Architectural principles for Hfq/Crc-mediated regulation of gene expression
Figures
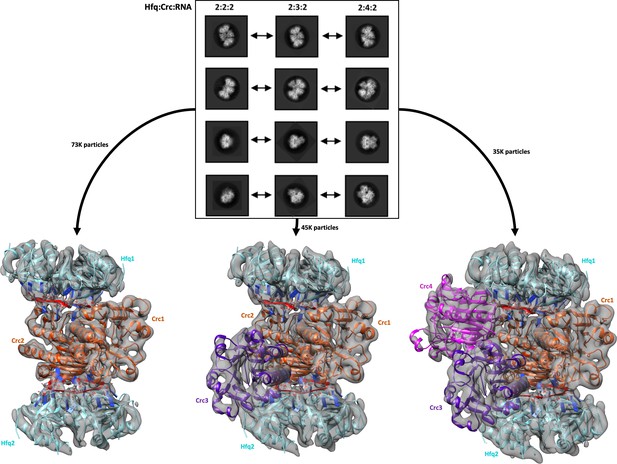
Reference free 2D classification and 3D classification of Hfq/Crc/RNA particles.
Three main classes of particles were observed after reference-free 2D classification (top), corresponding to Hfq/Crc/amiE6ARN assemblies with compositional stoichiometries of 2:2:2, 2:3:2 and 2:4:2. The amiE6ARN species (red) constitute the main interaction interface between Hfq and Crc, together forming the 2:2:2 core complex observable in all three models (bottom). Cyan: Hfq hexamer, orange purple and pink: Crc monomers, red: amiE6ARN. All cryoEM maps were low-pass filtered to 6 Å for clarity of presentation and the crystal structures were docked in as rigid bodies. Only a subset of the high quality 2D classes are shown in the panel.
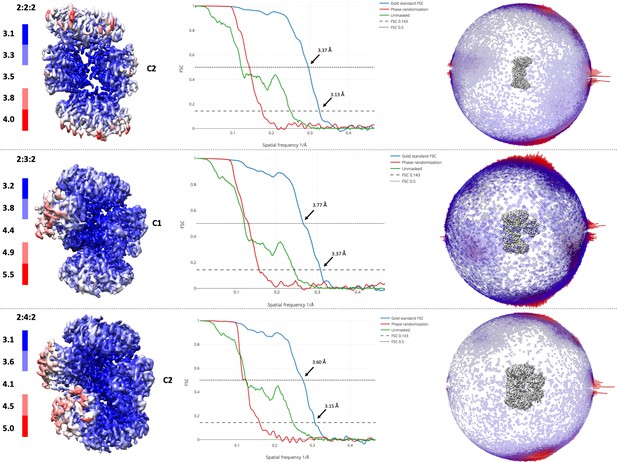
Resolution of maps and angular distributions of 2D images.
Local resolution estimates of the EM maps (left panel) and Gold standard Fourier shell correlation curves for all three reconstructions (central panel). FSC 0.143 and 0.5 are annotated. In the right panel, angular distributions of the 2D images are presented as a spherical bar plot, with red bars representing more preferred projections.
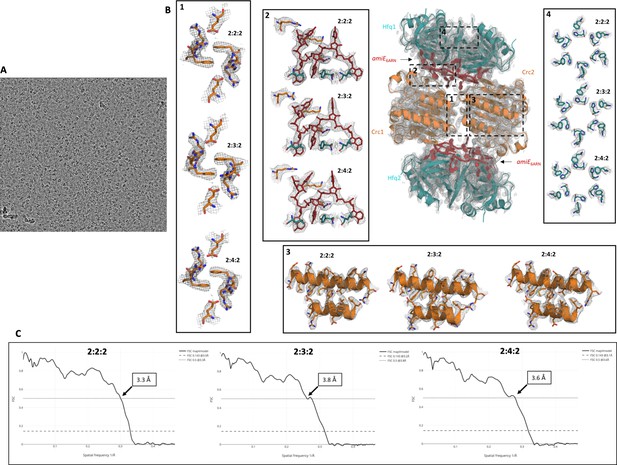
Raw micrograph image, fit of refined model into cryo-EM map, and correlation of experimental map and models.
