Dual control of NAD+ synthesis by purine metabolites in yeast
Figures
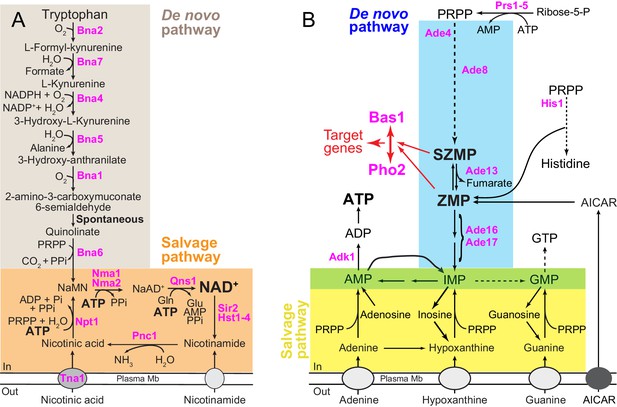
Schematic representation of the yeast pyridine and purine biosynthesis pathways.
(A) NAD+ de novo and salvage pathways in yeast. NaAD+: nicotinic acid adenine dinucleotide; NADP: nicotinamide adenine dinucleotide phosphate; NaMN: nicotinic acid mononucleotide; Pi: inorganic phosphate; PPi: pyrophosphate and PRPP: 5-phosphoribosyl-1-pyrophosphate. (B) Purine de novo and salvage pathways in yeast. AICAR: 5-amino-4-imidazole carboxamide ribonucleoside; IMP: inosine 5’-mono-phosphate; SZMP: succinyl-ZMP and ZMP: 5-amino-4-imidazole carboxamide ribonucleotide 5′-phosphate. Red arrows illustrate the (S)ZMP-dependent Bas1/Pho2 interaction leading to the transcriptional regulation of their target genes. IMP conversion onto either AMP or GMP is common to the de novo and salvage pathways (green box). (A–B) Only the proteins mentioned in the text are shown (in pink).
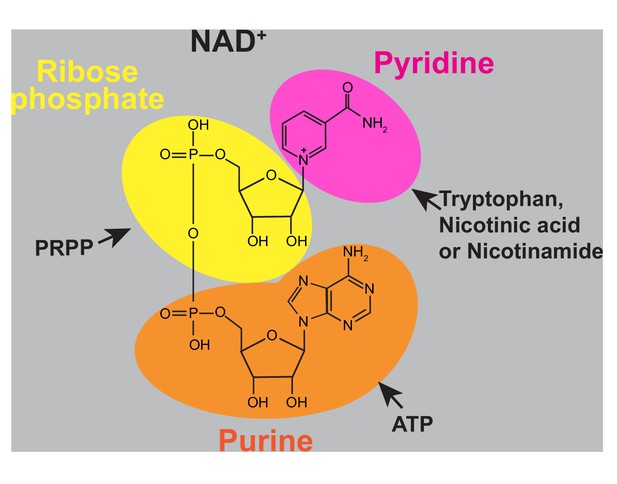
Chemical structure of NAD+.
Colored areas highlight the precursor metabolites in the NAD+ backbone. Pink area: Pyridine ring coming from tryptophan (via the pyridine de novo pathway), nicotinic acid and/or nicotinamide (via the pyridine salvage pathway). Yellow area: Ribose phosphate moiety resulting from 5-phosphoribosyl pyrophosphate (PRPP) incorporation by phosphoribosyl transferases. Orange area: Adenine nucleotide part arising from ATP incorporation at the step catalyzed by the nicotinic acid mononucleotide transferases.
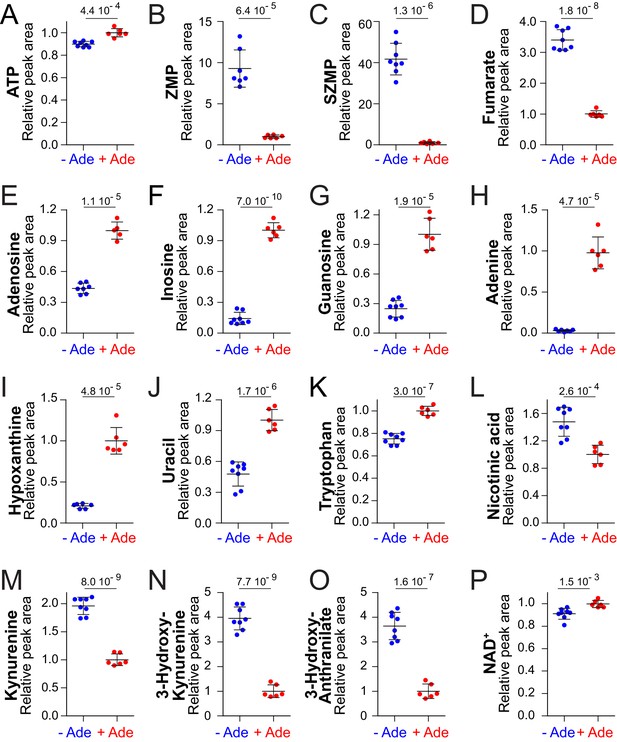
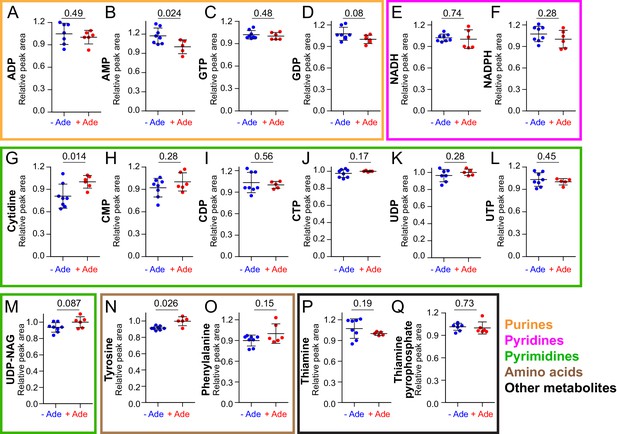
NAD+precursors and biosynthesis intermediates respond to variations in extracellular purine availability.
The prototrophic wild-type strain FY4 was kept in exponential phase for 24 hr in SDcasaWU medium supplemented (red dots) or not (blue dots) with extracellular adenine. Metabolites were then extracted and separated by liquid chromatography. Quantifications were determined on independent metabolite extractions (Biological replicates; N ≥ 5) and standard deviation is presented. For each metabolite, the amount measured in cells grown in the presence of adenine (red dots) was set at 1. Numbers on the top of each panel correspond to the p-value calculated from a Welch’s unpaired t-test.
-
Figure 2—source data 1
Highly significant metabolic variations for the FY4 prototrophic strain ±adenine.
- https://doi.org/10.7554/eLife.43808.013
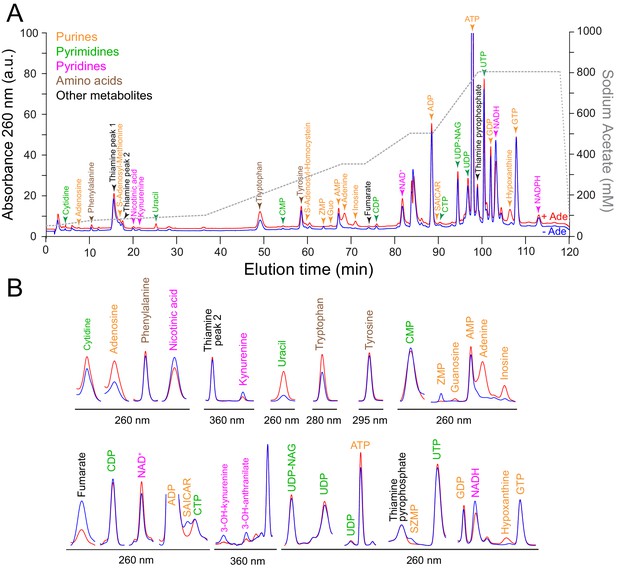
Separation of intracellular metabolites by high-performance ion chromatography.
