#66630–1-Ig; RRID: AB_2732886; Lot#10006062 | (1:1000) |
| Antibody | Rabbit Polyclonal anti-TRIM28 Antibody | Proteintech | Cat#15202–1-AP; RRID: AB_2209890; Lot#00051172 | (1:1000) |
| Antibody | Rabbit Polyclonal Anti-Histone H3 (tri methyl K4) Antibody | Abcam | Cat#ab8580; RRID: AB_306649; Lot#GR273043-3 | Use 2 µg for 25 µg of chromatin |
| Antibody | Rabbit Polyclonal Anti-Histone H3 (acetyl K9) Antibody | Abcam | Cat#ab4441; RRID: AB_2118292; Lot#GR270585-1 | Use 2 µg for 25 µg of chromatin |
| Antibody | Mouse Monoclonal Anti-Histone H3 (tri methyl K27) Antibody | Abcam | Cat#ab6002; Lot#GR275911-3 | Use 5 µg for 25 µg of chromatin |
| Antibody | Normal Rabbit Anti-IgG Antibody | CST | Cat#2729; RRID: AB_1031062 | Use 1 µg for 25 µg of chromatin |
| Antibody | Rabbit Polyclonal Anti-UBE2I Antibody | Abclonal | Cat#A2193; Lot#45473 | (1:1000) |
| Antibody | Rabbit Polyclonal Anti-UBA2 Antibody | Abclonal | Cat#A4363 | (1:1000) |
| Antibody | Rabbit Polyclonal Anti-SAE1 Antibody | Proteintech | Cat#10229–1-AP; RRID: AB_2182917; Lot#00040591 | (1:1000) |
| Antibody | Rabbit Monoclonal Anti-SUMO4 Antibody | Abcam | Cat#ab126606; RRID: AB_11128131; Lot#GR851138-12 | (1:1000) |
| Antibody | Rabbit Monoclonal Anti-CDK9 (C12F7) Antibody | CST | Cat#2316; Lot#6 | (1:1000) |
| Antibody | Rabbit Polyclonal Anti-SENP3 Antibody | Proteintech | Cat#17659–1-AP; RRID: AB_2301618; Lot#00025621 | (1:1000) |
| Antibody | Rabbit Polyclonal Anti-RNA polymerase II CTD repeat YSPTSPS (phosphor-Ser2) Antibody | Abcam | Cat#ab5095; RRID: AB_304749; Lot#GR278215-1 | Use 2 µg for 25 µg of chromatin |
| Antibody | Mouse Monoclonal Anti-Histone H3 (di methyl K9) Antibody | Abcam | Cat#ab1220; RRID: AB_449854 | Use 4 µg for 25 µg of chromatin |
| Antibody | Rabbit Polyclonal Anti-Histone H3 (tri methyl K9) Antibody | Abcam | Cat#ab8898; RRID: AB_306848 | Use 4 µg for 25 µg of chromatin |
| Antibody | Donkey Anti-Mouse IgG H and L (Alexa Fluor 647) Antibody | Abcam | Cat#ab150107; Lot#GR311164-3 | (1:200) |
| Antibody | Donkey Anti-Rabbit IgG H and L (Alexa Fluor 647) Antibody | Abcam | Cat#ab150075; Lot#GR3174006-4 | (1:200) |
| Antibody | Donkey Anti-Rabbit IgG (H + L), Highly Cross- Adsorbed, CF 568 Dye Conjugates, Single Label for STORM | Biotium | Cat#20803–500 μl; Lot#17C0626 | (1:200) |
| Antibody | Donkey Anti-Mouse IgG (H + L), Highly Cross- Adsorbed, CF 568 Dye Conjugates, Single Label for STORM | Biotium | Cat#20802–500 μl; Lot#17C1004 | (1:200) |
| Antibody | Rabbit Anti-DDDDK Tag Polyclonal Antibody, Unconjugated | MBL | Cat#PM020; RRID: AB_591224; Lot#026 | (1:1000) |
| Antibody | Mouse Monoclonal Anti-HA-Tag Antibody | MBL | Cat#M180-3; RRID: AB_10951811; Lot#008 | (1:10000) |
| Antibody | Mouse Monoclonal Anti-His-Tag Antibody | Proteintech | Cat#66005–1-Ig; RRID: AB_11232599; Lot#00083246 | (1:1000) |
