Mutationally-activated PI3’-kinase-α promotes de-differentiation of lung tumors initiated by the BRAFV600E oncoprotein kinase
Figures
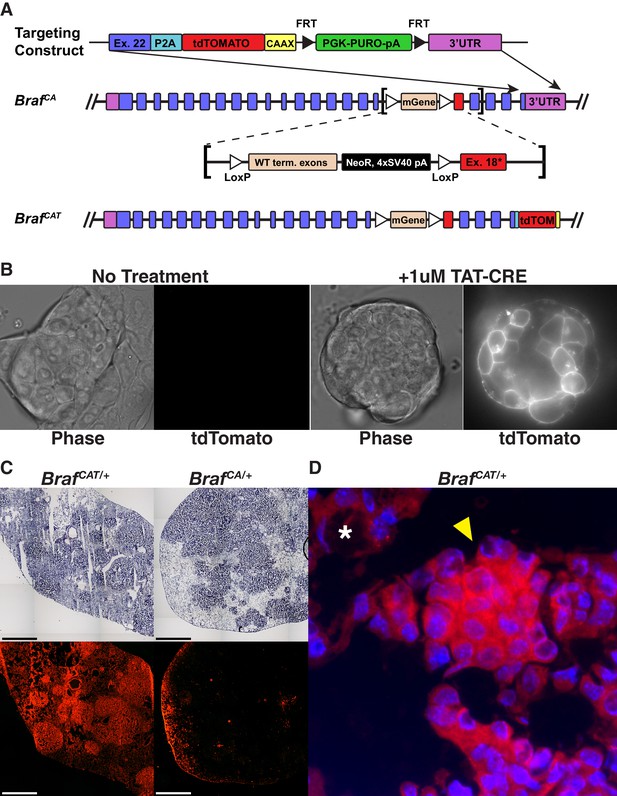
Engineering and validation of a novel genetically engineered mouse model of BRAFV600E driven cancer.
(A) The BrafCAT mouse builds upon the utility of the BrafCA mouse by tying expression of the oncogenic form of BRAF to expression of the red fluorescent protein, tdTomato. (B) Targeted BrafCAT ES cells display membrane associated red fluorescence only after the addition of TAT-CRE. (C) Comparison of Ad-CMV-CRE initiated lung tumor formation and fluorescence in frozen sections from lungs of BrafCAT/+ and BrafCA/+ animals. (D) Lung adenoma found in a BrafCAT animal showing fluorescence in the tumor (arrowhead) and not in the lung parenchyma (asterisk).
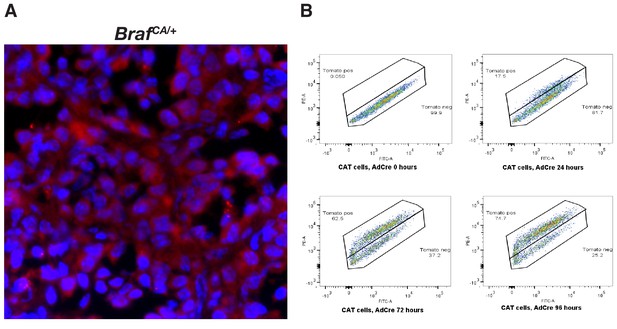
Engineering and validation of a novel genetically engineered mouse model of BRAFV600E driven cancer.
(A) Lung adenoma found in a BrafCA animal showing no specific fluorescence in the tumor. (B) BrafCAT MEFs display detectable red fluorescence within 24 hr of Ad-CMV-CRE addition, with the majority of cells displaying red fluorescence within 72 hr.
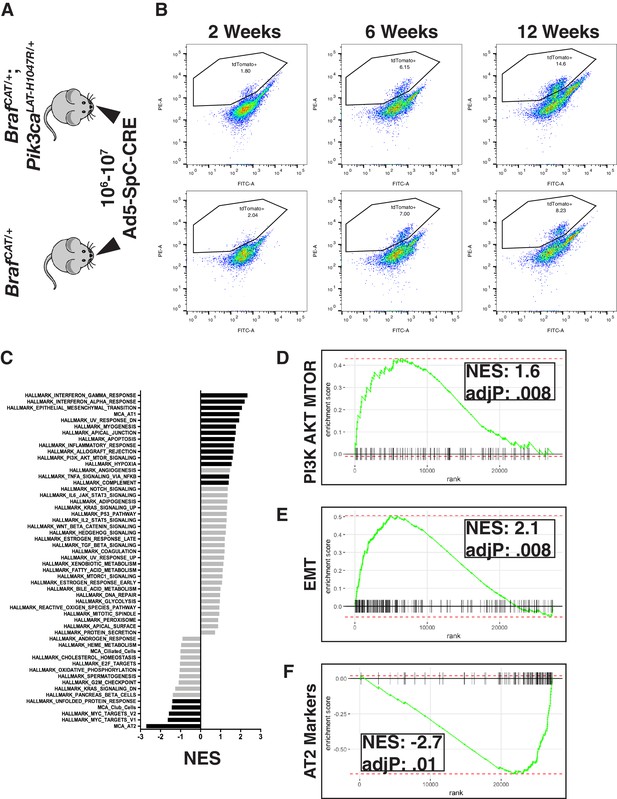
Examining global gene expression changes caused by combination of PI3K and MAPK activation using RNA-SEQ.
(A) Tumors were adenovirally induced in cohorts of BrafCAT/+ and BrafCAT/+;Pik3caLAT-H1047R/+ mice with AT2 specific CRE expression. Mice harvested at 2 or 6 weeks were induced with 107 PFU of Ad5-SpC-CRE, whereas mice harvested at 12 weeks were induced with 106 PFU of Ad5-SpC-CRE. (B) Tumor cells were harvested from each genotype at 2 weeks, 6 weeks, and 12 weeks post tumor induction via tissue dissociation and FACS. (C) GSEA analyses profiling cells sorted from BRAFV600E/PI3KαH1047R driven tumor bearing mice compared to BRAFV600E driven tumor bearing mice, showing all ‘Hallmark’ gene sets along with gene sets constructed from the most specific markers of the cell types of the distal lung epithelium. Black bars indicate adjP <.05, gray bars indicate Benjamini-Hochberg corrected enrichment statistic adjP ≥. 05. Here all time points combined within genotype. (D) GSEA mountain plot showing broad activation of PI3K signaling in BRAFV600E/PI3KαH1047R driven tumor bearing mice; adjP is Benjamini-Hochberg corrected enrichment statistic. (E) GSEA mountain plot showing broad activation of EMT in BRAFV600E/PI3KαH1047R driven tumor bearing mice; adjP is Benjamini-Hochberg corrected enrichment statistic. (F) GSEA mountain plot showing widespread loss of AT2 identity in BRAFV600E/PI3KαH1047R driven tumor bearing mice; adjP is Benjamini-Hochberg corrected enrichment statistic.
