Nitric oxide radicals are emitted by wasp eggs to kill mold fungi
Figures
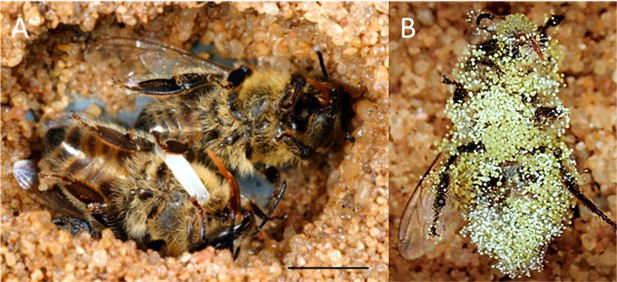
Paralyzed honeybees under different conditions.
(A) Brood cell of the European beewolf with two bees, one carrying an egg, in an observation cage. (B) Honeybee paralyzed by a beewolf female but immediately removed and kept in an artificial brood cell, heavily overgrown by mold fungi that have already developed conidia. Scale bar = 5 mm.
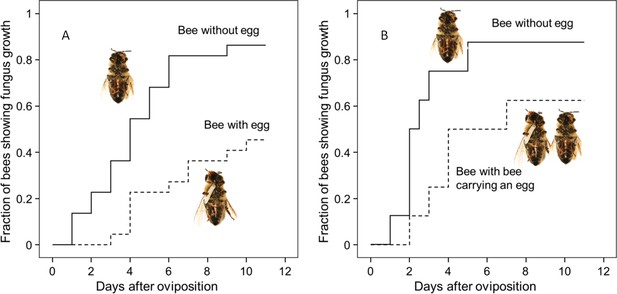
Onset of fungal growth on paralyzed honeybees taken from Philanthus triangulum nests and kept in artificial brood cells.
The fraction of bees showing first signs of fungal growth is shown as a function of days since oviposition. (A) Honeybees that either carried an egg (dashed line) or not (solid line) (N = 22 each, hazard ratio = 0.29, 95% confidence interval: 0.13–0.64). (B) Honeybees that were either kept alone (solid line) or shared a brood cell with a bee carrying an egg (dashed line) (N = 16 each, hazard ratio = 0.39, 95% confidence interval: 0.17–0.9).
-
Figure 2—source data 1
Effect of egg on fungus growth.
- https://doi.org/10.7554/eLife.43718.005
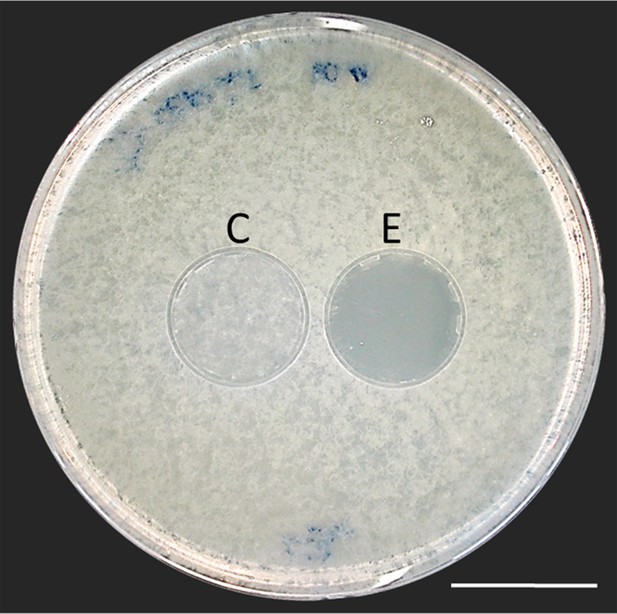
Bioassay demonstrating the inhibitory effect of a beewolf egg against Aspergillus flavus.
Two areas on the agar were covered by caps of a volume similar to natural beewolf brood cells. One cap, the control (C), was empty, while the experimental cap (E) contained a fresh beewolf egg attached to the ceiling of the cap. The caps were removed and the picture was taken after 24 hr of incubation at 25°C. The control area (C) shows dense whitish fungal hyphae similar to the surroundings. However, the area that was exposed to the volatiles from a beewolf egg (E) shows bare agar, indicating that the growth of this aggressive fungus was entirely inhibited. Scale bar = 2.5 cm.