| Antibody | Rabbit Polyclonal Anti-GAPDH Antibody | Proteintech | Cat#10494–1-AP; RRID: AB_2263076; Lot#00039889 | (1:10000) |
| Antibody | IRDye 680RD Goat anti-Mouse IgG (H + L), 0.5 mg Antibody | LI-COR Biosciences | Cat#926–68070; RRID: AB_10956588; Lot#C70613-15 | (1:10000) |
| Antibody | IRDye 800CW Goat Anti-Rabbit IgG, Conjugated Antibody | LI-COR Biosciences | Cat#926–32211; RRID: AB_621843; Lot#C70620-05 | (1:10000) |
| Antibody | PerCP-Cy 5.5 Mouse Anti-Human CD45RO | BD Biosciences | Cat#560607; RRID: AB_1727500; Lot#5338941 | (1:1000) |
| Antibody | APC/Cy7 anti- human CD45RA | BioLegend | Cat#304127; RRID: AB_10708419; Lot#B164612 | (1:1000) |
| Antibody | Anti-Human CD69 PE-Cy7 | ThermoFisher | Cat#25-0699-42; RRID: AB_1548714; Lot#E10154-1635 | (1:1000) |
| Antibody | Anti-Human CD62L PE-Cyanine7 | ThermoFisher | Cat#25-0629-42; RRID: AB_1257142; Lot#4291471 | (1:1000) |
| Antibody | Anti-Human CD4 FITC | ThermoFisher | Cat#11-0048-42; RRID: AB_1633390; Lot#E10526-1631 | (1:1000) |
| Antibody | PE-Cy5 Conjugated Amti-human CD25 (IL-2R) | ThermoFisher | Cat#15-0259-42; RRID: AB_1944361; Lot#E11289-102 | (1:1000) |
| Recombinant DNA reagent | VSV-G glycoprotein- expression vector | PMID: 9306402 | Addgene Plasmid #12259 | Dr. Didier Trono (School of Life Sciences, Ecole Polytechnique Fédérale de Lausanne, Lausanne, Switzerland) |
| Recombinant DNA reagent | Lentiviral packaging construct pCMVΔR8.2 | PMID: 9306402 | Addgene Plasmid #12263 | Dr. Didier Trono (School of Life Sciences, Ecole Polytechnique Fédérale de Lausanne, Lausanne, Switzerland) |
| Recombinant DNA reagent | Lentiviral construct vector pLKO.3G-RFP | This paper | N/A | Progenitor: pLKO.3G |
| Recombinant DNA reagent | Lentiviral construct vector lentiCRISPRv2 | PMID: 25075903 | Addgene Plasmid #52961 | Dr. Feng Zhang (Broad Institute of MIT and Harvard) |
| Recombinant DNA reagent | Plasmid: 10His- SUMO1-Q92R | This paper | Supplementary file 3 | Progenitor: pcDNA3.1(+) |
| Recombinant DNA reagent | Plasmid: 10His -SUMO2-Q88R | This paper | Supplementary file 3 | Progenitor: pcDNA3.1(+) |
| Recombinant DNA reagent | Plasmid: 10His- SUMO4-Q88R | This paper | Supplementary file 3 | Progenitor: pcDNA3.1(+) |
| Recombinant DNA reagent | Plasmid: 3HA- CDK9-KKR | This paper | Supplementary file 3 | Progenitor: pcDNA3.1(+) |
| Recombinant DNA reagent | Plasmid: 3HA- CDK9-RRK | This paper | Supplementary file 3 | Progenitor: pcDNA3.1(+) |
| Recombinant DNA reagent | Plasmid: 3HA -CDK9-RKK | This paper | Supplementary file 3 | Progenitor: pcDNA3.1(+) |
| Recombinant DNA reagent | Plasmid: 3HA- CDK9-KRR | This paper | Supplementary file 3 | Progenitor: pcDNA3.1(+) |
| Recombinant DNA reagent | Plasmid: 3HA- CDK9-KRK | This paper | Supplementary file 3 | Progenitor: pcDNA3.1(+) |
| Recombinant DNA reagent | Plasmid: 3HA- CDK9-RKR | This paper | Supplementary file 3 | Progenitor: pcDNA3.1(+) |