A prototrophic yeast strain (FY4) was grown in SDcasaWU medium ±Adenine. Metabolite extraction and separation by liquid chromatography were performed as described in the Materials and methods section. (A) Typical entire chromatograms obtained at 260 nm in the presence (red line) or the absence (blue line) of external adenine (Ade). Similar patterns were found in all metabolic extractions presented in Figure 2. Identified peaks are indicated with a color code corresponding to metabolite families. (B) Zooms of absorbance signals obtained from chromatogram at indicated wavelengths. ADP: adenosine 5’-diphosphate, AMP: adenosine 5’-monophosphate, ATP: adenosine 5’-triphosphate, CDP: cytidine 5’-diphosphate, CMP: cytidine 5’-monophosphate, CTP: cytidine 5’-triphosphate, GDP: guanosine 5’-diphosphate, GTP: guanosine 5’-triphosphate, NAD+: nicotinamide adenine dinucleotide, NADH: nicotinamide adenine dinucleotide reduced form, NADPH: phosphorylated form of NADH, SAICAR: succinyl-aminoimidazole carboxamide ribonucleoside, UDP: uridine 5’-diphosphate, UDP-NAG: uridine 5’-diphosphate-N-acetyl glucosamine, UTP: uridine 5’-triphosphate, ZMP: 5-amino-4-imidazole carboxamide ribonucleotide 5′-phosphate and SZMP: succinyl-ZMP.
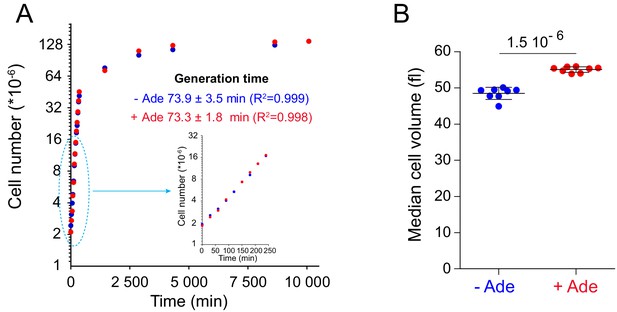
Adenine supplementation affects cell growth but not cell proliferation.
(A) A prototrophic yeast strain (FY4) was exponentially grown for 24 hr in SDcasaWU medium ±Adenine as in Figure 2. Cultures were then diluted at 2 106 cells/ml and cell number was followed by using a Multisizer IV (Beckman Coulter). Generation time was determined with a non-linear growth fit (GraphPad Prism) on cells grown in the presence (red dots) or the absence (blue dots) of adenine (N = 2). (B) Median cell volume was determined on the independent exponentially grown cells (N = 8) used for the metabolic extraction presented in Figure 8A–D. Number on the top corresponds to the p-value calculated from a Welch’s t-test.
-
Figure 2—figure supplement 2—source data 1
Median cell volume for the FY4 prototrophic strain grown in glucose medium ±adenine.
- https://doi.org/10.7554/eLife.43808.007
Quantification of metabolites not or poorly significantly affected by extracellular purine availability.
Results were obtained on the prototrophic wild-type strain FY4 as in Figure 2. Quantifications were determined on independent metabolite extractions (N ≥ 5) and standard deviation is presented. For each metabolite, the amount measured in cells grown in the presence of adenine (red dots) was set at one and p-values indicated on the top of each panel were calculated from a Welch’s t-test.
-
Figure 2—figure supplement 3—source data 1
Non- or lowly-significant metabolic variations in the FY4 prototrophic strain ±adenine.
- https://doi.org/10.7554/eLife.43808.009
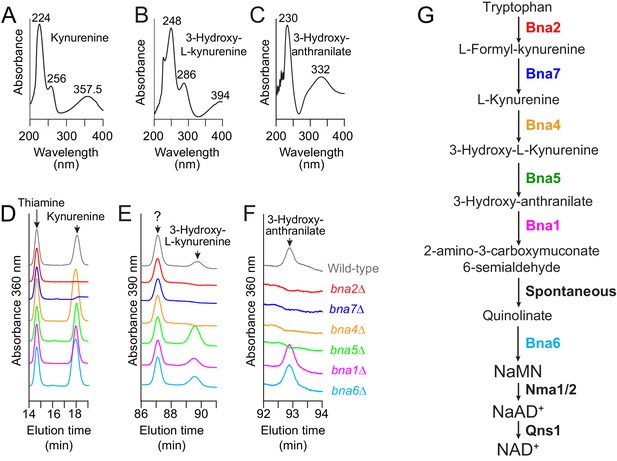
Identification of NAD+ de novo pathway intermediates.
(A–C) Absorption spectra observed for some NAD+-de novo pathway intermediates in the ion chromatography conditions used. Values indicate the wavelength of absorption maxima (in nm). (D–F) Identification of the peaks corresponding to kynurenine, 3-hydroxy-L-kynurenine and 3-hydroxy-anthranilate using bna knock-out mutants. Wild-type (BY4742) and bna-mutant (bna2: Y10838; bna7: Y10904; bna4: Y5822; bna5: Y10903; bna1: Y10901 and bna6: Y5891) strains were exponentially grown in SDcasaWAU medium for 24 hr, harvested by filtration and shifted for 7 hr in SDcasaWAU medium lacking nicotinic acid (see Materials and methods section). Metabolite extraction and separation by liquid chromatography were performed as in Figure 2. Panels correspond to the chromatogram sections containing peaks of interest. (G) Simplified representation of the NAD+ de novo pathway showing the block occasioned by each bna mutant. NaMN: nicotinic acid mononucleotide. NaAD+: nicotinic acid adenine dinucleotide. Of note, L-Formyl-kynurenine and quinolinic acid could not be detected in our experimental setup.
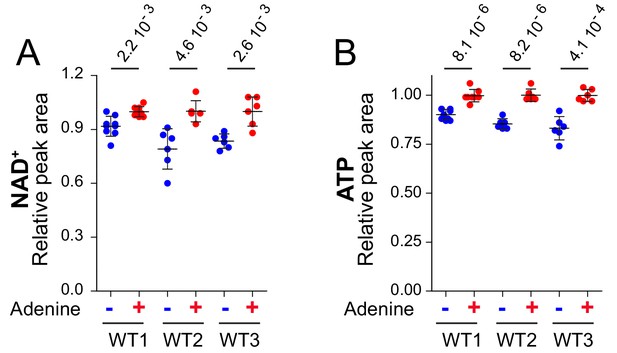
NAD+ and ATP variation in response to adenine availability in wild-type strains.
Wild-type (WT1: FY4; WT2: Y286; WT3: BY4742) strains were maintained in exponential growth phase for 24 hr in SDcasaWU supplemented (red dots) or not (blue dots) with adenine. Metabolite extraction, separation and quantification were done as in Figure 2. Quantifications were determined on independent metabolite extractions (N ≥ 5) and standard deviation is presented. The amount of each metabolite measured in cells grown in the presence of adenine (red dots) was set at one and p-values indicated on the top of each panel were determined by a Welch’s t-test.