-
Figure 2—source code 1
R script to perform gene set enrichment analysis on Figure 2—source data 1–2, as well as plot these results.
- https://doi.org/10.7554/eLife.43668.007
-
Figure 2—source data 1
DEseq2 output of differentially expressed genes comparing BRAFV600E/PI3KαH1047R and BRAFV600E driven tumors – all weeks pooled.
- https://doi.org/10.7554/eLife.43668.008
-
Figure 2—source data 2
DEseq2 output of differentially expressed genes comparing BRAFV600E/PI3KαH1047R and BRAFV600E driven tumors – weeks separated.
- https://doi.org/10.7554/eLife.43668.009
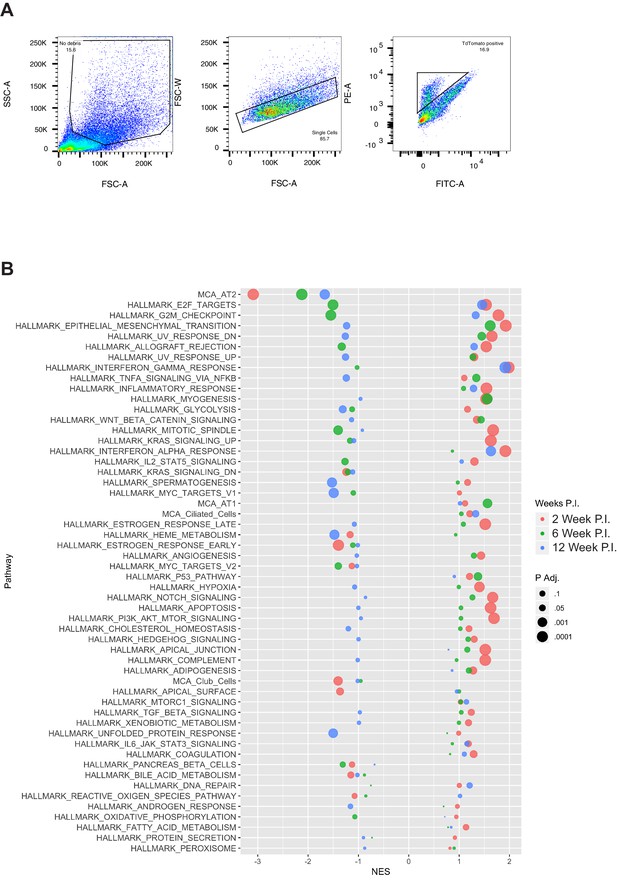
Examining global gene expression changes caused by combination of PI3K and MAPK activation using RNA-SEQ.
(A) Expanded view of GSEA analyses comparing results at each individual time point. Point size inversely proportional to adjusted enrichment statistic p value, color indicates time point at which comparison was performed. (B) In depth gating strategy used for isolation of all FACS sorted tumor cells in this study.
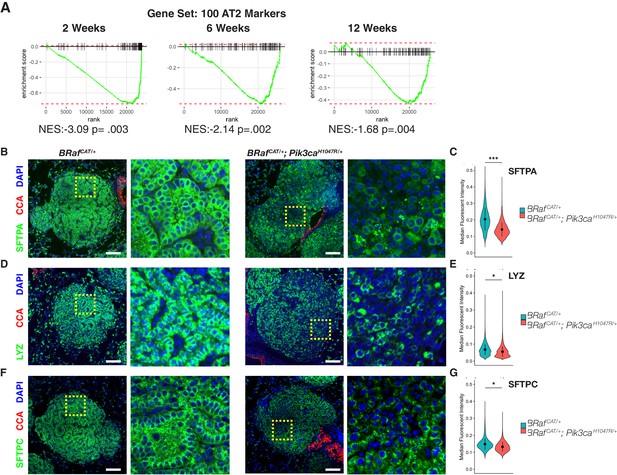
BRAFV600E/PI3KαH1047R Driven Tumors Display Widespread Heterogeneous Loss of AT2 Marker Expression, Whereas BRAFV600E Driven Tumors Maintain AT2 marker expression.
(A) GSEA mountain plots comparing gene expression profiles of cells sorted from BrafCAT/+;Pik3caLAT-H1047R/+ mice to those from BrafCAT/+ mice, using 100 specific markers of AT2 identity as the gene set. Analyses show comparison of tumors from the two genotypes at each time point analyzed; P value is enrichment statistic. (B) Tumors found in BrafCAT/+;Pik3caLAT-H1047R/+ mice have widespread variegated loss of expression of the functional AT2 marker, SFTPA, compared to tumors found in BrafCAT/+ mice. Expression of CCA is seen in airways but not in tumors of either genotype. Dashed boxes highlight areas of increased magnification. Scale bars = 100 um. (C) CellProfiler based quantitation of SFTPA immunofluorescence. Wilcoxon rank sum p value = 0.00013 (D) Tumors found in BrafCAT/+;Pik3caLAT-H1047R/+ mice have widespread loss of expression of the functional AT2 marker, LYZ, compared to tumors found in BrafCAT/+ mice. (E) CellProfiler based quantitation of LYZ immunofluorescence. Wilcoxon rank sum p value = 0.02224 (F) Tumors found in BrafCAT/+;Pik3caLAT-H1047R/+ mice have widespread loss of expression of the functional AT2 marker, SFTPC, compared to tumors found in BrafCAT/+ mice. (G) CellProfiler based quantitation of SFTPC immunofluorescence. Wilcoxon rank sum p value = 0.02323.
-
Figure 3—source code 1
R script to perform gene set enrichment analyses on Figure 2—source data 2, as well as plot these results.
- https://doi.org/10.7554/eLife.43668.012
-
Figure 3—source code 2
R script to perform statistics on Figure 3—source data 1–3, as well as plot these results.
- https://doi.org/10.7554/eLife.43668.013
-
Figure 3—source code 3
Cellprofiler pipeline to quantify raw images, producing Figure 3—source data 1–3.
- https://doi.org/10.7554/eLife.43668.014
-
Figure 3—source data 1
Cellprofiler output quantifying SFTPA immunofluorescence in BRAFV600E/PI3KαH1047R and BRAFV600E driven tumors.
