HDAC3 restrains CD8-lineage genes to maintain a bi-potential state in CD4+CD8+ thymocytes for CD4-lineage commitment
Figures
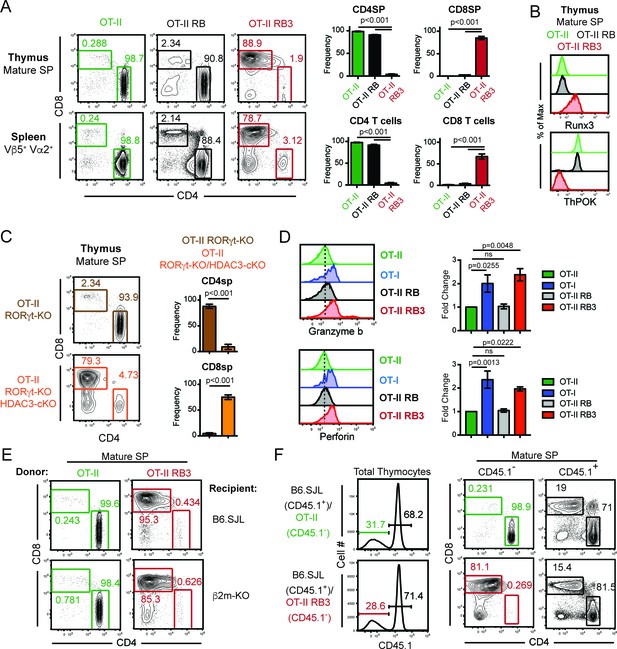
MHC class II restricted thymocytes are redirected to the CD8-lineage when HDAC3 is absent.
(A) CD4/CD8 profile of mature thymic SP (Vβ5+Vα2+H2K+CD24lo) and splenic T cells (Vβ5+Vα2+) from OT-II, OT-II RORγt-KO Bcl-xl tg (RB) and OT-II RB3 mice. Graphs depict frequency of gated cells from at least three independent experiments (n = 3–5/group) (B) Runx3 and ThPOK expression in mature thymic SP cells from OT-II, OT-II RB, and OT-II RB3 mice (n = 4/group from four independent experiments). (C) CD4/CD8 profile of mature thymic SP from OT-II RORγt-KO and OT-II RORγt-KO HDAC3-cKO mice. Graphs depict frequency of gated cells from at least three independent experiments (n = 3–4/group). (D) Granzyme b and perforin expression in SP thymocytes from OT-II, OT-I, OT-II RB and OT-II RB3 mice. FACS plots were gated on CD4SP cells for OT-II and OT-II RB mice and CD8SP cells for OT-I and OT-II RB3 mice. Bar graphs depict mean ± SEM (n = 3/group from three independent experiments) of the fold change in MFI between unstimulated and TCR/CD2 stimulated conditions. (E) Representative FACS plots of the proportion of CD4SP and CD8SP mature thymocytes from straight BMCs, where bone marrow from OT-II or OT-II RB3 mice were transplanted into B6.SJL or β2m-KO recipients. Mature SP thymocytes were gated as Vβ5+Vα2+H2K+CD24lo. Mice (n = 3–5/group from three independent experiments) were analyzed 8–10 weeks after transfer. (F) Representative FACS plots of the proportion of CD4SP and CD8SP mature thymocytes from mixed BMCs from OT-II (CD45.1-)/B6.SJL (CD45.1+) and OT-II RB3 (CD45.1-)/B6.SJL (CD45.1+) mice. Mature SP thymocytes from CD45.1+ cells were gated as H2K+CD24lo; mature SP thymocytes from CD45.1- cells were gated as Vβ5+Vα2+H2K+CD24lo. Mice were analyzed 8–10 weeks after transfer. (n = 5/group from three independent experiments).
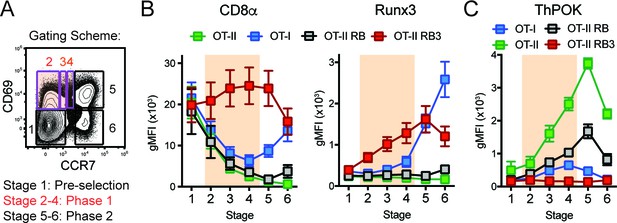
Acceleration of CD8-lineage commitment in HDAC3-deficient thymocytes.