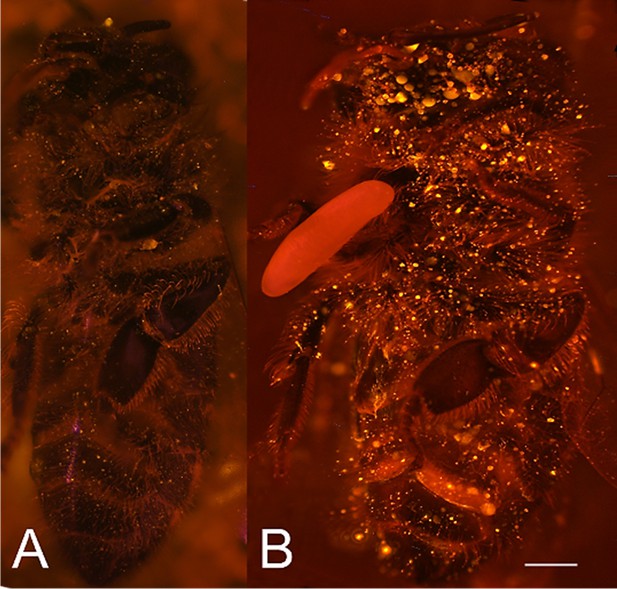
Visualization of NO⋅ emission by beewolf eggs using fluorescence imaging.
(A) Honeybee from a brood cell without an egg and (B) honeybee with egg. Both bees were sprayed with a solution of the NO⋅ specific fluorescence probe DAR4M-AM. Only the droplets on the bee with the egg (B) show a bright yellow and orange fluorescence indicating the presence of NO⋅. Images are composites of multiple pictures of the x/y plane and z-axis. Scale bar = 1 mm.
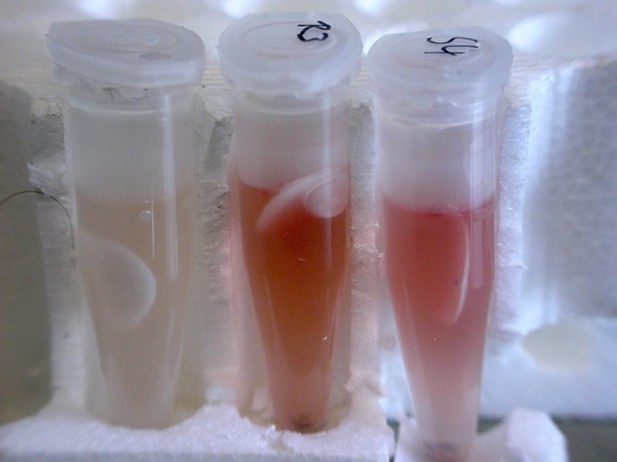
Results of the Griess assay with beewolf eggs.
The left reaction vial is a control without beewolf egg, the other two vials containd eggs that were placed in the lid within 1 hr of oviposition. Vials were incubated at 25°C for 24 hr.
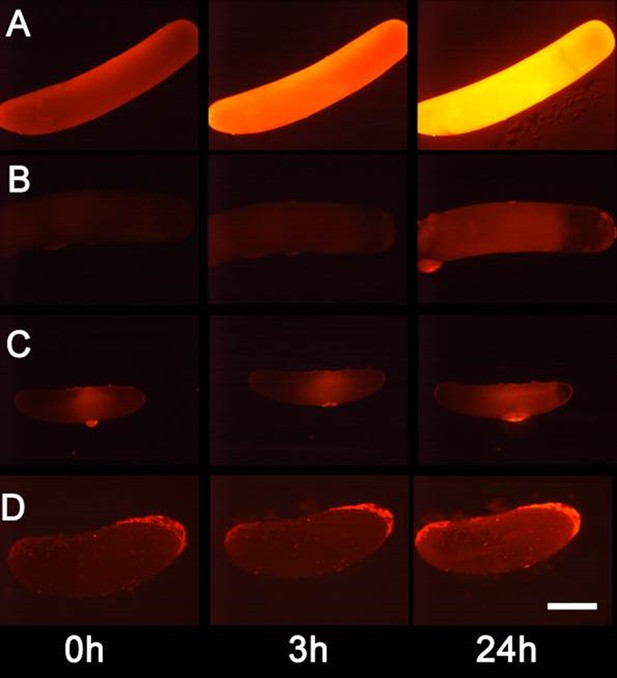
Detection of nitric oxide (NO⋅) in beewolf eggs.