- https://doi.org/10.7554/eLife.43668.015
-
Figure 3—source data 2
Cellprofiler output quantifying LYZ immunofluorescence in BRAFV600E/PI3KαH1047R and BRAFV600E driven tumors.
- https://doi.org/10.7554/eLife.43668.016
-
Figure 3—source data 3
Cellprofiler output quantifying SFTPC immunofluorescence in BRAFV600E/PI3KαH1047R and BRAFV600E driven tumors.
- https://doi.org/10.7554/eLife.43668.017
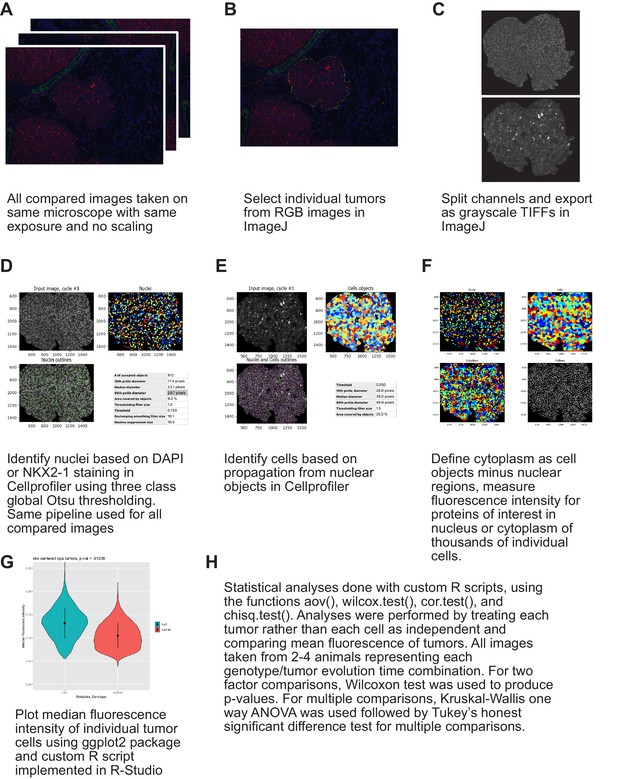
Cellprofiler based immunofluorescence quantification strategy.
(A) Images were only compared when they had been immunostained in parallel, imaged on the same microscope with the same exposure settings, and not been processed or scaled post image acquisition. (B) Individual tumors were traced in NIH imageJ. (C) Color images were split in to component channels in NIH imageJ. (D) CellProfiler was used to identify tumor nuclei based on NKX2-1 staining when done, or based on DAPI. (E) Cells were identified based on propagation from nuclear objects. (F) Cytoplasm was defined as Cell objects minus nuclei. (G) Measurements taken always used median fluorescent intensity with no outlier removal. Data were imported and graphed using a custom R script in RStudio. (H) Statistical detail of comparisons done with a custom R script in RStudio.
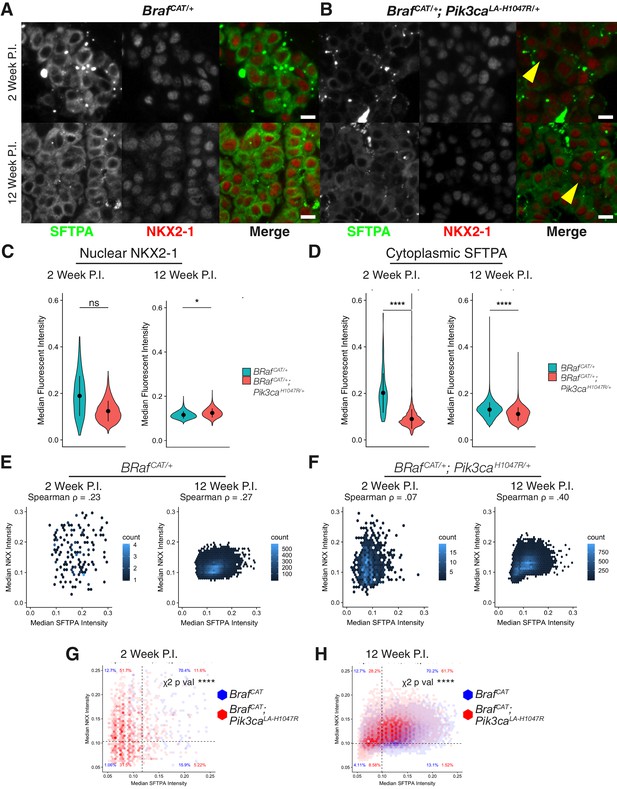
Expression levels and localization of lung lineage survival transcription factors are maintained in BRAFV600E/PI3KαH1047R driven tumors, Including those cells which have lost expression of markers of AT2 identity.
(A) BRAFV600E driven hyperplasia and tumors display widespread expression of both SFTPA and nuclear localization of the lung lineage transcription factor, NKX2-1, at 2 and 12 weeks post initiation. Scale bar = 10 um. (B) BRAFV600E/PI3KαH1047R driven hyperplasia and tumors show decreased SFTPA expression at 2 and 12 weeks post initiation. These tumors maintain broad nuclear expression of NKX2-1, including those cells with decreased SFTPA expression (yellow arrowheads). (C) Quantitation showing no significant difference in NKX2-1 immunoreactivity at 2 weeks post initiation, but a slight increase in nuclear NKX2-1 at 12 weeks post initiation. Wilcoxon rank sum p values = 0.2,. 02 respectively. (D) Significant reduction of SFTPA immunoreactivity seen in BRAFV600E/PI3KαH1047R driven hyperplasia and tumors at both 2 and 12 weeks post initiation. Wilcoxon rank sum p values = 5e-5, 4e-5 respectively. (E) Cytoplasmic SFTPA immunoreactivity plotted versus nuclear NKX2-1 immunoreactivity in BRAFV600E driven hyperplasia and tumors at 2 and 12 weeks post initiation. Similar association seen at both time points (Rho = 0.23,. 27 respectively). (F) Cytoplasmic SFTPA immunoreactivity plotted versus nuclear NKX2-1 immunoreactivity in BRAFV600E/PI3KαH1047R driven hyperplasia and tumors at 2 and 12 weeks post initiation. Relatively lower association seen at 2 weeks compared to 12 weeks (Rho = 0.07,. 40 respectively). (G) Overlay of BRAFV600E/PI3KαH1047R and BRAFV600E driven hyperplasia 2 weeks post initiation. Dashed line for each marker drawn at mean - one standard deviation of BRAFV600E driven tumors. BRAFV600E/PI3KαH1047R driven tumors show fewer SFTPA+, NKX2−1 + cells, most strongly accounted for by an increase in SFTPA-, NKX2−1 + cells. Chi square test associates genotype with distribution, p val <1e-5. (H) Overlay of BRAFV600E/PI3KαH1047R and BRAFV600E driven tumors 12 weeks post initiation. Dashed line for each marker drawn at mean - one standard deviation of BRAFV600E driven tumors. BRAFV600E/PI3KαH1047R driven tumors show fewer SFTPA+, NKX2−1 + cells, most strongly accounted for by an increase in SFTPA-, NKX2−1 + cells. Chi square test associates genotype with distribution, p val <1e-5.