-
Figure 2—figure supplement 5—source data 1
Metabolic analyses for different wild-type strains grown in ±adenine.
- https://doi.org/10.7554/eLife.43808.012
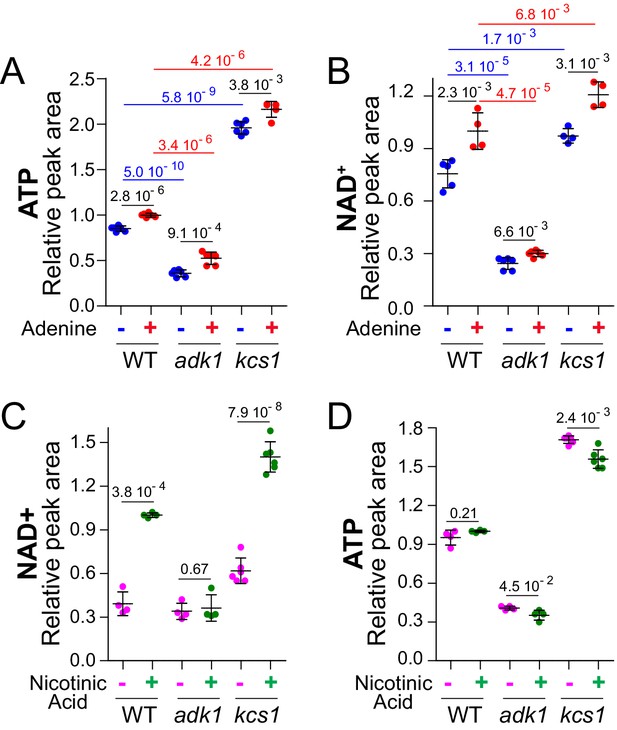
ATP controls NAD+level in yeast.
(A–B) Intracellular NAD+ varies concomitantly with ATP. Wild-type (BY4742), adk1 (Y10991) and kcs1 (Y2337) strains were grown in exponential phase for 24 hr in SDcasaWU medium supplemented (red dots) or not (blue dots) with adenine. The amount of metabolites measured in wild-type cells grown in the presence of adenine (red dots) was set at 1. (C–D) Nicotinic acid (NA) supplementation leads to an increase in NAD+ amount in wild-type cells. Cells (wild-type (WT): BY4742, adk1: Y10991 and kcs1: Y2337) were exponentially grown for 24 hr in SDcasaWU-NA supplemented (green dots) or not (pink dots) with nicotinic acid (400 µg/l). Metabolite amounts measured in wild-type cells grown in the presence of nicotinic acid were set at 1. A-D, Metabolite extraction, separation and quantification were performed as in Figure 2. Quantifications were determined on independent metabolite extractions (N ≥ 4) and standard deviation is presented. Numbers on the top of each panel correspond to the p-values determined by a Welch’s t-test.
-
Figure 3—source data 1
Metabolic analyses for wild-type, adk1 and kcs1 mutant strains grown ±adenine.
- https://doi.org/10.7554/eLife.43808.027
-
Figure 3—source data 2
Metabolic analyses for wild-type, adk1 and kcs1 mutant strains grown ±nicotinic acid.
- https://doi.org/10.7554/eLife.43808.028
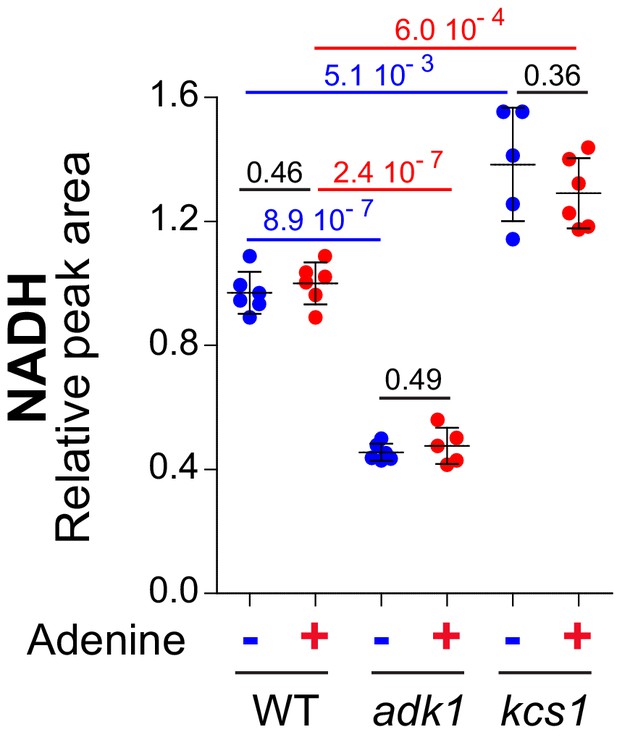
NADH varies concomitantly with ATP in adk1 and kcs1 knock-out mutants.
Results were obtained from the metabolic extraction and separation described in Figure 3A–B (N ≥ 4). The amount measured in cells grown in the presence of adenine (red dots) was set at 1. Error bars correspond to standard deviation and indicated p-values were calculated from a Welch’s t-test.
-
Figure 3—figure supplement 1—source data 1
NADH determination in wild-type and mutant strains grown ±adenine.
- https://doi.org/10.7554/eLife.43808.016
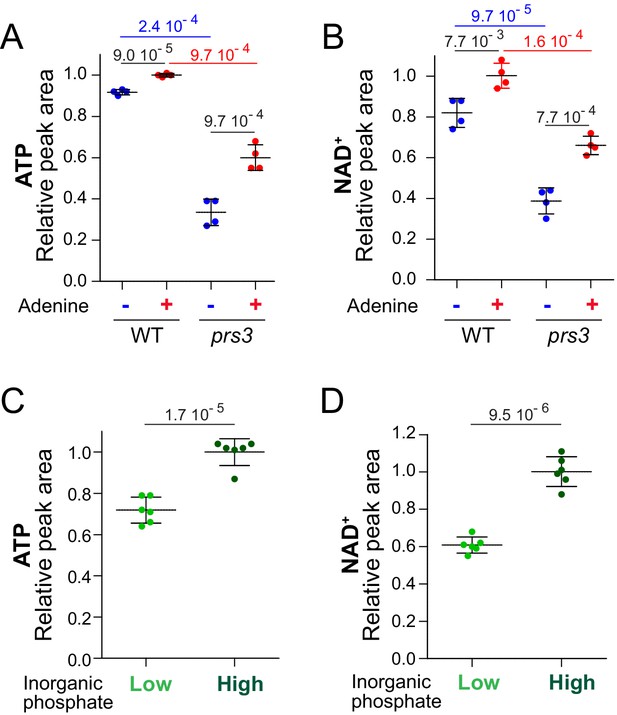
Robust correlation between ATP and NAD+variations in diverse ATP-limiting conditions.
ATP and NAD+ variations were measured in a wild-type strain (BY4742; A–D) and a prs3 (Y4835; A–B) mutant. (A–B) Strains were grown in exponential phase for 24 hr in SDcasaWU medium supplemented (red dots) or not (blue dots) with adenine. The amount measured in cells grown in the presence of adenine (red dots) was set at 1. (C–D) A prototrophic strain (FY4) was exponentially grown for 24 hr in SD medium containing two concentrations of inorganic phosphate (100 µM: Low, light-green dots; 7.3 mM: High, dark-green dots). The amount measured in cells grown in 7.3 mM phosphate (dark green dots, concentration commonly supplied in yeast defined media) was set at 1. (A–D) Independent metabolic extraction (N ≥ 4) and separation were performed as in Figure 2. Error bars correspond to standard deviation and indicated p-values were calculated using a Welch’s t-test.