Newly laid eggs of beewolves, Philanthus triangulum, of the cockroach wasp Ampulex compressa and of the Red Mason bee, Osmia bicornis were injected with the NO⋅ sensitive fluorescence probe DAR4M-AM. Control beewolf eggs were injected with phosphate buffer. Images were obtained by fluorescence microscopy 0, 3 and 24 hr after injection. Row (A) DAR4M-AM injected beewolf egg showing strong increase in fluorescence; (B) Buffer-injected control beewolf egg showing the level of autofluorescence; (C) DAR4M-AM injected egg of A. compressa; (D) DAR4M-AM injected egg of O. bicornis. Scale bar: 1 mm.
-
Figure 5—source data 1
Eggs injected with DAR4M-AM.
- https://doi.org/10.7554/eLife.43718.011
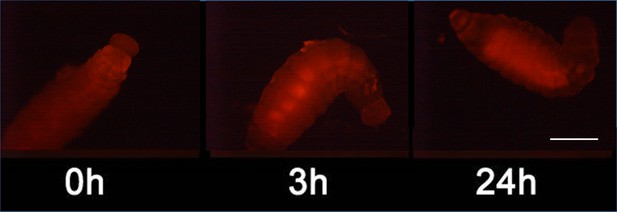
Detection of nitric oxide (NO⋅) in beewolf larva.
Newly hatched beewolf larva, Philanthus triangulum, was injected with the NO⋅ sensitive fluorescence probe DAR4M-AM. Images were obtained by fluorescence microscopy 0, 3 and 24 hr after injection. Scale bar: 1 mm.
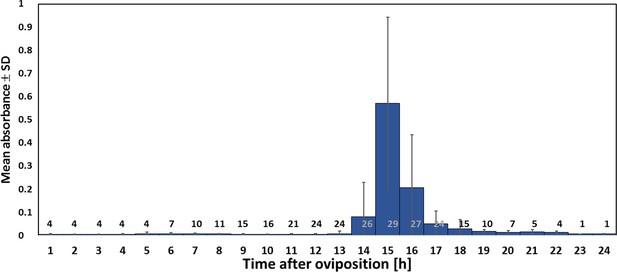
Timing of NO⋅ emission from beewolf eggs (kept at 28°C).
The photometrically determined absorbance at 590 nm (mean ± SD) is shown as a function of time after oviposition for iodide-starch solutions successively exposed to beewolf eggs for one hour. Sample size (number of eggs measured) at each one hour interval is indicated above the x-axis.
-
Figure 6—source data 1
Timing of NO emssion.
- https://doi.org/10.7554/eLife.43718.014
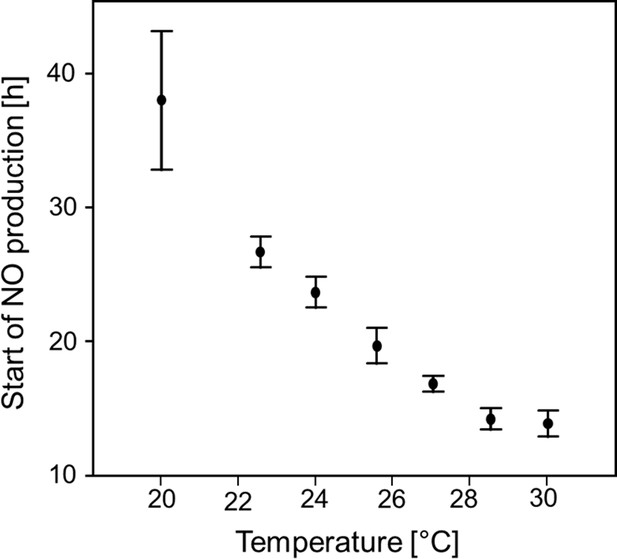
Start of NO⋅ emission (h after oviposition) as a function of temperature.