(A) Gating scheme outlining two phases of CD4/CD8-lineage commitment—Phase one includes CD69+ CCR7- cells (Stage 2), CD69+ CCR7lo cells (Stage 3), and CD69+ CCR7int cells (Stage 4); Phase two identifies CD69+ CCR7+ cells (Stage 5) and CD69- CCR7+ cells (Stage 6). Orange shading highlights Phase 1. (B) Expression of Runx3 and CD8α during stages of lineage commitment (Stages 1–6), as outlined in (A) from OT-I, OT-II, OT-II RB and OT-II RB3 mice. CD69-versus-CCR7 plots were gated from DN-removed, CD45.2+ cells. (C) Expression of ThPOK during stages of lineage commitment, gated as in (B). Plots in B and C show mean ±SEM of MFI from 5 to 7 mice per group from five independent experiments.
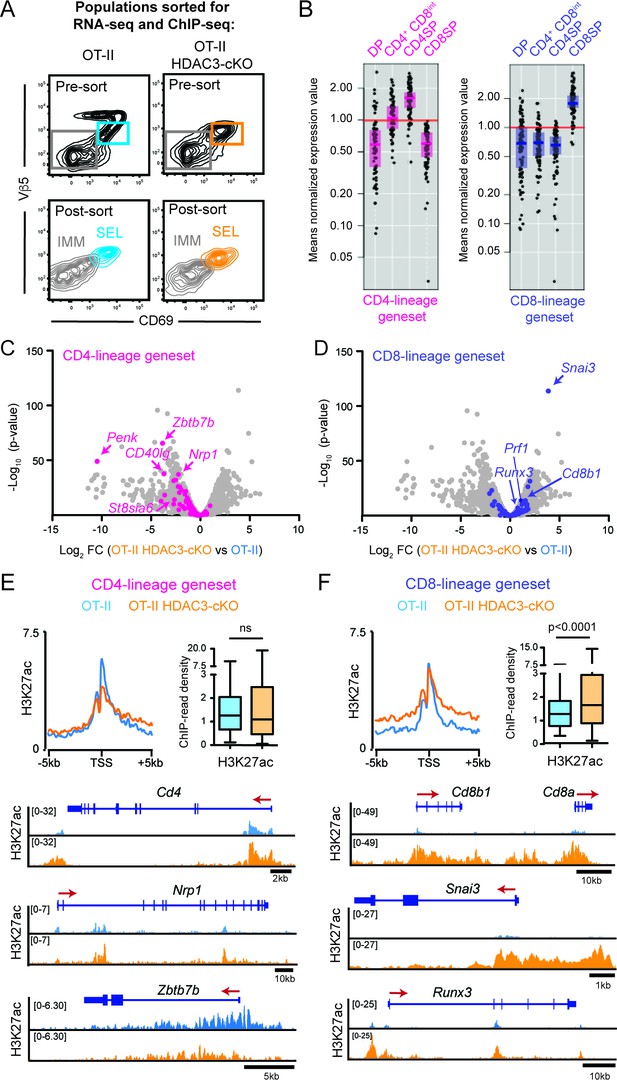
HDAC3-deficient thymocytes fail to induce the CD4-lineage program.
