The genetic requirements of fatty acid import by Mycobacterium tuberculosis within macrophages
Figures
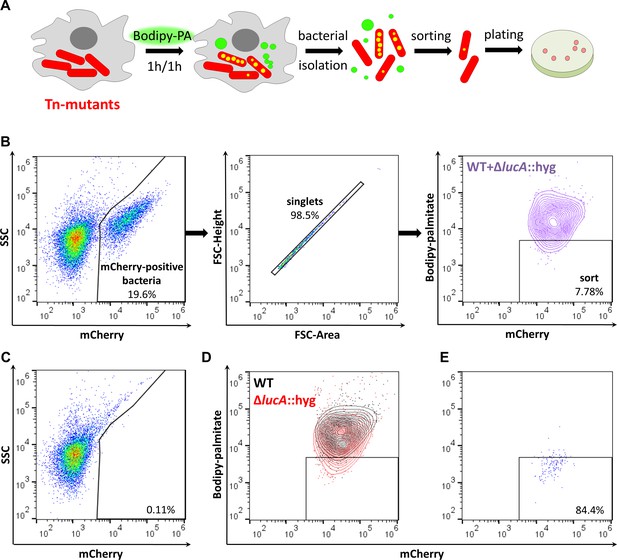
Developing a screening approach to identify fatty acid import mutants in Mtb.
(A) Schematic of the genetic screen. Macrophages are infected with mCherry-Mtb for three days. Intracellular transposon mutant library (Tn-mutants) is pulse labeled with Bodipy-palmitate (Bodipy-PA). Intracellular bacteria are isolated, sorted, and plated on selective agar. Transposon insertion sites were determined for the mutant pools recovered from agar. (B) Gating strategy to isolate single Bodipy-low Mtb cells. (C) Analysis of uninfected macrophage lysates that have been pulse labeled with Bodipy-palmitate. (D) Flow analysis of a 1:1 mixture containing mCherry-positive WT and ∆lucA::hyg bacteria. (E) Purity of sorted low Bodipy-low population.
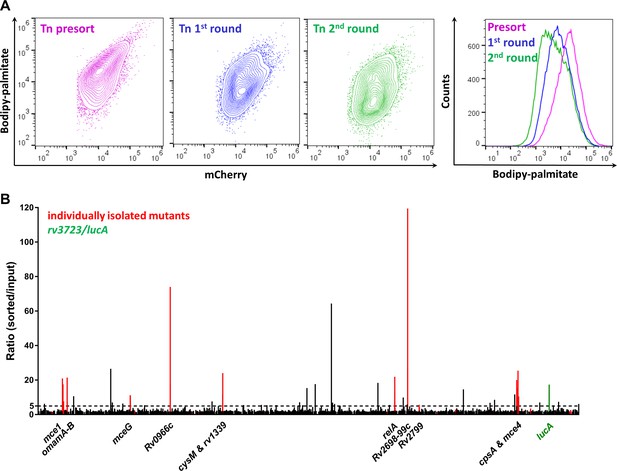
Enrichment and identification of mutants with a reduced ability to import Bodipy-palmitate.
(A) Flow analysis of Bodipy-palmitate incorporation by input mutant pool (Tn presort) and subsequent enriched mutant pools. (B) Relative abundance of transposon insertion sites following the second enrichment sort which are overrepresented >1.5 fold with p < 0.05. The x-axis represents position of the transposon insertion in the Mtb chromosome and peak height represents relative level of overrepresentation in the analysis. Individually isolated mutants are indicated in red. The rv3723/lucA mutation is highlighted in green and the dotted line indicates fivefold cut-off. Full gene lists and probe sequences used in the TraSH analysis are supplied in Figure 2—source data 1. Fold-change distribution of insertion sites shown on Figure 2B (>1.5 fold overrepresented with p < 0.05 after two rounds of enrichment) among the first and second rounds of sorting is supplied in Figure 2—figure supplement 1. A summary of the isolated mutants is provided in Figure 2—source data 2.
-
Figure 2—source data 1
Summary of TraSH analysis results.
- https://doi.org/10.7554/eLife.43621.005
-
Figure 2—source data 2
Spreadsheet of all individually isolated mutants.
- https://doi.org/10.7554/eLife.43621.006
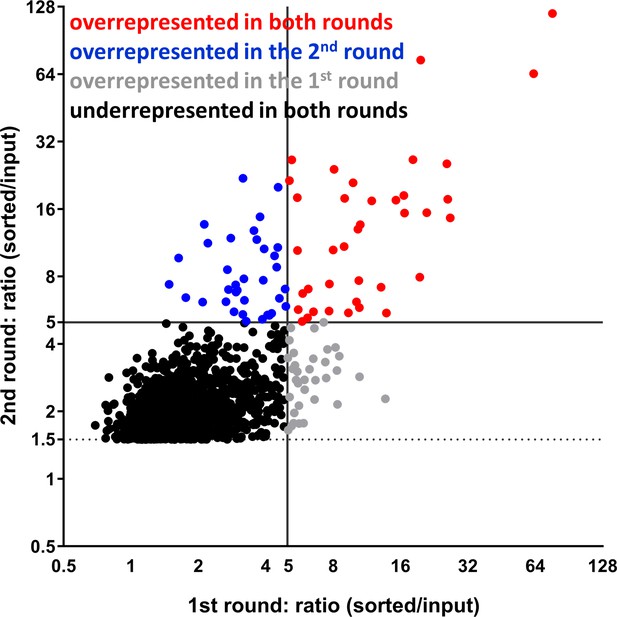
Fold-change distribution of insertion sites from the enrichment screens.