-
Figure 3—figure supplement 2—source data 1
Metabolic analyses for wild-type and prs3 mutant strains grown ±adenine.
- https://doi.org/10.7554/eLife.43808.018
-
Figure 3—figure supplement 2—source data 2
Metabolic analyses for the FY4 strain grown in low and high phosphate medium.
- https://doi.org/10.7554/eLife.43808.019
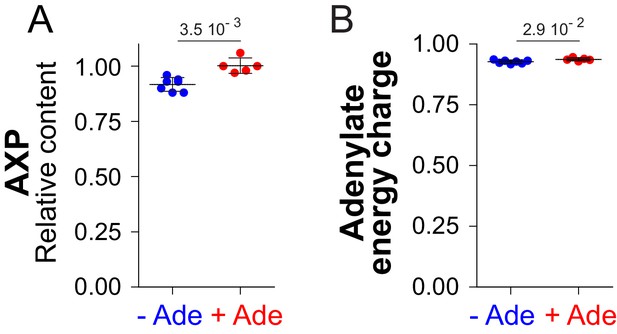
Variation of AXP and adenylate energy charge in response to adenine availability.
AXP and adenylate energy charge were calculated, as described in the Material and methods section, from metabolic analyses presented in Figure 2 and Figure 2—figure supplement 3. For both parameters, values obtained in the presence of adenine (red dots) were set at one and indicated p-values were calculated using a Welch’s t-test (N ≥ 5).
-
Figure 3—figure supplement 3—source data 1
AXP and adenylic energy charge (AEC) determination for the FY4 prototrophic strain ±adenine.
- https://doi.org/10.7554/eLife.43808.021
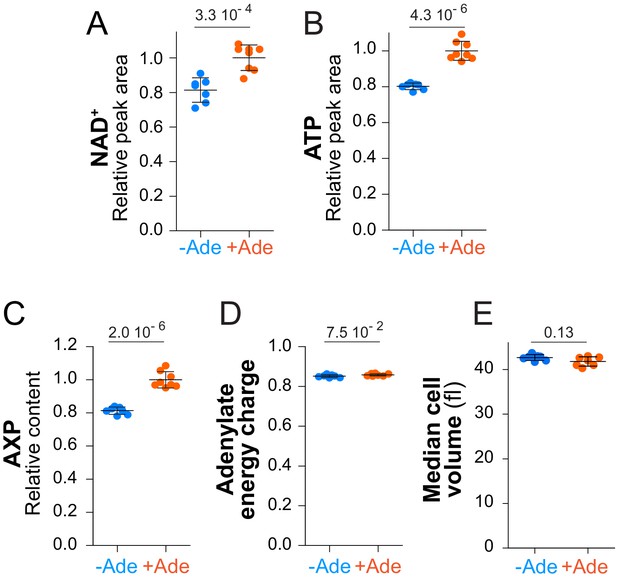
NAD+, ATP, AXP, adenylate energy charge and median cell volume variations in response to adenine availability under respiratory conditions.
(A–D) The prototrophic wild-type strain (FY4) was exponentially grown for 24 hr in SGEcasaWU supplemented (orange dots) or not (blue dots) with adenine. Metabolite extraction, separation and quantification were done as in Figure 2. Quantifications were determined from independent metabolite extractions (N ≥ 7) and standard deviation is presented. The amount of each metabolite measured in cells grown in the presence of adenine (orange dots) was set at 1. Indicated p-values were calculated using a Welch’s t-test. AXP and adenylate energy charge were determined as described in the Materials and methods section. (E) Median cell volume was determined with a Multisizer IV (Beckman coulter) on the independent cell cultures used for the metabolic extraction presented in this figure (N ≥ 7).
-
Figure 3—figure supplement 4—source data 1
Metabolic analyses for the FY4 prototrophic strain grown in glycerol/ethanol medium ±adenine.
- https://doi.org/10.7554/eLife.43808.023
-
Figure 3—figure supplement 4—source data 2
Median cell volume for the FY4 prototrophic strain grown in glycerol/ethanol medium ±adenine.
- https://doi.org/10.7554/eLife.43808.024
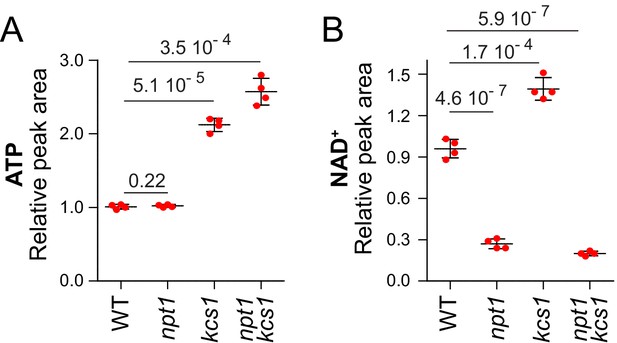
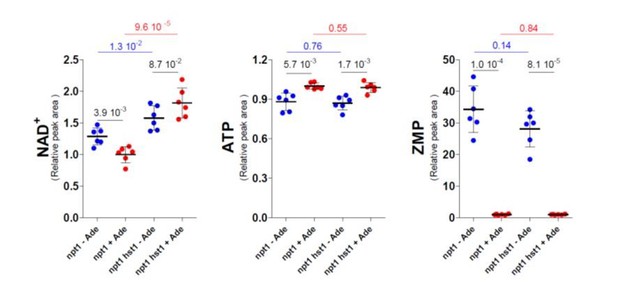
Stimulation of NAD+synthesis by ATP is abolished in npt1 mutants.
(A–B) Wild-type (WT, Y286), npt1 (Y5581), kcs1 (Y2337), and kcs1 npt1 (Y11017) cells exponentially grown for 24 hr in SDcasaWAU medium. Quantifications (set at one for the amount measured in WT cells) were performed on independent metabolic extractions (N = 4). Error bars correspond to standard deviation and indicated p-values were calculated using a Welch’s t-test.
-
Figure 3—figure supplement 5—source data 1
Metabolic analyses for wild-type npt1, kcs1 and npt1 kcs1 mutant strains grown in the presence of adenine.
- https://doi.org/10.7554/eLife.43808.026
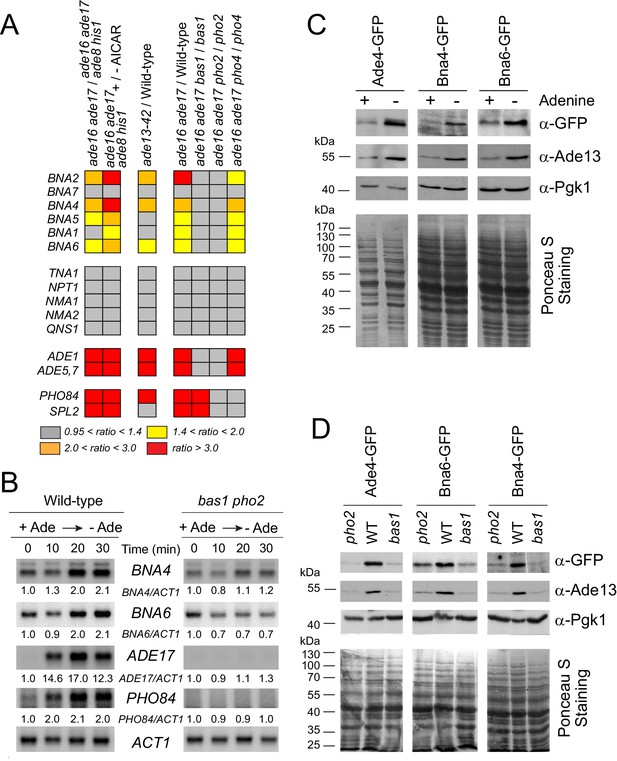
Bas1 and Pho2 transcriptionally upregulate the BNA genes in response to (S)ZMP.