-
Figure 4—source code 1
R script to perform statistics on Figure 4—source data 1–2, as well as plot these results.
- https://doi.org/10.7554/eLife.43668.020
-
Figure 4—source code 2
Cellprofiler pipeline to quantify raw images from BRAFV600E/PI3KαH1047R and BRAFV600E driven tumors, producing Figure 4—source data 1.
- https://doi.org/10.7554/eLife.43668.021
-
Figure 4—source code 3
Cellprofiler pipeline to quantify raw images from KRASG12D/PIK3CAH1047R and KRASG12D driven tumors, producing Figure 4—source data 2.
- https://doi.org/10.7554/eLife.43668.022
-
Figure 4—source data 1
Cellprofiler output quantifying immunofluorescence of SFTPA and NKX2-1 in BRAFV600E/PI3KαH1047R and BRAFV600E driven tumors.
- https://doi.org/10.7554/eLife.43668.023
-
Figure 4—source data 2
Cellprofiler output quantifying immunofluorescence of SFTPA and NKX2-1 in KRASG12D/PIK3CAH1047R and KRASG12D driven tumors.
- https://doi.org/10.7554/eLife.43668.024
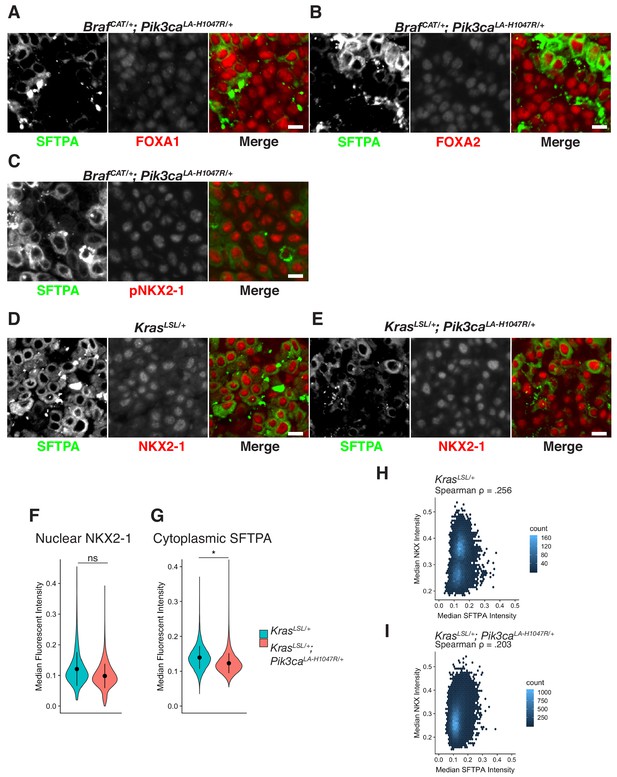
Expression levels and localization of lung lineage survival transcription factors are maintained in BRAFV600E/PI3KαH1047R and KRASG12D/PI3KαH1047R Driven Tumors, including those cells which have lost expression of markers of AT2 identity.
(A) Tumor cells with decreased SFTPA maintain expression and nuclear localization of the lung lineage transcription factor, FOXA1. Scale bars = 10 um. (B) Tumor cells with decreased SFTPA maintain expression and nuclear localization of the lung lineage transcription factor, FOXA2. (C) Tumor cells with decreased SFTPA maintain phosphorylation of the lung lineage transcription factor, NKX2-1. (D) Expression of SFTPA and NKX2.1 are maintained in KRASG12D driven tumors, 16 weeks post initiation. (E) Activation of PI3K in KRASG12D driven tumors causes a decrease of SFTPA immunoreactivity not apparently associated with a decrease in NKX2-1 immunostaining. (F) No significant difference in NKX2-1 immunoreactivity upon PI3K activation in KRASG12D driven tumors. (G) Significant reduction in cytoplasmic staining of SFTPA upon PI3K activation in KRASG12D driven tumors. Wilcoxon rank sum p=0.0354. (H) Modest association between immunoreactivity of SFTPA and NKX2-1 in KRASG12D driven tumors (Spearman Rho = 0.256). (I) Modest association between immunoreactivity of SFTPA and NKX2-1 in in KRASG12D/PI3KαH1047R driven tumors (Spearman Rho = 0.203).
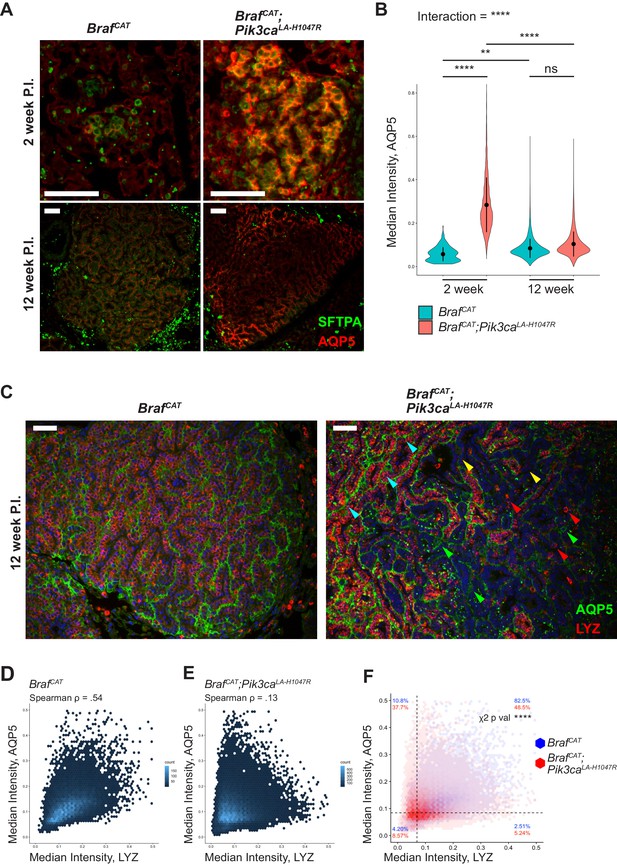
BRAFV600E/PI3KαH1047R and BRAFV600E driven tumors both show effects on differentiation status, with BRAFV600E/PI3KαH1047R driven tumors displaying more profound effects on identity.