Beewolf eggs were kept at different temperatures and the onset of NO⋅ release was assessed using the color change of an iodide starch solution as monitored by a digital camera at 30 min intervals. Symbols are means ± SD (Quadratic regression: R2 = 0.98, N = 33, p<0.001; Q10 = 2.74). Source data file: Figure 6—figure supplement 1—source data 1.
-
Figure 6—figure supplement 1—source data 1
Start of NO emission.
- https://doi.org/10.7554/eLife.43718.015
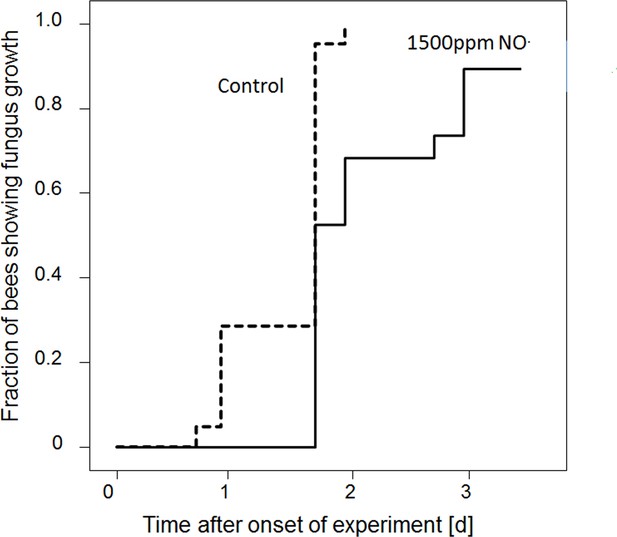
Onset of fungal growth (time after onset of experiment) on honeybees that were not embalmed in artificial brood cells.
Brood cells were either injected with synthetic NO⋅ to a concentration of 1500ppm (solid line) or were injected with nitrogen (dashed line) (N = 20 each, hazard ratio = 0.41, 95% confidence interval: 0.198–0.845).
-
Figure 7—source data 1
Effect of synthetic nitric oxide on fungus growth.
- https://doi.org/10.7554/eLife.43718.017
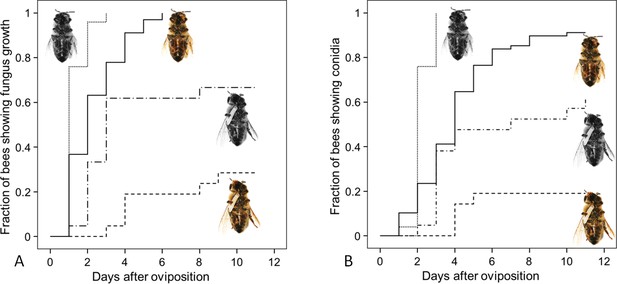
Fungus growth on honeybees of four different treament groups.
Timing of occurrence of (A) fungal hyphae and (B) conidia on paralyzed honeybees that were (1) not embalmed by beewolf females and did not carry an egg (n = 25, colorless bee, point line), (2) embalmed but did not carry an egg (n = 68, colored bee, solid line), (3) not embalmed but carried an egg (n = 21, colorless bee with egg, dash-point line) or (4) embalmed and carried an egg (n = 21, colored bee with egg, dashed line). See Appendix 1—table 2 for hazard ratios.
-
Figure 8—source data 1
Combined effect of embalming and fumigation.
- https://doi.org/10.7554/eLife.43718.019
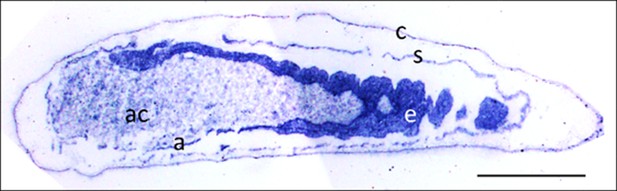
Micrograph of a longitudinal section of a beewolf egg fixed 15–16 hr after oviposition showing fixation insensitive NADPH-diaphorase activity.