(A) Heat-map representation of the expression of the pyridine pathway genes. Results were extracted from our previously reported microarray analyses (Hürlimann et al., 2011; Pinson et al., 2009). Raw data are available at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc= GSE13275 and https://www.ncbi.nlm.nih.gov/geo/queryacc.cgi?acc=GSE29324. (B) Kinetic analysis of ADE17, PHO84 and BNA genes expression in a wild-type and a bas1 pho2 double mutant upon adenine depletion. Wild-type (BY4742) and bas1 pho2 mutant (Y1487) cells were grown in SDcasaWU + adenine medium, centrifuged for 2 min at 3500 g, washed twice with SDcasaWU and re-suspended in the same medium lacking adenine (time 0). Aliquots were taken at indicated times, total RNA were extracted and gene expression was monitored by Northern blotting (N ≥ 2). Images shown correspond to one representative experiment. (C–D) Bna4 and Bna6 proteins are more abundant in adenine-depleted conditions in wild-type cells (C) but not in the absence of the Bas1 and Pho2 transcription factors (D). (C) Wild-type cells harboring either ADE4 (Y11325), BNA4 (Y11328), or BNA6 (Y11327) -GFP fusion at the corresponding gene locus were exponentially grown for 24 hr in SDcasaWU medium supplemented (+) or not (-) with adenine. (D) Wild-type, bas1 and pho2 yeast strains harboring either ADE4 (Y11325, Y11885 and Y11879), BNA4 (Y11328, Y11894 and Y118887) or BNA6 (Y11327, Y11885 and Y11890) -GFP fusion at the corresponding gene locus were exponentially grown for 24 hr in SDcasaWAU medium, filtered and then shifted for 2 hr in SDcasaWU medium. (C–D) Total proteins were extracted, separated by SDS-PAGE, electro-transferred and revealed by western-blotting using anti-GFP (1/500 (C); 1/2,500 (D)), anti-Ade13 (1/1200,000) and anti-Pgk1 (1/50,000) antibodies. Ade4-, Bna4- and Bna6-GFP fusions proteins were revealed at 84, 79 and 60 kDa, respectively. Images shown correspond to one representative experiment (N ≥ 2).
-
Figure 4—source data 1
Northern blot quantification for wild-type and bas1 pho2 mutant strains shifted in adenine-depleted medium.
- https://doi.org/10.7554/eLife.43808.032
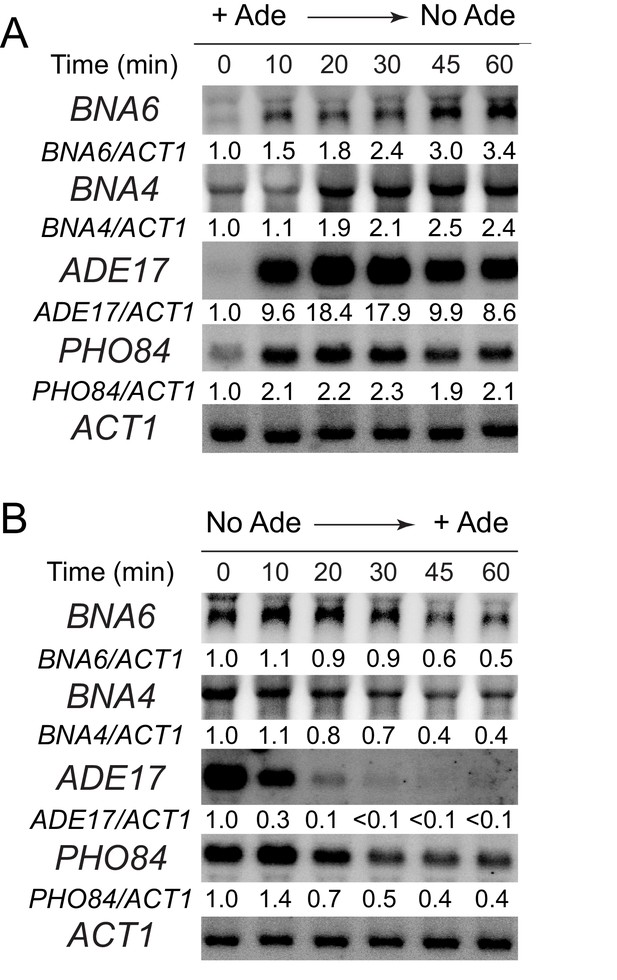
Kinetic analysis of ADE17, PHO84 and BNA genes expression upon either external adenine depletion (A) or addition (B).
(A) Wild-type cells (BY4742) were grown in SDcasaWU + adenine medium, centrifuged for 2 min at 3500 g, washed twice with SDcasaWU and re-suspended in the same medium lacking adenine (time 0). Aliquots were taken at indicated times and gene expression was monitored as in A (N = 1). (B) Wild-type cells (BY4742) were exponentially grown in SDcasaWU medium and adenine was added (time 0). Total RNA were extracted and gene expression was monitored by northern blotting on aliquots collected at indicated time (N = 1).
-
Figure 4—figure supplement 1—source data 1
Northern blot quantification for the wild-type strain shifted in either adenine-depleted or adenine-replete medium.
- https://doi.org/10.7554/eLife.43808.031
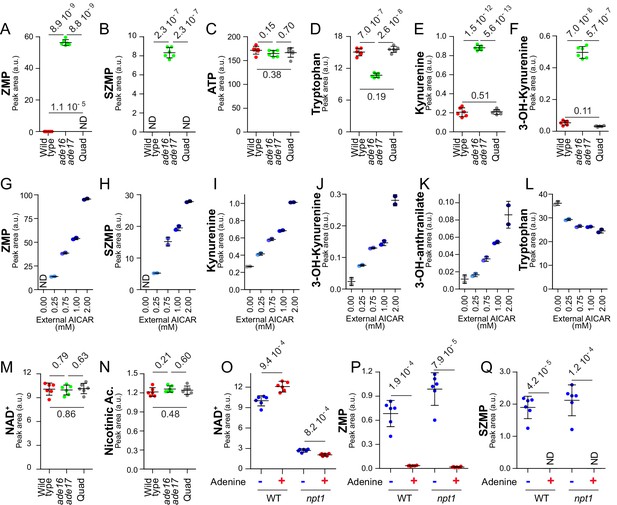
(S)ZMP-increase is sufficient to stimulate the NAD+ de novo pathway.
(A–F) (S)ZMP accumulation in the ade16 ade17 mutant is associated to a significant increase in NAD+ de novo pathway intermediates. Wild-type (BY4742, red dots), ade16 ade17 (Y1162, green dots) and ade16 ade17 ade8 his1 (quad; Y2950, grey dots) were grown in SDcasaWAU medium. (G– L) Accumulation of ZMP achieved by external AICAR addition correlates with increasing amounts of NAD+ de novo pathway intermediates. The ade16 ade17 ade8 his1 quadruple mutant strain (Y2950) was grown in SDcasaWAU medium and incubated for 24 hr with indicated amounts of AICAR prior to metabolite extraction. (M–N) (S)ZMP accumulation has no significant effect on NAD+ and nicotinic acid levels when the salvage pathway is active. NAD+ and nicotinic acid levels were determined from the experiment described in Figure 5A–F. (O–Q) Upregulation of the purine de novo pathway in the absence of adenine, when (S)ZMP is high, results in higher intracellular NAD+ only when the pyridine salvage pathway is inactivated. Wild-type (BY4742) and npt1 knock-out (Y5581) strains were exponentially grown for 24 hr in SDcasaWU medium ±Adenine. (A–F, M–Q) Quantifications were determined from independent metabolite extractions (N = 5). Error bars correspond to standard deviation and indicated p-values were calculated from a Welch’s t-test.