(A) BRAFV600E/PI3KαH1047R and BRAFV600E driven hyperplasia both show immunoreactivity of the AT1 marker, AQP5, 2 weeks post initiation with BRAFV600E/PI3KαH1047R driven hyperplasia showing enhanced immunostaining. BRAFV600E/PI3KαH1047R and BRAFV600E driven tumors also show immunoreactivity of AQP5 12 weeks post initiation with BRAFV600E/PI3KαH1047R driven tumors showing a more variable pattern of immunostaining. Scale bars = 100 um. (B) Quantitation demonstrating significant effect of PI3KαH1047R on AQP5 immunoreactivity in BRAFV600E driven tumors 2 weeks post initiation. No difference seen in AQP5 immunoreactivity between BRAFV600E/PI3KαH1047R and BRAFV600E driven tumors 12 weeks post initiation. This appears to be the result of a slight increase in AQP5 immunoreactivity between 2 and 12 weeks in BRAFV600E driven tumors and a more dramatic decrease in AQP5 immunoreactivity between 2 and 12 weeks in BRAFV600E/PI3KαH1047R driven tumors. ANOVA p<1e-5, multiple comparisons done by Tukey’s Honest Significant Difference test, ****: p<1 e −5, **p=0.0014. (C) BRAFV600E driven tumors display widespread immunoreactivity to both AQP5 and the AT2 marker, LYZ, 12 weeks post initiation. BRAFV600E/PI3KαH1047R driven tumors show cells with widely varied expression of differentiation markers, including AQP5+, LYZ+ (Cyan arrows); AQP5-, LYZ+ (Red arrows); AQP5+, LYZ- (Green arrows); and AQP5-, LYZ- (Yellow arrows) cells. Scale bars = 100 um. (D) BRAFV600E driven tumors show relatively high association between AQP5 and LYZ immunoreactivity (Rho = 0.54). (E) BRAFV600E/PI3KαH1047R driven tumors show relatively low association between AQP5 and LYZ immunoreactivity (Rho = 0.13) (F) Overlay of BRAFV600E/PI3KαH1047R and BRAFV600E driven tumors 12 weeks post initiation. Dashed line for each marker drawn at mean - one standard deviation of BRAFV600E driven tumors. BRAFV600E/PI3KαH1047R driven tumors show fewer AQP5+, LYZ + cells, most strongly accounted for by an increase in AQP5+, LYZ- cells, but with increases also seen in AQP5-, LYZ + and AQP5-, LYZ- cells. Chi square test associates genotype with distribution, p val <1e-5.
-
Figure 5—source code 1
R script to perform statistics on Figure 4—source data 1, as well as plot these results.
- https://doi.org/10.7554/eLife.43668.027
-
Figure 5—source code 2
Cellprofiler pipeline to quantify raw images from BRAFV600E/PI3KαH1047R and BRAFV600E driven tumors, producing Figure 5—source data 1.
- https://doi.org/10.7554/eLife.43668.028
-
Figure 5—source data 1
Cellprofiler output quantifying AQP5 and LYZ immunofluorescence in BRAFV600E/PI3KαH1047R and BRAFV600E driven tumors.
- https://doi.org/10.7554/eLife.43668.029
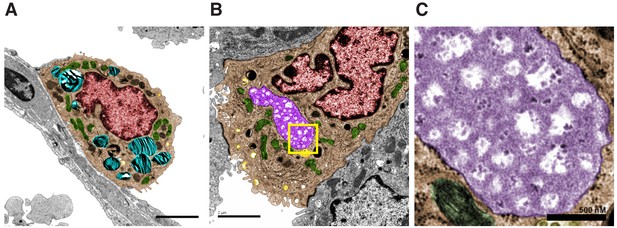
BRAFV600E-driven lung tumors show characteristics of bipotent progenitor cells.
(A) Electron micrograph showing a normal AT2 cell in the lung epithelium. Cyan structures are surfactant rich lamellar bodies, red structure is nucleus, green structures are mitochondria. Scale bar = 2 um. (B) Electron micrograph showing an abnormal AT2 cell in a BRAFV600E driven tumor harboring a large vacuole (purple). Scale bar = 2 um. (C) Enhanced magnification of vacuole showing rough electron dense pattern characteristic of glycogen storage. Scale bar = 500 nm. (D) Human lung adenocarcinomas harboring mutations in either PIK3CA, PTEN, or AKT show a significant reduction in PPARGC1A transcript levels when compared to tumors not shown to be harboring any of these three mutations (p=0.0295 by unpaired t-test with Welch’s correction). (E) 5 kb promoter regions of AT2 specific genes show enrichment of novel DNA motifs. The most significantly enriched of these motifs (E,1) is represented in 71 of 99 promoters scanned and significantly matches the known motif of NR5A2. (F) Alignment of the consensus binding motif for NR5A2 with the novel conserved motifs found in E,1 and E,4.
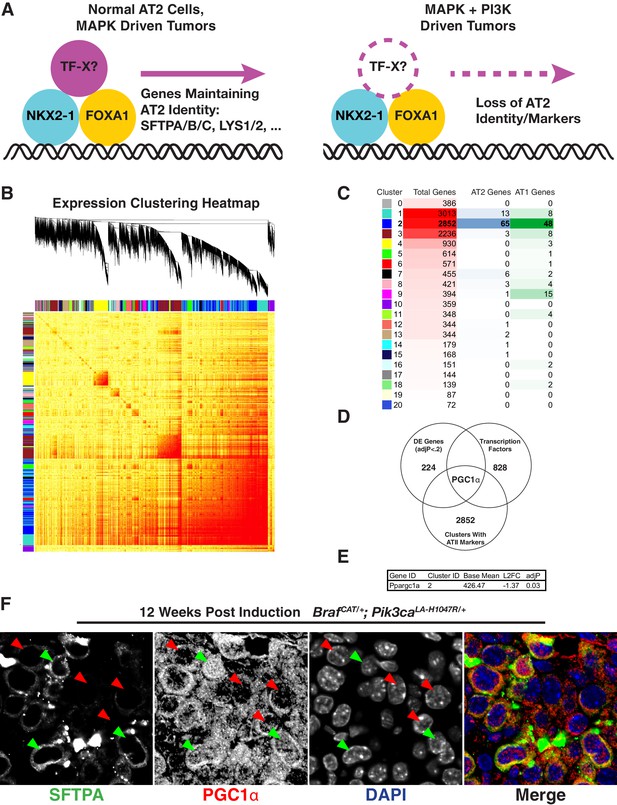
Loss of PGC1α Expression Correlates With Change in Expression of Lung Markers.