Distribution of TraSH identified transposon insertion sites that are overrepresented >1.5 fold p < 0.05 after second round of enrichment. Mutants are considered overrepresented if the fold increase is ≥5 in each enrichment.
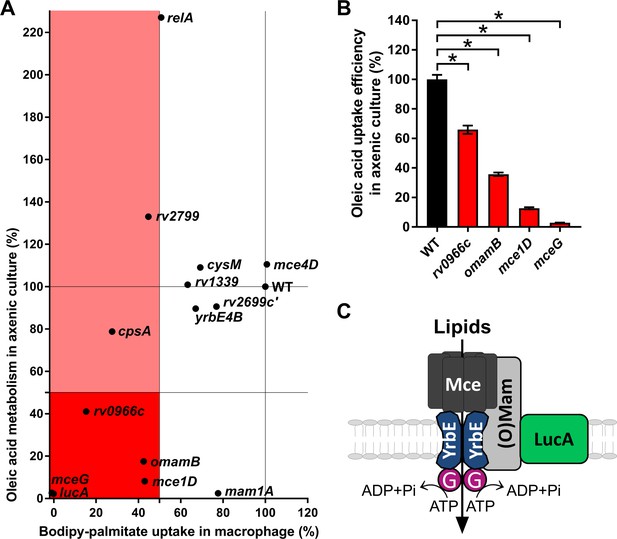
Phenotypic analysis of selected mutants.
(A) Bodipy-palmitate incorporation by Mtb during macrophage infection in the selected mutants (X axis) and catabolic metabolism of [1-14C]-oleic acid in axenic culture (Y axis). (B) Uptake of [1-14C]-oleic acid by selected mutants in axenic culture. Data are means ± SD (n = 4). *p < 0.0001 (Student’s t test). (C) Hypothetical model for Mce transporter configuration. Mce subunits are substrate specific and deliver lipids to the YrbE permease subunits. ATP is hydrolyzed by Rv0655/MceG (G) to transport substrates into the cytosol. LucA interacts with Mce transporters through accessory subunits Mam’s or Omam’s.
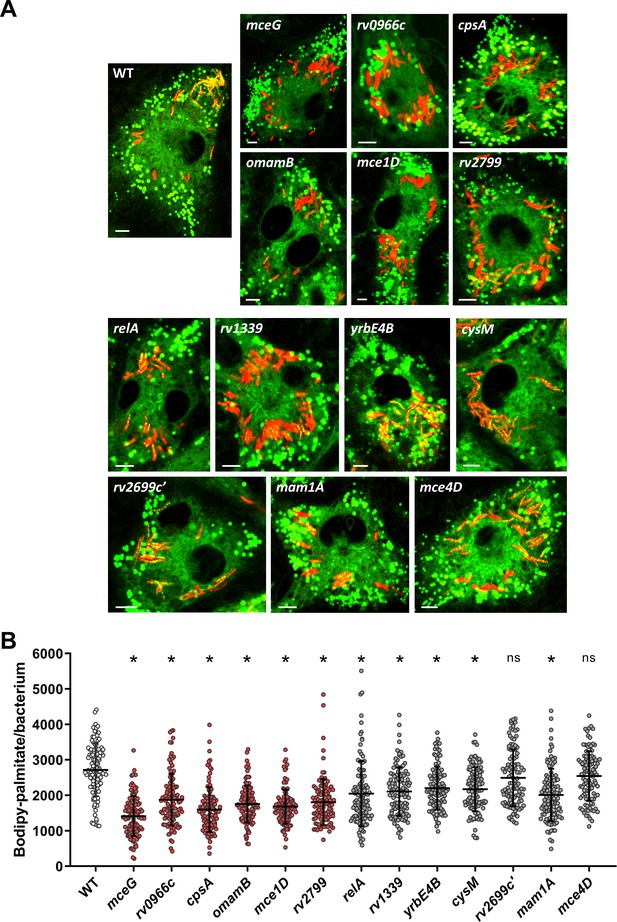
Confocal microscopy of selected mutants.
(A) Representative confocal images of Bodipy-palmitate labeled macrophages infected with selected mutants. Red = mCherry Mtb, green = Bodipy palmitate. Scale bar 5.0 μm. (B) Quantification of the Bodipy-palmitate/µm2 signal for individual bacteria detected based on mCherry signal. Measurements were performed for multiple confocal images. Mean ± SD are shown (n = 107). *p < 0.0001, compared to WT (Student’s t test).
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.43621.009