-
Figure 5—source data 1
Metabolic analyses for wild-type and ade16 ade17-derived mutant strains grown in +adenine.
- https://doi.org/10.7554/eLife.43808.036
-
Figure 5—source data 2
Metabolic analyses for the ade16 ade17 ade8 his1 mutant strain grown in various amount of AICAR.
- https://doi.org/10.7554/eLife.43808.037
-
Figure 5—source data 3
Metabolic analyses for wild-type and npt1 mutant strains grown in ±adenine.
- https://doi.org/10.7554/eLife.43808.038
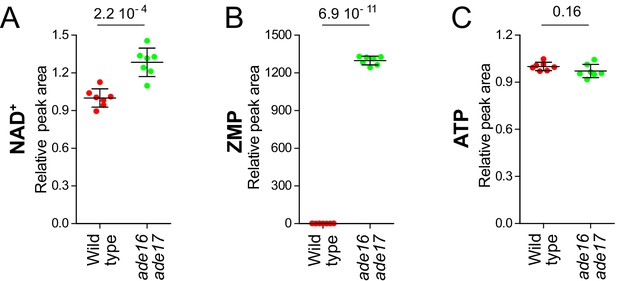
NAD+ level is significantly increased in the ade16 ade17 (S)ZMP accumulating mutant in the absence of external nicotinic acid.
Wild-type (BY4742, red dots) and ade16 ade17 (Y1162, green dots) were exponentially grown for 24 hr in SDcasaWAU medium lacking nicotinic acid. Metabolite extraction, separation and quantification were done as in Figure 2. Quantifications were determined from independent metabolite extractions (N ≥ 7) and standard deviation is presented. Values measured in wild-type cells (red dots) were set at 1. Indicated p-values were calculated using a Welch’s t-test.
-
Figure 5—figure supplement 1—source data 1
Metabolic analyses for wild-type and ade16 ade17 mutant strains grown in NA-free medium.
- https://doi.org/10.7554/eLife.43808.035
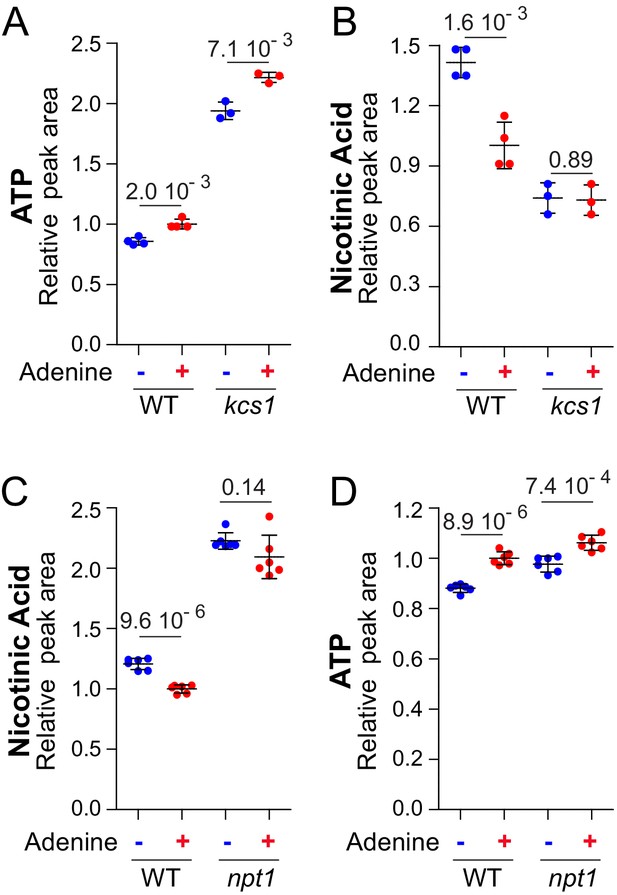
Metabolization of nicotinic acid is tightly connected to the amount of ATP.
Nicotinic acid utilization is increased when ATP is higher (A–B) and not anymore affected by extracellular adenine in a npt1 mutant (C–D). (A–D) Wild-type (BY4742; (A–D) and either kcs1 (Y2337; A–B) or npt1 (Y5581; C–D) knock-out mutant strains were exponentially grown for 24 hr in SDcasaWU medium supplemented (red dots) or not (blue dots) with adenine. Quantifications were determined from independent metabolite extractions (N ≥ 3) and standard deviation is presented. For each metabolite, the amount measured in wild-type cells grown in the presence of adenine was set at one and indicated p-values were calculated from a Welch’s t-test.
-
Figure 6—source data 1
Metabolic analyses for wild-type and kcs1 mutant strains grown in ±adenine.
- https://doi.org/10.7554/eLife.43808.044
-
Figure 6—source data 2
Nicotinic acid and ATP determination for wild-type and npt1 mutant strains grown in ±adenine.
- https://doi.org/10.7554/eLife.43808.045
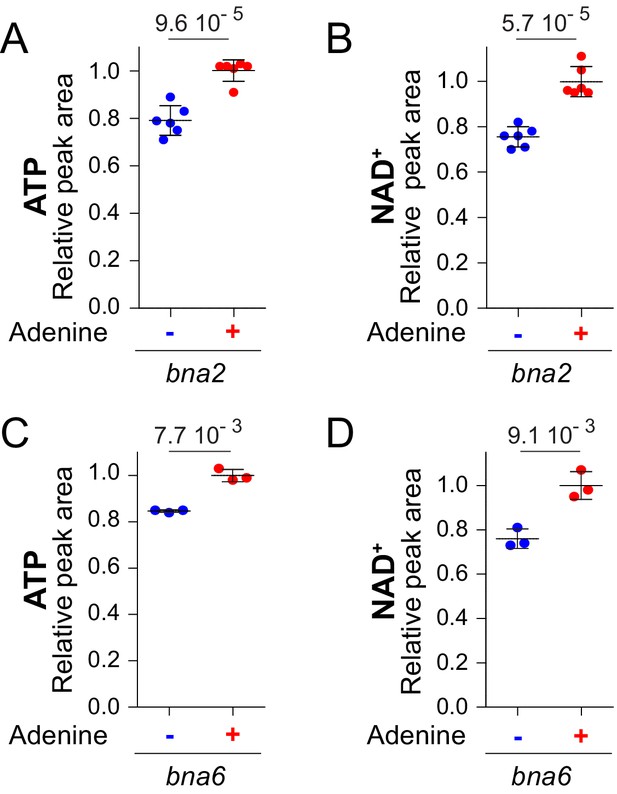
NAD+variations in response to external adenine availability do not require a functional pyridine de novo pathway.
ATP (A–C) and NAD+ (B–D) levels were determined from bna2 (Y10838) and bna6 (Y5891) knock-out mutants exponentially grown for 24 hr in SDcasaWU supplemented (red dots) or not (blue dots) with adenine. Quantifications were determined from independent metabolite extractions (A-B: N = 6; C-D: N = 3) and standard deviation is presented. Metabolite amount measured in cells grown in the presence of adenine (red dots) was set at 1. A Welch’s t-test was used to determine the indicated p-values.