(A) Hypothetical model showing an additional factor cooperating with lung lineage transcription factors that is downregulated upon dual arm mutational activation of growth factor signaling. (B) Weighted gene correlation network analysis (WGCNA) heat map identifies 21 correlated gene expression modules. Gene tree shows relationship of individual genes, where multi-color bars adjacent to heat map identify individual clusters. (C) Table summarizing result of WGCNA analysis and AT1/AT2 memberships. Cluster number is listed adjacent to color corresponding to cluster in (B). For each cluster, shown is the total number of genes along with the number of AT1 and AT2 marker genes from AT1-100 and AT2-100. Cluster two contains the majority of both AT1 and AT2 marker genes. (D) A three factor approach to identify novel regulators of pneumocyte identity. Within the intersection of differentially expressed genes, genes co-regulated with the majority of AT1 and AT2 specific genes, and known transcription factors, lies a single gene, PGC1α. (E) PGC1α is significantly downregulated in BRAFV600E/PI3Kα H1047R driven tumors compared to BRAFV600E driven tumors; adjP is Benjamini-Hochberg corrected P value from DESeq2. (F) Decreased nuclear PGC1α immunoreactivity (red arrows) correlates with loss of AT2 identity on a cell by cell basis. AT2 identity is maintained in those cells which maintain nuclear PGC1α immunoreactivity (green arrows).
-
Figure 6—source code 1
R script to perform weighted correlation network (WGCNA) on Figure 6—source data 1.
- https://doi.org/10.7554/eLife.43668.033
-
Figure 6—source data 1
DEseq2 normalized RNA-seq count output of all BRAFV600E/PI3KαH1047R and BRAFV600E driven tumors.
- https://doi.org/10.7554/eLife.43668.034
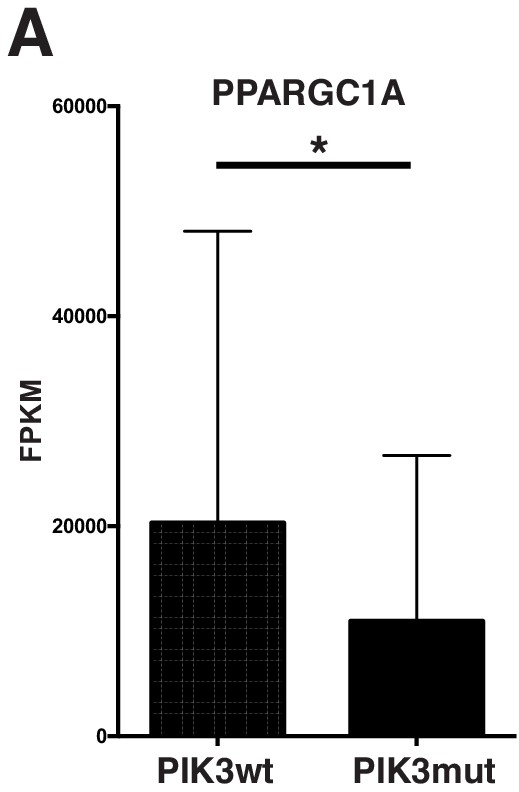
Mutations predicted to affect the PI3' lipid signaling pathway correlate with decreased PPARGC1A in human lung adenocarcinoma.
(A) PI3' pathway signaling is predicted to be activated by mutations inPIK3CA,PTEN, andAKT1. Segregating human lung adenocarcinoma by presence (PIK3mut) or absence (PIK3wt) of any of these mutations, shows that predicted PI3' lipid pathway activation correlates with decreased Ppargc1a transcript levels.
-
Figure 6—figure supplement 1—source data 1
Ppargc1a FPKM values from human tumors.
- https://doi.org/10.7554/eLife.43668.032
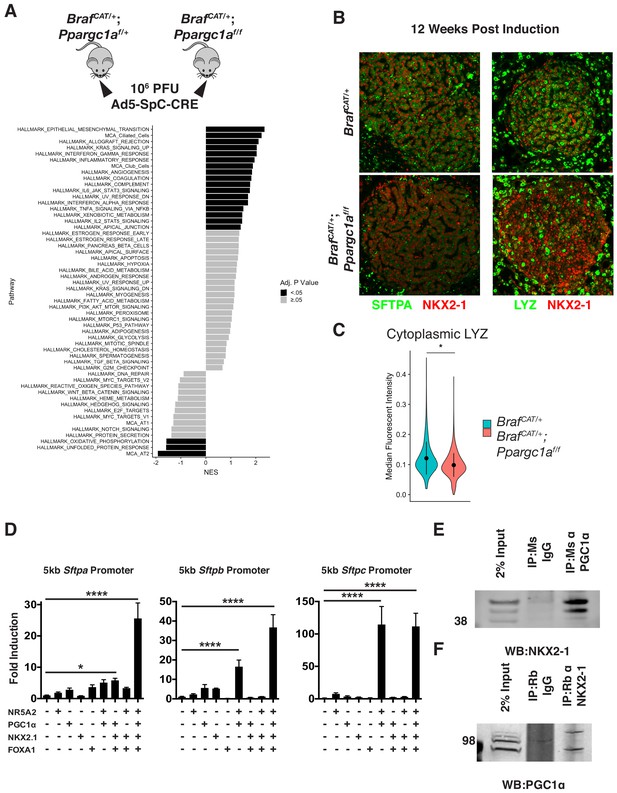
PGC1α is required for maintenance of lung identity in BRAFV600E driven tumors.
(A) Tumors were induced in cohorts of BrafCAT/+ and BrafCAT/+;Ppargc1af/f mice via intranasal instillation of 106 PFU Ad5-SpC-CRE and harvested from each genotype 12 weeks post tumor induction via tissue dissociation and FACS. GSEA analyses of hallmark pathways and lung identity gene sets. Black bars indicate adjP <.05, gray bars indicate Benjamini-Hochberg corrected enrichment statistic adjP ≥. 05. (B) Immunostaining confirms decreased expression of the AT2 markers SFTPA and LYZ in BRAFV600E/PGC1αNULL tumors. (C) Quantitation demonstrating a significant decrease of LYZ immunoreactivity in BRAFV600E/PGC1αNULL tumors. Wilcoxon rank sum p val. = 0.0288. (D) Luciferase assays in HEK293T cells demonstrating the cooperation of NKX2-1, FOXA1, PGC1α, and NR5A2 in transactivation of surfactant promoters. All three promoters showed significant induction by ordinary one-way ANOVA (p<0.0001). Comparison of individual groups to mock transfected controls by Dunnett’s test for multiple comparisons: (*) p=0.0189, (****) p<0.0001. (E) Co-Immunoprecipitation of NKX2-1 by immunoprecipitation with a mouse monoclonal antibody recognizing PGC1α but not with IgG. (F) Co-Immunoprecipitation of PGC1α by immunoprecipitation with a mouse monoclonal antibody recognizing NKX2-1 but not with mouse IgG.