| Recombinant DNA reagent | Plasmid: 3HA- CDK9-K0R | This paper | Supplementary file 3 | Progenitor: pcDNA3.1(+) |
| Recombinant DNA reagent | Plasmids: 3HA-CDK9-K0R-RXK (X represent mutation position) | This paper | Supplementary file 3 | Progenitor: pcDNA3.1(+) |
| Sequence- based reagent | siRNA Library | RiboBio | Supplementary file 1; http://www.ribobio.com/ | |
| Sequence- based reagent | ChIP-qPCR Primers | This paper | Supplementary file 2 | |
| Sequence- based reagent | siRNA targeting TRIM28 3’UTR:5’- GCTCTGTTCTCTGTCCTGT-3’ | RiboBio | http://www.ribobio.com/ | |
| Sequence- based reagent | shRNA targeting Luciferase:5’- ACCGCCTGAAGTCTCTGATTAA-3’ | PMID: 29863470 | N/A | |
| Sequence- based reagent | shRNA targeting TRIM28 CDS:5’- CCAGCCAACCAGCGGAAATGTGA-3’ | PMID: 18082607 | N/A | |
| Sequence- based reagent | sgRNA targeting Dummyguide (sgNT):5’- ACGGAGGCTAAGCGTCGCAA-3’ | PMID: 25075903 | N/A | Dr. Feng Zhang (Broad Institute of MIT and Harvard) |
| Sequence- based reagent | sgRNA targeting TRIM28 CDS:5’- CACCGATTGAGCTGGCAGTCTCGGC-3’ | PMID: 25075903 | N/A | Dr. Feng Zhang (Broad Institute of MIT and Harvard) |
| Sequence- based reagent | β-Actin qPCR Forward Primer:5’- GCATGGAGTCCTGTGGCA-3’ | PMID: 27291871 | N/A | |
| Sequence- based reagent | β-Actin qPCR Reverse Primer:5’- CAGGAGGAGCAATGATCTTGA-3’ | PMID: 27291871 | N/A | |
| Sequence- based reagent | TRIM28 qPCR Forward Primer:5’- CTACTCAAGTGCAGAGCCCC-3’ | This paper | N/A | |
| Sequence- based reagent | TRIM28 qPCR Reverse Primer:5’- GGGAAGACCTTGAAGACGGG-3’ | This paper | N/A | |
| Sequence- based reagent | HIVTotRNA Forward Primer:5’- CTGGCTAACTAGGGAACCCACTGCT-3’ | PMID: 27291871 | N/A | |
| Sequence- based reagent | HIVTotRNA Reverse Primer:5’- GCTTCAGCAAGCCGAGTCCTGCGTC-3’ | PMID: 27535056 | N/A | |
| Sequence- based reagent | 1 st round Nest PCR Forward Primer (E00):5’- TAGAAAGAGCAGAAGACAGTGGCAATGA-3’ | PMID: 27434587 | N/A | |
| Sequence- based reagent | 1 st round Nest PCR Reverse Primer (ES8B):5’- CACTTCTCCAATTGTCCCTCA-3’ | PMID: 27434587 | N/A | |
| Sequence- based reagent | 2nd round Nest PCR Forward Primer (E20):5’- GGGCCACACATGCCTGTGTACCCACAG-3’ | PMID: 27434587 | N/A | |
| Sequence- based reagent | 2nd round Nest PCR Reverse Primer (E115):5’- AGAAAAATTCCCCTCCACAATTAA-3’ | PMID: 27434587 | N/A | |
| Chemical compound, drug | (+)-JQ-1 | Selleckchem | Cat#S7110 | |
| Chemical compound, drug | Vorinostat (SAHA) | Selleckchem | Cat#S1047 | |
| Chemical compound, drug | Formaldehyde solution | Sigma-Aldrich | Cat#F8775-25ML | |
| Chemical compound, drug | TRIzol Reagent | ThermoFisher | Cat#15596018 | |
| Chemical compound, drug | 4',6-Diamidino-2- Phenylindole, Dihydrochloride (DAPI) | ThermoFisher | Cat#D1306 | |
| Chemical compound, drug | Cysteamine (MEA) | Sigma-Aldrich | Cat#30070–10G | |
| Chemical compound, drug | Glucose Oxidase from Aspergillus niger, Type VII, lyophilized powder, ≥100,000 units/g solid | Sigma-Aldrich | Cat#G2133-250KU | |