-
Figure 6—figure supplement 1—source data 1
Metabolic analyses for bna2 and bna6 mutant strains grown in ±adenine.
- https://doi.org/10.7554/eLife.43808.041
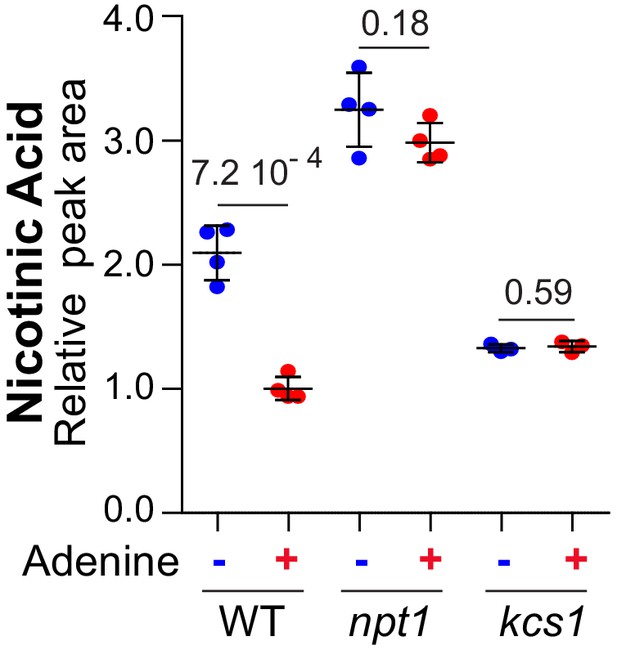
Nicotinic acid variations in response to external adenine availability are abolished in npt1 and kcs1 mutants fed with nicotinamide.
Wild-type (BY4742) and knock-out mutant strains (npt1: Y5581 and kcs1: Y2337) were exponentially grown for 24 hr in SDcasaWU-NA medium (lacking nicotinic acid), supplemented with nicotinamide, as an external pyridine source, and containing (red dots) or not (blue dots) adenine. Quantifications were determined from independent metabolite extractions (N ≥ 3) and standard deviation is presented. The amount measured in wild-type cells grown in the presence of adenine was set at one and indicated p-values were calculated from a Welch’s t-test.
-
Figure 6—figure supplement 2—source data 1
Metabolic analyses for wild-type, npt1 and kcs1 mutant strains grown in NA-free medium supplemented with nicotinamide ±adenine.
- https://doi.org/10.7554/eLife.43808.043
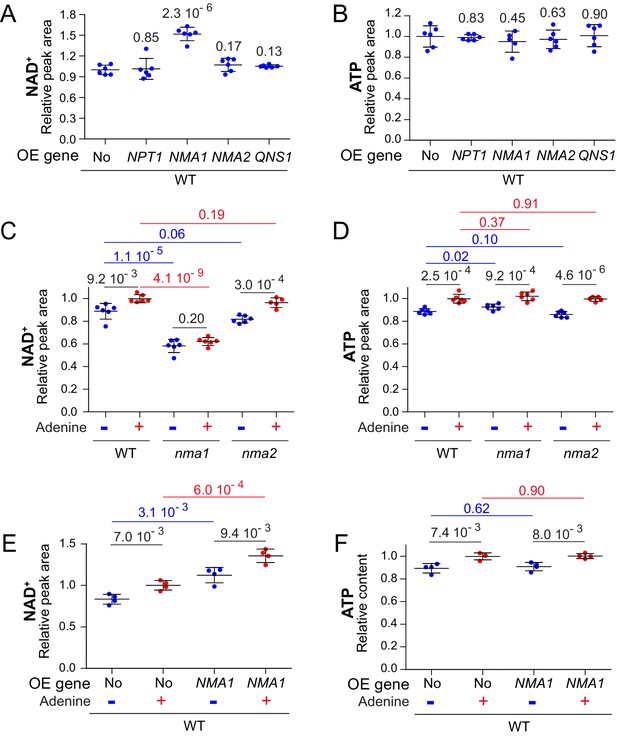
Nicotinic acid mononucleotide adenylyl transferase activity limits NAD+ synthesis.
(A–B) Overexpression of NMA1 is sufficient to increase NAD+ content when cells are grown in the absence of adenine. Wild-type cells (BY4742) were transformed with plasmid allowing overexpression (OE) of the indicated pyridine salvage pathway genes or the empty vector (No). Transformants were grown in SDcasaWU medium lacking adenine. Values obtained with the empty vector were set at 1. (C–D) NAD+ does not respond to adenine availability in cells lacking NMA1. Wild-type cells (BY4742) and nma mutants (nma1: Y5662; nma2: Y5663) strains were grown in SDcasaWU medium lacking (blue dots) or not (red dots) adenine. Values obtained in the wild-type strain grown in the presence of adenine were set at 1. (E–F) Overexpression of NMA1 leads to increased intracellular NAD+. Wild-type cells (BY4742) were transformed with a plasmid allowing overexpression (OE) of NMA1 gene or the empty vector (No). Transformants were grown in SDcasaWU medium lacking (blue dots) or not (red dots) adenine. Values obtained with the empty vector in the presence of adenine were set at 1. (A–F) Quantifications were determined independent metabolite extractions (N ≥ 4) and standard deviation is presented. Indicated p-values were calculated from a Welch’s t-test.
-
Figure 7—source data 1
Metabolic analyses for the wild-type strain overexpressing various NAD+-synthesis genes and grown in - adenine.
- https://doi.org/10.7554/eLife.43808.051
-
Figure 7—source data 2
Metabolic analyses for wild-type, nma1 and nma2 mutant strains grown in ±adenine.
- https://doi.org/10.7554/eLife.43808.052
-
Figure 7—source data 3
Metabolic analyses for the wild-type strain overexpressing NMA1 and grown in ±adenine.
- https://doi.org/10.7554/eLife.43808.053
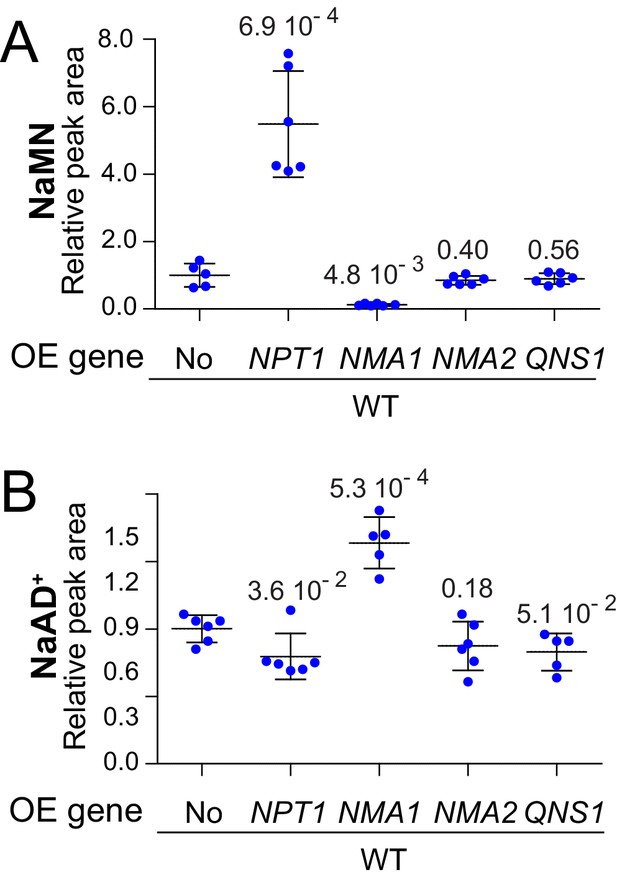
NaMN and NaAD+are increased in response to NPT1 and NMA1 genes overexpression, respectively.