-
Figure 7—source code 1
R script to perform gene set enrichment analysis on Figure 7—source data 1, as well as plot these results.
- https://doi.org/10.7554/eLife.43668.038
-
Figure 7—source code 2
R script to perform statistics on Figure 7—source data 2, as well as plot these results eLife’s transparent reporting form.
- https://doi.org/10.7554/eLife.43668.039
-
Figure 7—source data 1
DEseq2 output of differentially expressed genes comparing BRAFV600E/PGC1αNULL and BRAFV600E/PGC1αHET driven tumors.
- https://doi.org/10.7554/eLife.43668.040
-
Figure 7—source data 2
Cellprofiler output quantifying immunofluorescence of LYZ in BRAFV600E/PGC1αNULL and BRAFV600E/PGC1αWT driven tumors.
- https://doi.org/10.7554/eLife.43668.041
-
Figure 7—source data 3
Data from luciferase assays looking for transactivation of Sftpa, Sftpb, and Sftpc promoters.
- https://doi.org/10.7554/eLife.43668.042
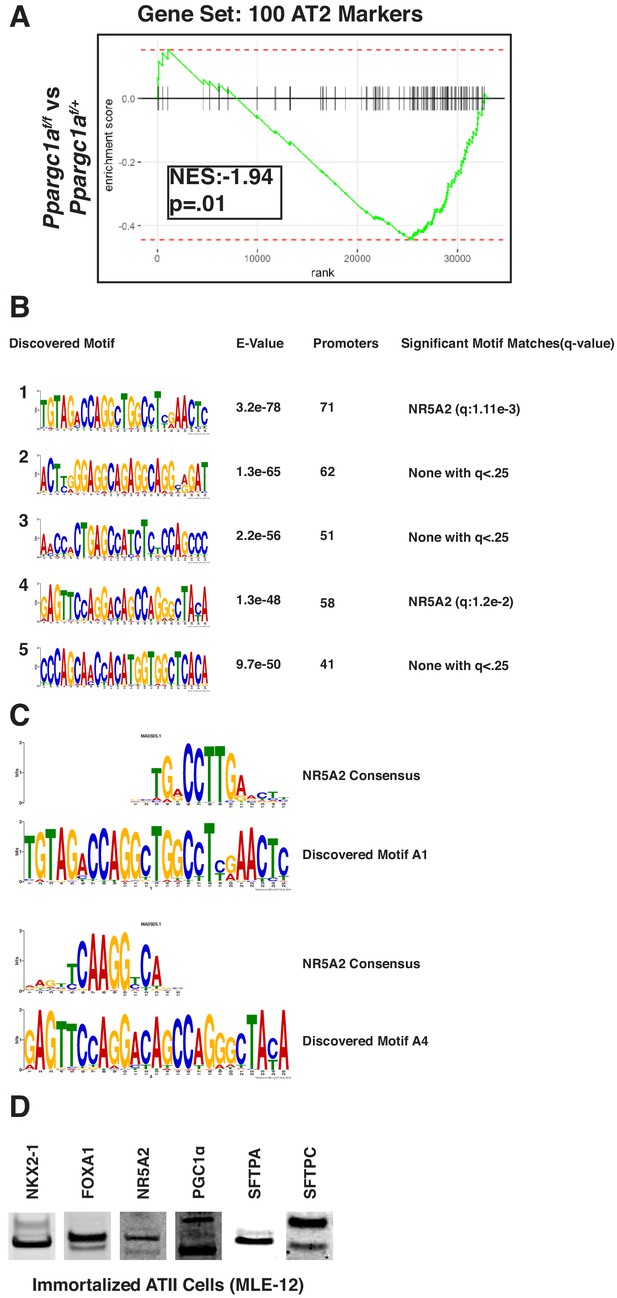
PGC1α is required for maintenance of lung identity in BRAFV600E driven tumors.
(A) GSEA mountain plot comparing BRAFV600E/PGC1αNULL driven tumors to BRAFV600E/PGC1αHET driven tumors indicates that reduced PGC1α expression results in a widespread decrease in markers of AT2 pneumocyte identity, P is enrichment statistic. (B) Western blot showing expression of NKX2-1, FOXA1, NR5A2, PGC1α, SFTPA, and SFTPC in the immortalized AT2 cell line, MLE-12.
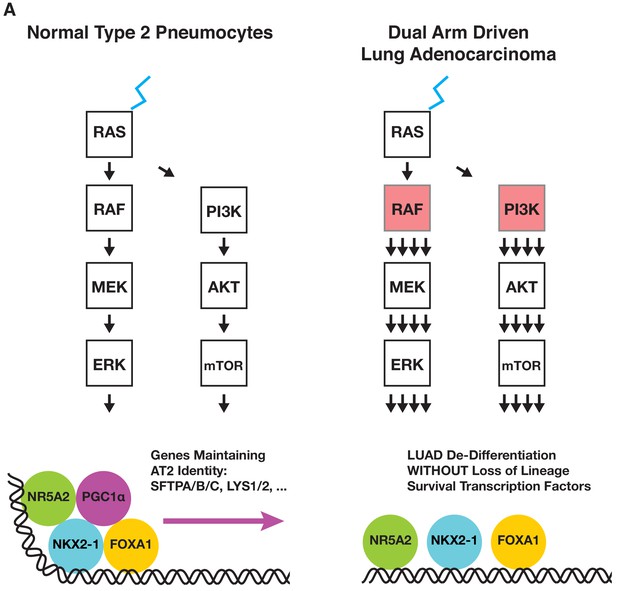
Proposed mechanism of PI3’-kinase-α promoting de-differentiation of lung tumors initiated by the BRAFV600E oncoprotein kinase.