| Chemical compound, drug | Catalase from bovine liver , lyophilized powder, ≥10,000 units/mg protein | Sigma-Aldrich | Cat#C40-1G | |
| Chemical compound, drug | Sodium borohydride (NaBH4) | Sigma-Aldrich | Cat#213462–25G | |
| Chemical compound, drug | 16% Paraformaldehyde (formaldehyde) Aqueous Solution | Electron Microscopy Sciences | Cat#15710 | |
| Chemical compound, drug | 8% Glutaraldehyde Aqueous Solution | Electron Microscopy Sciences | Cat#16019 | |
| Chemical compound, drug | Normal Donkey Serum (NDS) | Jackson ImmunoResearch | Cat#017-000-121 | |
| Chemical compound, drug | Triton X-100 | Sigma-Aldrich | Cat#T8787-50ML | |
| Chemical compound, drug | Protease Inhibitor Cocktail (PIC) | Sigma-Aldrich | Cat#P8340-1ML | |
| Chemical compound, drug | N-Ethylmaleimide (NEM) | Selleckchem | Cat#S3692 | |
| Chemical compound, drug | EZview Red Anti-HA Affinity Gel | Sigma-Aldrich | Cat#E6779-1ML | |
| Chemical compound, drug | EZview Red Anti- FLAG M2 Affinity Gel | Sigma-Aldrich | Cat#F2426-1ML | |
| Chemical compound, drug | Anti-His-tag Agarose | Abcam | Cat#ab1231 | |
| Chemical compound, drug | Penicillin-Streptomycin, Liquid | ThermoFisher | Cat#15140122 | |
| Chemical compound, drug | L-Glutamine, 200 mM Solution | ThermoFisher | Cat#25030081 | |
| Chemical compound, drug | Fetal Bovine Serum (FBS) | ThermoFisher | Cat#10270–106 | |
| Chemical compound, drug | Phytohemagglutinin -M (PHA-M) | Sigma-Aldrich | Cat#11082132001 | |
| Peptide, recombinant protein | Recombinant Human TNF-α | PeproTech | Cat#300-01A | |
| Peptide, recombinant protein | Recombinant Human IL-2 | R&D Systems | Cat#202-IL-500 | |
| Peptide, recombinant protein | Recombinant Human SUMO Activating Enzyme E1 (SAE1/UBA2) | R&D Systems | Cat#E-315 | |
| Peptide, recombinant protein | Recombinant Human UBE2I/Ubc9 | R&D Systems | Cat#E2-645-100 | |
| Peptide, recombinant protein | Recombinant Human CDK9 | Abcam | Cat#ab85603 | |
| Peptide, recombinant protein | Recombinant Human SUMO4 | This paper | N/A | |
| Peptide, recombinant protein | Recombinant Human TRIM28 | Abcam | Cat#ab131899 | |
| Commercial assay or kit | SUMO Conjugation Reaction Buffer Kit | R&D Systems | Cat#SK-15 | |
| Commercial assay or kit | Human Lymphocyte Separation Kit | TBDsciences | Cat#LTS1077 | |
| Commercial assay or kit | BD IMag Human CD4 + T Lymphocyte Enrichment Set-DM | BD Biosciences | Cat#557939 | |
| Commercial assay or kit | Luciferase Assay System | Promega | Cat#E4550 | |
| Commercial assay or kit | SimpleChIP Enzymatic Chromatin IP Kit (Magnetic Beads) | CST | Cat#9003S | |
| Commercial assay or kit | TruePrep DNA Library Prep Kit V2 for Illumina | Vazyme | Cat#TD501 | |
| Commercial assay or kit | HIV-1 p24 ELISA Kit | Abcam | Cat#ab218268 | |