(A) Raw micrograph after motion correction at three microns under focus, lowpass filtered to 20 Å. (B) High resolution cryo-EM maps with refined atomic models for all complexes showing the quality of the EM reconstructions. All maps and models were generated and refined independently of each other or a high-resolution reference structure, showing well defined and highly reproducible densities for all side chains in these signature regions. Even at the periphery the map density is of good quality, maintaining the six-fold symmetry of the Hfq components (inset 4). (C) Model versus map Fourier shell correlation (FSC) show a good correlation between the individual atomic models and the experimental cryo-EM maps. FSC 0.5 is annotated on the graph, whereas FSC 0.143 is annotated in the legends.
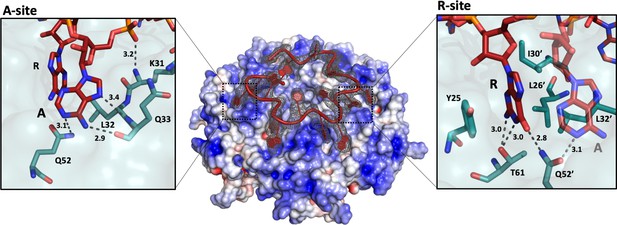
The 'A-R-N crown' in the Hfq/amiE6ARN RNA complex.
6 RNA triplets are partially embedded in six binding pockets on the Hfq distal side, forming a weaving, crown-like pattern. The A and R sites are occupied by adenine and a purine, respectively, whereas the RNA entry/exit site has no discriminatory preferences and is therefore referred to as the ‘N’ site. The Cryo-EM density for amiE6ARN is depicted as a grey mesh, with the RNA ‘crown’ modelled in red. Positively charged protuberances (blue) guide the RNA to fold into a star-shaped conformation to maximize the surface interaction between the negatively charged RNA backbone, and the positively charged Hfq surface pattern. An atomic model of the A-R-N occupation pattern. Left panel: Adenosine specificity site. Right panel: Purine specificity site. amiE nucleotide carbon atoms are depicted in red, Hfq carbon atoms are in green.
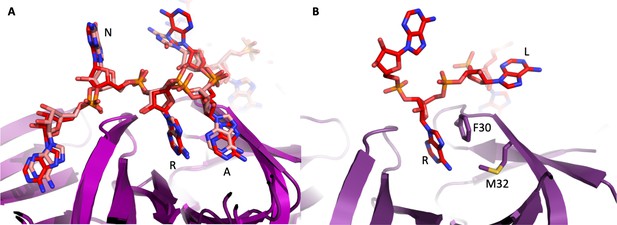
Interactions of RNA with the distal face of Hfq.
(A) Similarity of interactions between E. coli and P. aeruginosa Hfq and polyA18 and amiE6ARN RNA, respectively, on the distal face. The crystal structure of E. coli Hfq in complex with polyA18 reveals the A-R-N motif (red), which is strikingly congruent with the interactions between P. aeruginosa Hfq and amiE6ARN RNA (pink) in the complex with Crc. (B) The A-R-N motif is not supported by the Hfq of the Gram-positive bacteria Staphylococcus aureus and Bacillus subtilis, which instead use a R-L (purine, linker) motif (Horstmann et al., 2012). Based on this divergence, it seems unlikely that Hfq in the Gram-positive species form assemblies that resemble the P. aeruginosa Hfq/Crc assembly.