Strong blue staining in the embryonic tissue indicates the presence of reduced nitroblue tetrazolium demonstrating NOS activity (c = cuticle, s = serosa, e = embryo, a = amnion, ac = amnion cavity, scale bar = 1 mm, image composed from two separate photos of the left and right parts of the egg.).
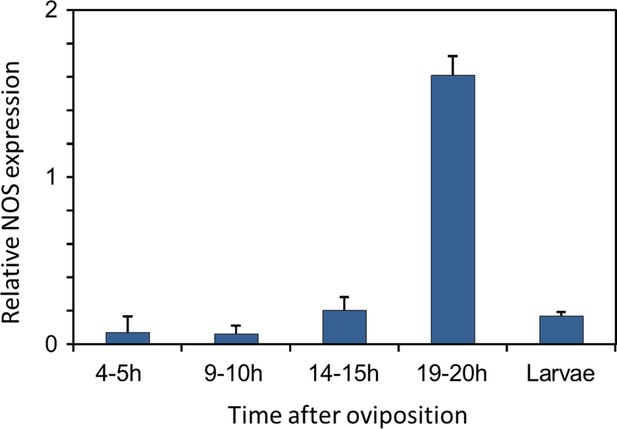
Gene expression of NOS relative to ß-actin in beewolf eggs at different times after oviposition and in freshly hatched larvae.
Two trials were conducted, each with 25 pooled eggs or larvae per time interval. Mean ratios of NOS-mRNA to ß-Actin-mRNA are shown (with standard deviations), as determined by Q-RT-PCR.
-
Figure 10—source data 1
NOS gene expression.
- https://doi.org/10.7554/eLife.43718.022
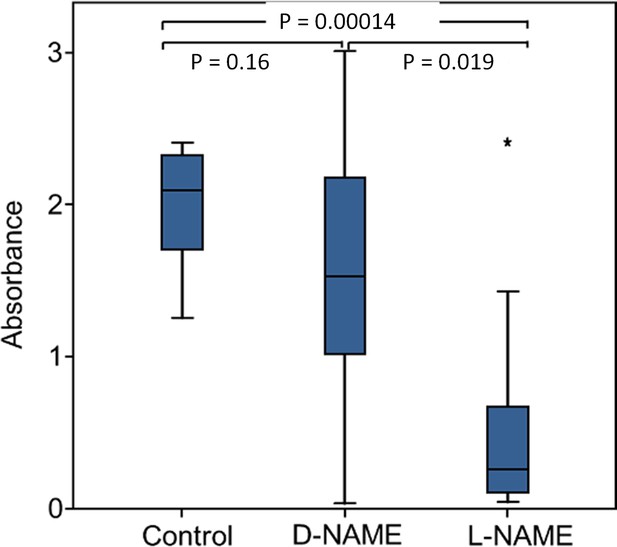
Effect of NOS inhibition on NO⋅ production.
Amount of NO⋅ and/or NO2⋅ emanating from non-injected beewolf eggs (control; N = 14) and those injected with D-NAME (a non-inhibiting enantiomer of L-NAME, N = 9) or L-NAME (a NOS inhibiting L-arginine analog, N = 14). The photometrically determined absorbance at 590 nm is shown for iodide-starch solutions that were exposed for 24 hr to the headspace of eggs of the indicated treatment group (shown are median, quartiles and range, * indicates an outlier, included in the analysis). P-values are for Holm-corrected Mann-Whitney U-tests.
-
Figure 11—source data 1
NOS inhibition.
- https://doi.org/10.7554/eLife.43718.026
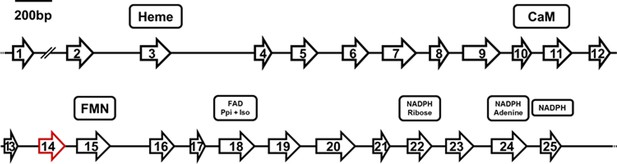
Structure of the Pt-NOS gene indicating position and length of exons.