| Commercial assay or kit | ProteoSilver Plus Silver Stain Kit | Sigma-Aldrich | Cat#PROTSIL2 -1KT | |
| Commercial assay or kit | CDK9/CyclinK Kinase Enzyme System | Promega | Cat#V4104 | |
| Commercial assay or kit | ADP-GloTM Kinase Assay | Promega | Cat#V6903 | |
| Commercial assay or kit | Cell Counting Kit-8 | Dojindo | Cat#CK04; Lot#KT793 | |
| Commercial assay or kit | Zombie Violet Fixable Viability Kit | BioLegend | Cat#423113; Lot#B256957 | |
| Commercial assay or kit | CellTrace CFSE Cell Proliferation Kit - For Flow Cytometry | ThermoFisher | Cat#C34554 | |
| Software, algorithm | Prism 5 | GraphPad | https://www.graphpad.com/scientific-software/prism/ | |
| Software, algorithm | MEGA 7 | MEGA | https://www.megasoftware.net/ | |
| Software, algorithm | Cytoscape (3.6.1) | Cytoscape Consortium | RRID:SCR_015784 | |
| Software, algorithm | STRING | Cytoscape Consortium | RRID:SCR_005223 | |
| Software, algorithm | MCODE | Cytoscape Consortium | RRID:SCR_015828 | |
| Software, algorithm | BD LSRFortessa cell analyzer | BD Biosciences | http://www.bdbiosciences.com/in/instruments/lsr/index.jsp | |
| Software, algorithm | FlowJo V10 | Tree Star | https://www.flowjo.com/ | |
| Software, algorithm | Odyssey CLX Imager | LI-COR Biosciences | https://www.licor.com/bio/products/imaging_systems/odyssey/ | |
| Software, algorithm | Image Studio Lite Ver 4.0 | LI-COR Biosciences | https://www.licor.com/bio/products/software/image_studio_lite/ | |
| Software, algorithm | CFX Manager | BIO-RAD | http://www.bio-rad.com/ | |
| Software, algorithm | GloMax 96 Microplate Luminometer Software (version 1.9.3) | Promega | https://www.promega.com/resources/software-firmware/detection-instruments-software/promega-branded-instruments/glomax-96-microplate-luminometer/ | |
| Software, algorithm | SkanIt SW for Microplate Readers | ThermoFisher | https://www.thermofisher.com/order/catalog/product/5187139?SID=srch-srp-5187139 | |
| Software, algorithm | NIS-Elements Advanced Research microscope imaging software | Nikon | https://www.nikoninstruments.com/Products/Software | |
| Software, algorithm | PyMOL | Schrödinger | RRID:SCR_000305 | |
| Software, algorithm | FastQC | Babraham Institute | RRID:SCR_014583 | |
| Software, algorithm | Hisat2 | PMID: 25751142 | RRID:SCR_015530 | |
| Software, algorithm | DEGseq | Bioconductor | RRID:SCR_008480 | |
| Software , algorithm | gplots | R Foundation | https://www.rdocumentation.org/packages/gplots/versions/3.0.1 | |
| Software, algorithm | Bowtie2 | PMID: 22388286 | RRID:SCR_016368 | |
| Software, algorithm | Samtools | PMID: 19505943 | RRID:SCR_002105 | |
| Software, algorithm | igvtools | Broad Institute | https://software.broadinstitute.org/software/igv/igvtools | |
| Software, algorithm | Imaris (Version 9.2) | BITPLANE | RRID:SCR_007370 |
Additional files
-
Supplementary file 1
SiRNA library used to screen HIV-1 suppression and latency contributors.