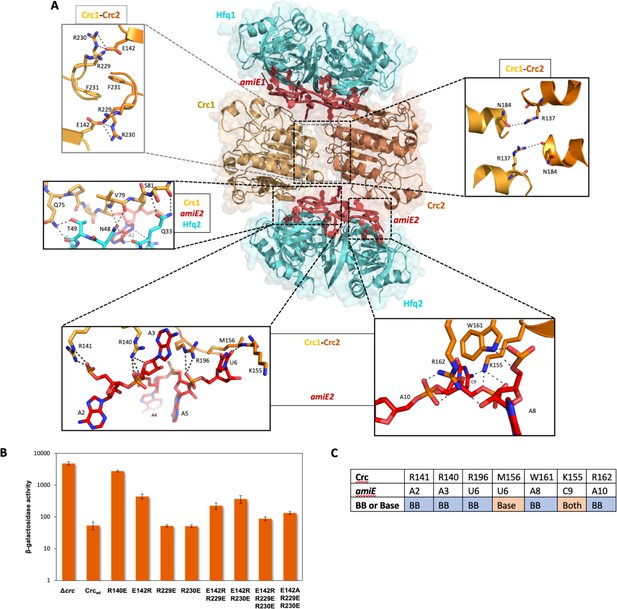
Model of the 2:2:2 Hfq/Crc/RNA complex and validation of interactions.
(A) Atomic model of the 2:2:2 Hfq/Crc/RNA complex. The view is along the C2 molecular symmetry axis which passes through the homodimeric Crc interface. Hfq hexamers flank the Crc dimer and present the amiE6ARN RNA to form two different interfaces with the Crc protomers, which form an anti-parallel dimer. The Crc protomers form strong polar contacts with mainly the backbone phosphate groups and exposed ribose rings (bottom insets). Two small C2 symmetric binding interfaces constitute the Crc dimerisation (top insets). A single short stretch on each Crc monomer binds a Hfq monomer (middle left panel). Dimeric Crc in yellow and orange, amiE6ARN RNA in red, Hfq hexamers in cyan. (B) Translational regulation of an amiE::lacZ reporter gene by Crc variants. P. aeruginosa strain PAO1Δcrc(pME9655) harboring plasmids pME4510 (vector control), pME4510crcFlag (Crcwt) or derivatives thereof encoding the respective mutant proteins was grown to an OD600 of 2.0 in BSM medium supplemented with 40 mM succinate and 40 mM acetamide. The β-galactosidase values conferred by the translational amiE::lacZ fusion encoded by plasmid pME9655 in the respective strains are indicated. The results represent data from two independent experiments and are shown as mean and range. (C) Table of Crc-amiE contacts in the 2:2:2 assembly. BB: Crc contacts with the RNA backbone; Base: Crc contacts with RNA bases.
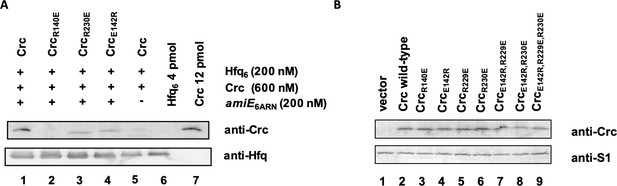
Association of Hfq and Crc in the presence of RNA.