Exon 14 (red) is missing in the NOS mRNA in beewolf eggs compared to adults. Presumed cofactor-binding domains as deduced from homologous sequences of the NOS of Anopheles stephensi (Luckhart et al., 1998; Luckhart and Li, 2001) are indicated for heme, calmodulin (CaM), FMN, FAD pyrophosphate (FAD PPi) and FAD isoalloxazine (FAD Iso), NADPH ribose, NADPH adenine, and NADPH.
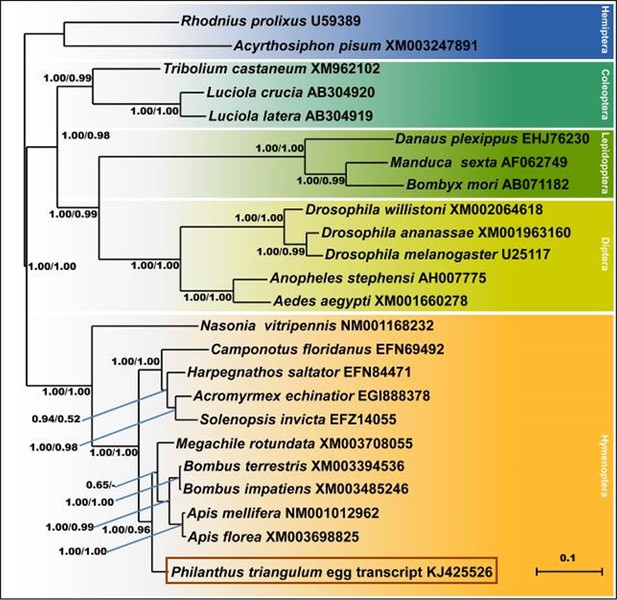
Consensus tree obtained from Bayesian analysis of NOS amino acid sequences from five orders of insects (distinguished by different colors), including the NOS sequences of P. triangulum eggs (lowermost entry).
Values at the nodes represent Bayesian posterior probabilities and local support values (FastTree analysis), respectively. Scale bar represents 0.1 changes per site.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Biological sample | European beewolf, Philanthus triangulum | Field caught or laboratory reared F1 of field caught females | ||
| Biological sample | Emerald cockroach wasp, Ampulex compressa | Laboratory reared | ||
| Biological sample | Red mason bee, Osmia bicornis | Field caught | ||
| Biological sample | Aspergillus flavus | Strain I: Isolated from beewolf brood cells, Strain II: Department of Hygiene and Microbiology of the University Hospital, Würzburg, Germany | na | |
| Biological sample | Penicillium roquefortii | Department of Hygiene and Microbiology of the University Hospital, Würzburg, Germany | na | |
| Biological sample | Candida albicans | Department of Hygiene and Microbiology of the University Hospital, Würzburg, Germany | na | |
| Biological sample | Trichophyton rubrum | Department of Hygiene and Microbiology of the University Hospital, Würzburg, Germany | na | |
| Sequence-based reagent | Adapter + PolyT | 3'RACE, Molecular cloning protocol | See Supplementary file 1 | |
| Sequence-based reagent | Adapter | 3'RACE, Molecular cloning protocol | See Supplementary file 1 | |
| Sequence-based reagent | polyT | Reverse transcription protocol | See Supplementary file 1 | |
| Sequence-based reagent | NOS_qPCR_F2 | P. triangulum, this paper | See Supplementary file 1 | |
| Sequence-based reagent | NOS_qPCR_R2 | P. triangulum, this paper | See Supplementary file 1 | |
| Sequence-based reagent | Actin_qPCR_F1 | Apis mellifera, Gryllus bimaculatus, P. triangulum, this paper | See Supplementary file 1 | |
| Sequence- based reagent | Actin_qPCR_R1 | A. mellifera, G. bimaculatus, P. triangulum, this paper | See Supplementary file 1 | |
| Sequence- based reagent | NOS860fwd2 | A. mellifera, D. melanogaster, Anopheles stephensi, Rhodnius prolixus, Manduca sexta, this paper | See Supplementary file 1 | |
| Sequence- based reagent | NOS1571rev1 | A. mellifera, D. melanogaster, A. stephensi, R. prolixus, M. sexta, this paper | See Supplementary file 1 | |
| Sequence-based reagent | NOS_seq_F1_deg | A. mellifera, Nasonia vitripennis, this paper | See Supplementary file 1 | |
| Sequence-based reagent | NOS_seq_R1_deg | A. mellifera, N. vitripennis, this paper | See Supplementary file 1 | |