(A) Flow cytometric analysis of populations from OT-II and OT-II HDAC3-cKO mice sorted for RNA-seq and ChIP-seq. Plots show representative pre-sort and post-sort analysis of two FACS sorted populations—Immature thymocytes (Vβ5loCD69- CD4+CD8+) and Selecting thymocytes (Vβ5loCD69+). (B) Expression of CD4-lineage and CD8-lineage gene sets across DP, CD4+CD8lo, CD4SP, and CD8SP ImmGen expression data. (C–D) Volcano plot depicting the Log2fold change (FC) between OT-II HDAC3-cKO and OT-II Selecting (CD69+) cells. Grey dots show all genes; pink dots show CD4-lineage gene set (C); blue dots show CD8-lineage gene set (D). (E–F) Average H3K27ac ChIP-seq signal at the transcription start site (TSS) between OT-II and OT-II HDAC3-cKO Selecting (CD69+) cells for CD4-lineage gene sets (E) and CD8-lineage gene sets (F). Box-and-whisker plots depict H3K27ac signal at the TSS at CD4- or CD8-lineage genes between OT-II and OT-II HDAC3-cKO mice. Snapshots of example ChIP-seq tracks for each gene set are below. See also Figure 3—figure supplements 1–5 and Figure 3—source datas 1–2.
-
Figure 3—source data 1
Gene expression values (RNA-seq) of Immature and Selecting populations from OT-II and OT-II HDAC3-cKO mice.
Numerical data corresponding to the graphs of Figure 3C–D.
- https://doi.org/10.7554/eLife.43821.010
-
Figure 3—source data 2
Gene lists: CD4-lineage, CD8-lineage, silenced genes, housekeeping genes.
CD4-lineage and CD8-lineage gene lists corresponding to graphs of Figure 3B–F. Silenced genes and housekeeping gene lists corresponding to graphs of Figure 3—figure supplement 5.
- https://doi.org/10.7554/eLife.43821.011
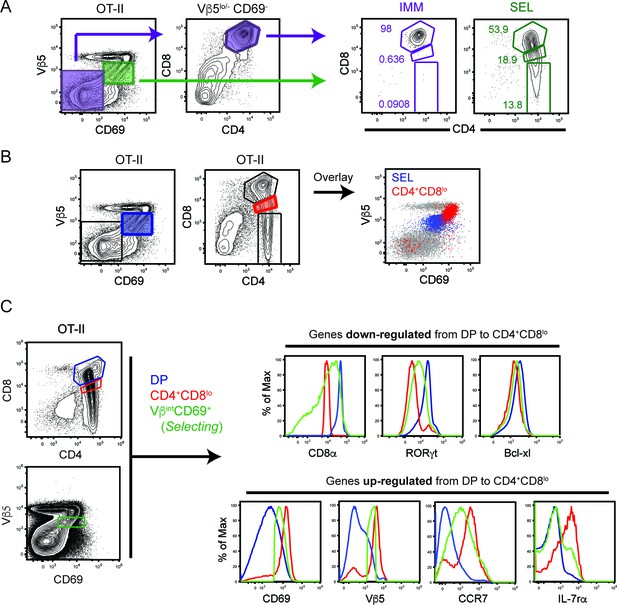
Characterization of Immature and Selecting thymic populations.
(A) CD4-versus-CD8 profile of Immature (purple) and Selecting (green) populations in OT-II mice. Immature thymocytes are gated as Vβ5-/loCD69- CD4+CD8+. Selecting thymocytes are gated as Vβ5intCD69+. (B) Overlay of Selecting (blue) and CD4+CD8lo (red) populations on a Vβ5-versus-CD69 profile in OT-II mice. Grey dots on the Vβ5/CD69 plot depict total thymocytes. Data is representative of 3 mice from three independent experiments. (C) Selecting cells show intermediary marker expression between DP and CD4+CD8lo thymocytes. Expression of ‘down-regulated’ genes (CD8α, RORγt, Bcl-xl) and ‘up-regulated’ genes (CD69, TCR, CCR7, IL-7Rα) on DP, CD4+CD8lo, and Selecting cells from OT-II mice. Data is representative of 3 mice from three independent experiments.
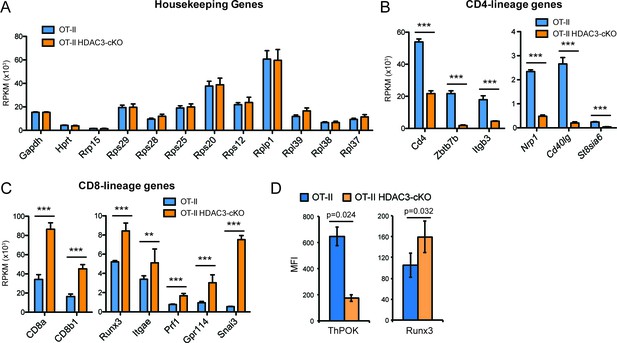
Gene expression of select housekeeping genes, CD4-lineage genes, and CD8-lineage genes.