(A) In vitro association of Hfq and Crc and Crc variants in the presence of RNA. The in vitro co-IP experiments were performed with Hfq and Crc and variants thereof in the presence (lanes 1–4) and absence (lane 5) of amiE6ARN RNA as indicated on top. Anti-Hfq specific antibodies and magnetic protein G beads were used for co-IP of Crc and Crc variants. The in vitro association of Hfq with Crc and variants thereof was visualized by western-blot analysis using anti-Crc or anti-Hfq specific antibodies as indicated at the right. Lanes 6 and 7, 4 pmol Hfq and 12 pmol Crc were loaded, respectively. The western-blot analyses were performed in triplicate. The result from one representative experiment is shown. (B) Crc variants are synthesized at comparable levels. Cultures of PAO1Δcrc(pME9655,pME4510) (lane 1), PAO1Δcrc(pME9655,pME4510crcFlag) (lane 2), PAO1Δcrc(pME9655, pME4510crc(R140E)Flag) (lane 3), PAO1Δcrc(pME9655,pME4510crc(E142R)Flag) (lane 4), PAO1Δcrc(pME9655,pME4510crc(R229E)Flag) (lane 5), PAO1Δcrc(pME9655,pME4510crc(R230E)Flag) (lane 6), PAO1Δcrc(pME9655, pME4510crc(E142R,R229E)Flag) (lane 7), PAO1Δcrc(pME9655, pME4510crc(E142R,R230E)Flag) (lane 8) and PAO1Δcrc(pME9655, pME4510crc(E142R,R229E,R230E)Flag) (lane 9), respectively, were grown to an OD600 of 2.0 in BSM medium supplemented with 40 mM succinate and 40 mM acetamide. The protein levels of Crc, the Crc variants (top) and of ribosomal protein S1 (loading control) were determined by quantitative western-blot analysis using anti-Crc and anti-S1 antibodies, respectively. The western-blot analyses were performed in triplicate. The result from one representative experiment is shown.
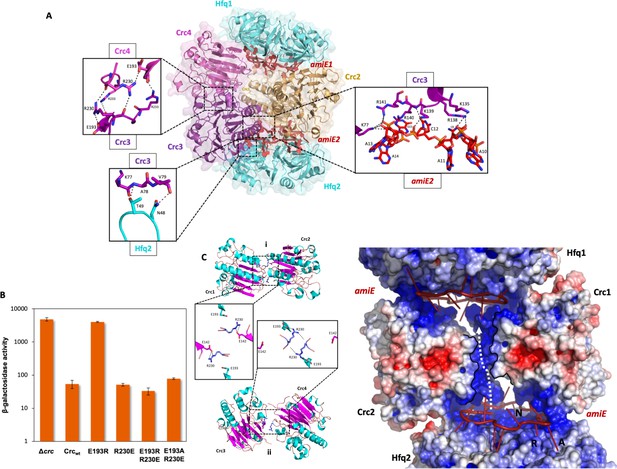
Model of the 2:4:2 Hfq/Crc/RNA complex and validation of interactions.
(A) Atomic model of the 2:4:2 Hfq/Crc/RNA complex. The insets show additional Hfq-Crc, Crc-Crc and Crc-RNA interactions not present in the 2:2:2 complex. The Crc3-4 dimer is formed by only one interface, with an R230-R230 interaction at the core, which globally overlaps with the dimer interface of the Crc1-Crc2 dimer (top left inset). Only one of two RNA-binding patches is presented to amiE6ARN in the Crc3-4 dimer, yet exploited more extensively (right inset). A small interface is formed between Crc3-4 and Hfq. Crc dimer in yellow, amiE6ARN RNA in red, Hfq hexamers in cyan, extra Crc dimer in magenta and purple. (B) Translational regulation of an amiE::lacZ reporter gene by Crc variants, as described in Figure 3B. The results represent data from two independent experiments and are shown as mean and range. (C) Two distinct dimeric Crc species are observed in the three complexes solved by cryoEM. i: The self-complementary interaction of the 2:2:2 core complex. ii: In the 2:4:2 complex, an alternative dimer is formed, showing a twisted dimer interface and more open configuration, with Arg230 serving as a dynamic hinge (bottom). (D) An electropositive half-channel runs along the dimer interface of the Crc1-2 dimer, and in the context of the Hfq/Crc/RNA assembly it could potentially serve as a conduit for RNA (dotted white arrow; see Figure 5). The A, R, and E RNA interaction sites of Hfq are annotated.