| Sequence-based reagent | NOS_seq_5-F1 | P. triangulum, this paper | See Supplementary file 1 | |
| Sequence- based reagent | NOS_seq_5-R1 | P. triangulum, this paper | See Supplementary file 1 | |
| Sequence-based reagent | NOS_seq_5-F2 | P. triangulum, this paper | See Supplementary file 1 | |
| Sequence- based reagent | NOS_seq_5-R2 | P. triangulum, this paper | See Supplementary file 1 | |
| Sequence-based reagent | NOS_seq_5-F3 | P. triangulum, this paper | See Supplementary file 1 | |
| Sequence-based reagent | NOS_seq_5-F6 | P. triangulum, this paper | See Supplementary file 1 | |
| Sequence-based reagent | NOS_seq_3-F1 | P. triangulum, this paper | See Supplementary file 1 | |
| Sequence-based reagent | NOS_seq_3-R1 | P. triangulum, this paper | See Supplementary file 1 | |
| Sequence-based reagent | NOS_seq_3-F2 | P. triangulum, this paper | See Supplementary file 1 | |
| Sequence- based reagent | NOS_seq_3-R2 | P. triangulum, this paper | See Supplementary file 1 | |
| Sequence-based reagent | NOS_seq_3-F3 | P. triangulum, this paper | See Supplementary file 1 | |
| Sequence- based reagent | NOS_seq_3-F6 | P. triangulum, this paper | See Supplementary file 1 | |
| Sequence-based reagent | NOS_RT_R1 | P. triangulum, this paper | See Supplementary file 1 | |
| Commercial assay or kit | Griess assay. Merck Spectroquant | Merck, Darmstadt, Germany | 114776 | |
| Commercial assay or kit | peqGOLD total RNA Kit | peqLab, Erlangen, Germany | 732–2867 | |
| Commercial assay or kit | GeneRacer Kit | Invitrogen, Carlsbad, CA, USA | L1502-01 | |
| Commercial assay or kit | BioScript One-Step RT-PCR-Kit | Bioline, London, UK | BIO-65033 | |
| Commercial assay or kit | peqGOLD Taq-DNA-Polymerase | peqLab, Erlangen, Germany | 01–1030 | |
| Commercial assay or kit | SensiMixPlus SYBR Mit | Quantace/Bioline, London, UK | QT615-05 | |
| Commercial assay or kit | Epicentre MasterPure Complete DNA and RNA purification Kit | Epicentre, now Lucigen, Middleton, WI, USA | MC85200 | |
| Commercial assay or kit | innuPREP RNA Mini Kit | Analytik Jena, Jena, Germany | 845-KS-2040050 | |
| Commercial assay or kit | PeqGOLD Mid- Range PCR System | peqLab, Erlangen, Germany | PEQL02-3020_P | |
| Chemical compound, drug | DNase I | Fermentas, Lithuania Now Thermo Fisher Scientific, Germany | EN0525 | |
| Chemical compound, drug | Oligo-dT primer | Fermentas, Lithuania Now Thermo Fisher Scientific, Germany | na | |
| Chemical compound, drug | L-NAME Hydrochloride | Axxora Deutschland, Lörrach, Germany | ALX-105–004 M250 | |
| Chemical compound, drug | D-NAME Hydrochloride | Axxora Deutschland, Lörrach, Germany | ALX-105–003 G005 | |
| Chemical compound, drug | DAR-4M AM | Axxora Deutschland, Lörrach, Germany | ALX-620–069 M001 | |
| Chemical compound, drug | 4-Nitro-m-Xylol | Merck, Darmstadt, Germany | 8415470025 | |
| Chemical compound, drug | NADPH- Tetranatriumsalz | Carl-Roth, Karlsruhe, Germany | AE14.1 | |
| Tools | Eppendorf Microinjector with Femtotips II | Eppendorf, Hamburg, Germany | 930000043 | |