Model in which dual arm mutational activation of growth factor signaling results in the loss of AT2 identity by repressing expression of PGC1α but without affecting expression of the lineage survival transcription factors NKX2-1 or FOXA1/FOXA2.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Mus musculus) | Braf | Ensembl: ENSMUSG00000002413 | ||
| Gene (Mus musculus) | Pik3ca | Ensembl: ENSMUSG00000027665 | ||
| Genetic reagent (Mus musculus) | BrafCA | McMahon lab stock | MGI:Braftm1Mmcm JAX:017837 RRID:IMSR_JAX:017837 | |
| Genetic reagent (Mus musculus) | Pik3caH1047R | Wayne Phillips | MGI:Pik3catm1.1Waph RRID:MGI:5427584 | |
| Genetic reagent (Mus musculus) | BrafCAT | This paper | N/A | New genetically engineered mouse reported in this paper |
| Genetic reagent (Mus musculus) | KrasLSL | The Jackson Lab | MGI:Krastm4Tyj JAX:008179 RRID:IMSR_JAX:008179 | |
| Cell line (Mus musculus) | MLE-12 | ATCC | CRL-2110 RRID:CVCL_3751 | |
| Cell line (Homo sapiens) | 293T | Lab Stock | RRID:CVCL_0063 | |
| Antibody | Mouse monoclonal PGC1α | Millipore | Cat# 1F3.9 RRID:AB_10806332 | Co-IP, 10 ug WB, 1:500 |
| Antibody | Rabbit polyclonal PGC1α | Millipore | Cat# AB3242 RRID:AB_2268462 | IHC, 1:50 WB, 1:500 |
| Antibody | Rabbit monoclonal NKX2-1 | Abcam | Cat# AB76013 RRID:AB_1310784 | Co-IP, 10 ug IHC, 1:250 WB, 1:1000 |
| Antibody | Rabbit polyclonal Phospho-S327-NKX2-1 | CST | Cat# 13608 RRID:AB_2798273 | IHC, 1:250 |
| Antibody | Rabbit monoclonal FOXA1 | Abcam | Cat# AB23738 RRID:AB_2104842 | IHC, 1:250 |
| Antibody | Rabbit monoclonal FOXA1 | CST | Cat# 58613 RRID:AB_2799548 | WB, 1:2500 |
| Antibody | Rabbit monoclonal FOXA2 | CST | Cat# D56D6 RRID:AB_10891055 | IHC, 1:250 WB, 1:1000 |
| Antibody | Rabbit monoclonal SFTPA1 + 2 | Abcam | Cat# AB206299 RRID:AB_2810211 | IHC, 1:250 WB, 1:1000 |
| Antibody | Goat polyclonal SFTPA | Santa Cruz | Cat# SC-7699 RRID:AB_661292 | IHC, 1:100 WB, 1:1000 |
| Antibody | Goat polyclonal SFTPA | Santa Cruz | Cat# SC-7700 RRID:AB_661293 | IHC, 1:100 WB, 1:1000 |
| Antibody | Rabbit polyclonal SFTPA | Santa Cruz | Cat# SC-13977 RRID:AB_661294 | WB, 1:1000 |
| Antibody | Rabbit monoclonal Lysozyme | Abcam | Cat# AB108508 RRID:AB_10861277 | IHC, 1:250 WB, 1:1000 |
| Antibody | Goat polyclonal SFTPC | Santa Cruz | Cat# SC-7705 RRID:AB_2185505 | IHC, 1:250 WB, 1:1000 |
| Antibody | Rabbit polyclonal SFTPC | Santa Cruz | Cat# SC-13979 RRID:AB_2185502 | WB, 1:1000 |
| Antibody | Goat polyclonal CCA | Santa Cruz | Cat# SC-9772 RRID:AB_2238819 | IHC, 1:1000 |
| Antibody | Goat polyclonal AQP5 | Santa Cruz | Cat# SC-9890 RRID:AB_2059877 | IHC, 1:50 |
| Antibody | Rabbit polyclonal NR5A2 | Abcam | Cat# AB153944 RRID:AB_2810212 | WB, 1:1000 |
| Recombinant DNA reagent | M50 Super 8x TOPFlash | Addgene | Plasmid #12456 RRID:Addgene_12456 | Vector used to build SFTP Luciferase reporters |
| Recombinant DNA reagent | pCDNA3-mLRH1 | Holly Ingraham | Ms NR5A2 expression vector | |
| Recombinant DNA reagent | pDTA-TK | Addgene | Plasmid #22677 RRID:Addgene_22677 | Empty targeting vector for mouse production |
| Recombinant DNA reagent | MSCV-NKX2.1 | Addgene | Plasmid #31271 RRID:Addgene_31271 | NKX2.1 expression vector |
| Recombinant DNA reagent | PCDH-FOXA1 | Eric Snyder | FOXA1 expression vector | |
| Recombinant DNA reagent | FUW-mKate | This paper | Fluorescent protein mKate expression vector used for assaying transfection efficiency | |
| Recombinant DNA reagent | GFP-PGC1 | Addgene | Plasmid #4 RRID:Addgene_4 | PGC1α expression vector |
| Recombinant DNA reagent | pWZL-Hygro | Addgene | Plasmid #18750 RRID:Addgene_18750 | Empty vector used to normalize amount of DNA transfected |
| Recombinant DNA reagent | SFTPA-LUC | This paper | 5 kb SFTPA promoter in luciferase reporter | |
| Recombinant DNA reagent | SFTPB-LUC | This paper | 5 kb SFTPB promoter in luciferase reporter | |
| Recombinant DNA reagent | SFTPC-LUC | This paper | 5 kb SFTPC promoter in luciferase reporter | |
| Peptide, recombinant protein | TAT-CRE | Excellgen | Cat# Eg-1001 | Cell permeant CRE protein |
| Commercial assay or kit | Dynabeads protein G IP kit. | Thermo | Cat# 10007D | |
| Commercial assay or kit | Pierce firefly luc one step glow assay kit | Thermo | Cat# 16196 | |
| Software, algorithm | MEME | http://meme-suite.org | Multiple Em Motif Elucidation | |
| Software, algorithm | FIMO | http://meme-suite.org | Find Individual MOtifs | |
| Software, algorithm | GSEA | http://gsea.org | Gene Set Enrichment Analysis | |
| Software, algorithm | R | http://rstudio.com | R programming language | |
| Software, algorithm | Custom R scripts | github.com/jevanveen /vanveen-elife | Referenced scripts hosted at GitHub | |
| Other | Antigen Retrieval | Syrbu and Cohen, 2011 | Antigen Retrieval Method for Immunostaining of Paraffin Sections | Method greatly aiding in immunostaining |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.43668.043