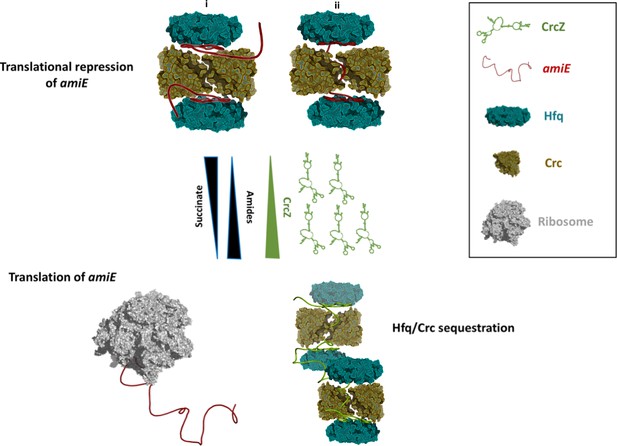
Schematic pathway of carbon catabolite repression.
When the preferred carbon source, succinate, is abundant, cellular CrcZ levels are low and Hfq and Crc occlude the amiE ribosome binding site by forming a higher order assembly, rendering CCR active and repressing synthesis of for example aliphatic amidase (top). We envision that translational repression may either occur on two mRNA molecules (i) or on a single mRNA molecule that comprises another Hfq binding site downstream of the first (ii). Upon depletion of succinate, CrcZ levels increase and CrcZ sequesters Hfq and Crc from amiE, potentially by occupying the multiple A-R-N patches on CrcZ and forming multicomponent ‘beads on a string’ (bottom). As such CCR is abolished, allowing metabolism of a secondary carbon source, for example amide conversion by aliphatic amidase.
Videos
A comparison of the Crc-Crc interface in the 2:2:2 Hfq:Crc:RNA complex (the core complex) and the second type of interface formed between the additional Crc components of the 2:4:2 complex.
https://doi.org/10.7554/eLife.43158.011Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Pseudomonas aeruginosa) | crc | NA | Pseudomonas genome database: PA5332 | |
| Gene (P. aeruginosa) | amiE | NA | Pseudomonas genome database: PA3366 | |
| Strain, strain background (P. aeruginosa) | PAO1 | PMID: 111024 | ||
| Strain, strain background (P. aeruginosa) | PAO1Δcrc | PMID: 20080802 | ||
| Strain, strain background (Escherichia coli) | XL1-Blue | Stratagene | ||
| Strain, strain background (E. coli) | BL21(DE3) | Novagen | ||
| Genetic reagent (P. aeruginosa) | amiE6ARN RNA | Microsynth, PMID: 29244160 | sequence: 5'-AAAAAUAACAACAAGAAG-3' | |
| Antibody | anti-Crc (rabbit polyclonal) | Pineda | (1:5000) | |
| Antibody | anti-Hfq (rabbit polyclonal) | Pineda | (1:10000) | |
| Antibody | anti-S1 (rabbit polyclonal) | Pineda | (1:10000) | |
| Antibody | anti-rabbit IgG (goat polyclonal) | Sigma | Sigma: A3687 | conjugated with alkaline phosphatase, (1:10000) |
| Recombinant DNA reagent | pME9655 (plasmid) | PMID: 20080802 | ||
| Recombinant DNA reagent | pETM14lic-His6Crc (plasmid) | PMID: 23717639 | ||
| Recombinant DNA reagent | pME4510crcFlag (plasmid) | PMID: 29244160 | ||
| Peptide, recombinant protein | Hfq | PMID: 21330354 | ||
| Peptide, recombinant protein | Crc | PMID: 23717639 | ||
| Software, algorithm | Relion3 | DOI: 10.7554/eLife.42166 | ||
| Software, algorithm | LocScale | DOI: 10.7554/eLife.27131 | ||
| Software, algorithm | LocalDeblur | DOI: 10.1101/433284 |
Additional files
-
Supplementary file 1
Resolution of maps and angular distributions of 2D images.
(A) Summary of the data collection parameters and the reconstruction and refinement statistics for the Hfq:Crc:RNA complexes. (B) List of the plasmids and strains used in this study. (C) Oligonucleotides used in this study.
- https://doi.org/10.7554/eLife.43158.013
-
Transparent reporting form
- https://doi.org/10.7554/eLife.43158.014