| Tools | Axiophot II Fluorescence microscope | Zeiss, Jena, Germany | ||
| Tools | Nikon DS-2 Mv | Nikon, Tokyo, Japan | ||
| Tools | Uvikon 860 spektrophotometer | Kontron, Augsburg, Germany | ||
| Tools | Cryostat microtome CM3000 | Leica, Wetzlar, Germany | ||
| Tools | Eppendorf Realplex Cycler | Eppendorf, Hamburg, Germany | ||
| Tools | NanoDrop TM1000 | peqLab, Erlangen, Germany | RRID: SCR_016517 | |
| Tools | Implen Nanophotometer Classic | Implen, Munich, Germany | ||
| Tools | Biometra T Gradient Thermocycler | Analytik Jena, Jena, Germany | ||
| Software | BioEdit | http://www.mbio.ncsu.edu/BioEdit/bioedit.html | RRID: SCR_007361 | |
| Software | Geneious | Biomatters, New Zealand | ||
| Software | SPSS | IBM, Armonk, NY, USA | RRID: SCR_002865 | |
| Software | FastTree | http://www.microbesonline.org/fasttree/ | RRID:SCR_015501 | |
| Software | MrBayes | http://mrbayes.sourceforge.net/ | RRID: SCR_012067 | |
| Software | Combine-ZP | www.hadleyweb.pwp.blueyonder.co.uk | ||
| Software | Photoshop Elements 5 | PSE5, Adobe Systems Inc, San José, CA, USA | ||
| Software | CLC genomics workbench | Qiagen, Hilden, Germany | RRID: SCR_011853 | |
| Database | NCBI | http://www.ncbi.nlm.nih.gov | RRID: SCR_006472 | |
| Database | Primer3 | http://primer3.ut.ee | RRID: SCR_003139 | |
| Dervice | Sanger Sequencing | Seqlab, Göttingen, Germany | ||
| Service | Transcriptome Sequencing on Illumina HiSeq TM2000 | Fasteris, Geneva, Switzerland |
Scores of fungus growth (0 = no fungus, 1 = very little fungus, 2 = little fungus, 3 = strong fungus) after 24 h of incubation at 25°C on Petri dishes inoculated with Aspergillus flavus conidia.
Scores are given for three areas of the petri dish that were covered with a cap under which either a 'Bee with egg', a 'Bee without egg' was placed as well as a 'Control' with no bee.
| Petri dish | Bee with egg | Bee without egg | Control |
|---|---|---|---|
| 1 | 0 | 3 | 3 |
| 2 | 0 | 2 | 2 |
| 3 | 0 | 2 | 3 |
| 4 | 1 | 2 | 3 |
| 5 | 1 | 3 | 3 |
| 6 | 0 | 3 | 3 |
| 7 | 0 | 3 | 3 |
| 8 | 0 | 3 | 3 |
| 9 | 0 | 3 | 3 |
| 10 | 0 | 3 | 3 |
| 11 | 0 | 3 | 3 |
| 12 | 0 | 3 | 3 |
Hazard ratios (and 95 % confidence intervals) for the comparison of timing of the onset of fungus growth on bees of four treatment groups: bees that carried an egg and were embalmed (+/+), bees that carried no egg but were embalmed (-/+), bees that carried no egg and were not embalmed (-/-) and bees that carried an egg but were not embalmed (+/-).
https://doi.org/10.7554/eLife.43718.031| Comparison | |||
|---|---|---|---|
| Egg/Embalming | Egg/Embalming | Hazard ratio | 95% conf. interval |
| +/+ | -/+ | 0.07 | 0.19 - 0.03 |
| +/+ | -/- | 0.13 | 0.36 - 0.05 |
| +/+ | +/- | 0.65 | 0.90 - 0.47 |
| -/+ | -/- | 0.48 | 0.79 - 0.29 |
| +/- | -/+ | 0.61 | 0.45 - 0.83 |
| +/- | -/- | 0.22 | 0.10 - 0.47 |
Additional files
-
Supplementary file 1
Primers used for sequencing of the Pt-NOS.
- https://doi.org/10.7554/eLife.43718.027
-
Transparent reporting form
- https://doi.org/10.7554/eLife.43718.028