Wild-type cells (BY4742) were transformed with plasmids allowing overexpression (OE) of the indicated pyridine salvage pathway genes or the empty vector (No). Transformants were grown in SDcasaWU medium lacking adenine. Values obtained with the empty vector were set at 1. Quantifications were determined from independent metabolite extractions (N ≥ 5), standard deviation is presented and p-values were calculated from a Welch’s t-test.
-
Figure 7—figure supplement 1—source data 1
NaMN and NaAD+ determination in the wild-type overexpressing various NAD+-synthesis genes and grown in - adenine.
- https://doi.org/10.7554/eLife.43808.048
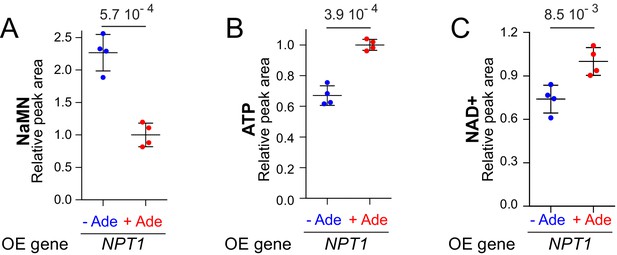
Accumulation of NaMN in response to NPT1 overexpression is higher when ATP is limiting.
Wild-type cells (BY4742) were transformed with a plasmid allowing overexpression (OE) of the NPT1 gene. Transformants were grown in SDcasaWU medium lacking (blue dots) or not (red dots) adenine. Values obtained with the empty vector were set at 1. Quantifications were determined from independent metabolite extractions (N = 4). Standard deviation is presented and indicated p-values were calculated using a Welch’s t-test.
-
Figure 7—figure supplement 2—source data 1
Metabolic analyses for the wild-type overexpressing NPT1 and grown in ±adenine.
- https://doi.org/10.7554/eLife.43808.050
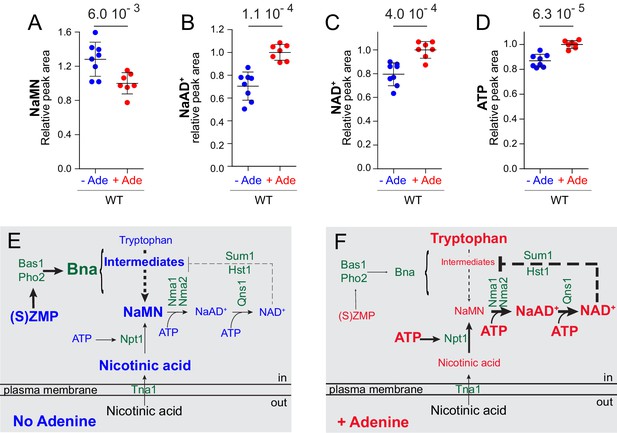
Yeast co-regulates purine and pyridine metabolism in response to adenine through two mechanisms.
(A–D) The Nma1-bottleneck for NAD+ synthesis operates under physiological conditions. A prototrophic strain (FY4) was grown in SDcasaWU medium lacking (blue dots) or not (red dots) adenine. Values obtained in the presence of adenine (red dots) were set at one for each metabolite. Quantifications were determined from independent metabolite extractions (N ≥ 7). Standard deviation is presented and a Welch’s t-test was used to calculate the indicated p-values. (E–F) Model of NAD+ synthesis rerouting in response to extracellular adenine availability. The thickness of lines and arrows refers to the intensity of signaling or flux in metabolic pathways. For each metabolite, the font-size reflects variation of its intracellular content.
-
Figure 8—source data 1
Determination of pyridine-pathway intermediates in the FY4 prototrophic strain ±adenine.
- https://doi.org/10.7554/eLife.43808.057
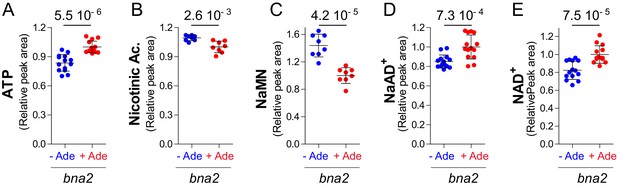
Modulation of adenine on NAD+biosynthesis operates in the absence of a functional de novo pathway.
Pyridine salvage intermediate levels were determined in a bna2 mutant strain (Y11838) exponentially grown for 24 hr in SDcasaWU medium lacking (blue dots) or not (red dots) adenine. Values obtained in the presence of adenine (red dots) were set at 1. Quantifications were determined from independent metabolite extractions (N ≥ 8). Standard deviation is presented and indicated p-values were calculated using a Welch’s t-test.
-
Figure 8—figure supplement 1—source data 1
Pyridine pathway intermediates determination for the bna2 mutant strain ±adenine.
- https://doi.org/10.7554/eLife.43808.056
Tables
List of the yeast strains used in this study.
https://doi.org/10.7554/eLife.43808.058| Strain name | Genotype |
|---|---|
| BY4742 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 |
| FY4 | MATa |
| Y286 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 |
| Y1162 | MATα ade16::kanMX4 ade17::kanMX4 his3Δ1 leu2Δ0 ura3Δ0 |
| Y2337 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 kcs1::kanMX4 |
| Y1487 | MATa his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 bas1::kanMX4 pho2::kanMX4 |
| Y2950 | MATα his3Δ1 leu2Δ0 ura3Δ0 ade16::kanMX4 ade17::kanMX4 ade8::kanMX4 his1::kanMX4 |
| Y4835 | MATα prs3::KanMX4 his3Δ1 leu2Δ0 ura3Δ0 |
| Y5581 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 npt1::kanMX4 |
| Y5662 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 nma1::kanMX4 |
| Y5663 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 nma2::kanMX4 |
| Y5731 | MATa his3Δ1 leu2Δ0 ura3Δ0 nma1::kanMX4 |
| Y5822 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 bna4::kanMX4 |
| Y5891 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 bna6::kanMX4 |
| Y10838 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 bna2::kanMX4 |
| Y10901 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 bna1::kanMX4 |
| Y10903 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 bna5::kanMX4 |
| Y10904 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 bna7::kanMX4 |
| Y10991 | MATα his3Δ1 leu2Δ0 ura3Δ0 adk1::kanMX4 |
| Y11017 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 npt1::kanMX4 kcs1::kanMX4 |
| Y11325 | MATa his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 ADE4-GFP-HIS3 |
| Y11327 | MATa his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 BNA6-GFP-HIS3 |
| Y11328 | MATa his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 BNA4-GFP-HIS3 |
| Y11879 | MATa pho2::KanMX4 his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 ADE4-GFP-HIS3 |
| Y11885 | MATa bas1::KanMX4 his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 ADE4-GFP-HIS3 |
| Y11887 | MATa pho2::KanMX4 his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 BNA4-GFP-HIS3 |
| Y11890 | MATa pho2::KanMX4 his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 BNA6-GFP-HIS3 |
| Y11894 | MATa bas1::KanMX4 his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 BNA4-GFP-HIS3 |
| Y11895 | MATa bas1::KanMX4 his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 BNA6-GFP-HIS3 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.43808.059