SiRNA library, which targeted several cellular pathways within the nucleus including chromatin binding, epigenetic modification, chromatin remodeling, ubiquitination, SUMOylation, and chromosome organization, was transfected into TZM-bl cells respectively. Both library and negative control siRNA were synthesized from RiboBio (Guangzhou, China).
- https://doi.org/10.7554/eLife.42426.034
-
Supplementary file 2
ChIP primers used to explore the enrichment of target proteins on HIV-1.
Eight ChIP-qPCR primers targeting integrated HIV-1 reporter provirus in TZM-bl cell line were designed. G5: Cellular DNA and viral 5’LTR junction; A: Nucleosome 0 assembly site; B: Nucleosome free region; C: Nucleosome one assembly site; V5: Viral 5’LTR and gag leader sequence junction; L: Luciferase region; V3: Viral poly purine tract and 3’LTR junction; G3: Viral 3’LTR and cellular DNA junction. For ChIP-qPCR conducted in J-Lat 10.6, G5’ represented cellular DNA and viral 5’LTR junction; E represented envelop; G3’ represented viral 3’LTR and cellular DNA junction; A, B, C, V5 and V3 represented as in TZM-bl cell lines.
- https://doi.org/10.7554/eLife.42426.035
-
Supplementary file 3
SUMO mutants used in SUMO-MS and CDK9 mutants used to identify SUMOylation sites.
The sequences of SUMO1-Q92R, SUMO2-Q88R and SUMO4-Q88R mutants, which mimicked yeast SUMO Smt3 to enable efficient identification of SUMO-acceptor lysines by MS, were represented below. Table also listed the major CDK9 mutants used in reversing mutation assay to identify SUMOylation sites on CDK9. All the sequences were verified by Sanger Sequencing to insure the accuracy.
- https://doi.org/10.7554/eLife.42426.036
-
Supplementary file 4
SUMOylated proteins at significance threshold below 10−7.
Table showed 1,329 SUMOylated proteins identified in global site-specific SUMO-MS at significance threshold below 10−7.
- https://doi.org/10.7554/eLife.42426.037
-
Supplementary file 5
Subclusters clustered by MCODE analysis.
Twelve highly interconnected functional subclusters were extracted from STRING network by MCODE analysis. Interconnectivity scores ranged from 14 to 96. Genes from each cluster were listed.
- https://doi.org/10.7554/eLife.42426.038
-
Supplementary file 6
Go analysis of SUMOylated proteins.
Biological process analysis, molecular function analysis, cellular component analysis and protein class analysis were conducted for the identified SUMOylated proteins. Table showed gene numbers and percentages of each group.
- https://doi.org/10.7554/eLife.42426.039
-
Supplementary file 7
SUMOylated proteins at significance threshold below 10−8.
Table showed 715 SUMOylated proteins identified in global site-specific SUMO-MS at significance threshold below 10−8.
- https://doi.org/10.7554/eLife.42426.040
-
Transparent reporting form
- https://doi.org/10.7554/eLife.42426.041