RPKM values of indicated genes of Selecting cells from OT-II and OT-II HDAC3-cKO mice. (A) Housekeeping genes, (B) CD4-lineage genes, and (C) CD8-lineage genes. Bar graphs depict mean ± SEM of three mice. P values (***, p < 0.001, **, p < 0.01) were calculated using the exactTest with edgeR software. Housekeeping genes did not show a significant difference between mice. (D) Protein expression of ThPOK and Runx3 in Selecting cells from OT-II and OT-II HDAC3-cKO. Bar graphs depict mean ± SEM of 3–6 mice per group from at least three independent experiments. P values were calculated using paired t test.
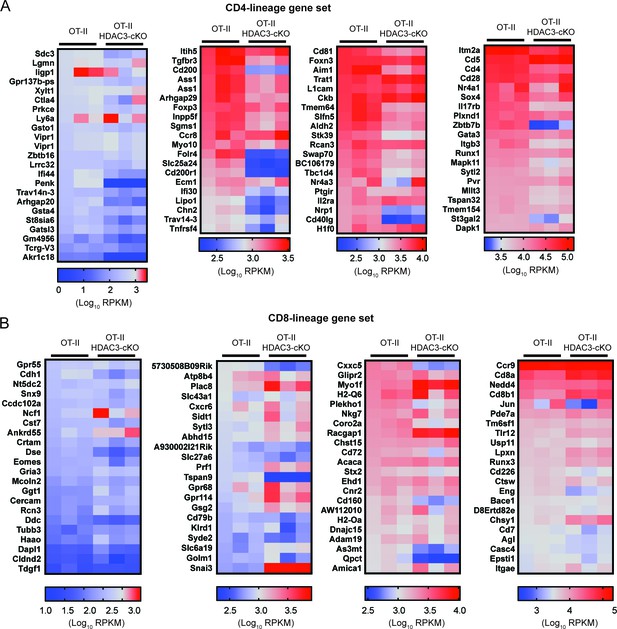
Heatmaps of CD4-lineage and CD8-lineage genes.
Heatmaps depict Log10 RPKM of genes in CD4-lineage (A) and CD8-lineage (B) gene sets in Selecting (CD69+) thymocytes from OT-II and OT-II HDAC3-cKO. Heatmaps show three biological replicates per mouse group. Scale bar is below each heatmap. Refer to Figure 3—source data 1 for Log2fold change between OT-II and OT-II HDAC3-cKO mice.
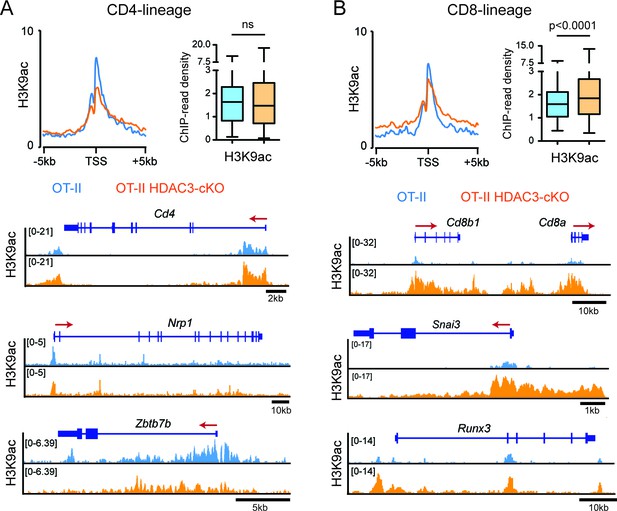
H3K9ac signal at CD4-lineage and CD8-lineage genes.
(A–B) Average H3K9ac ChIP-seq signal at the transcription start site (TSS) between OT-II and OT-II HDAC3-cKO Selecting cells for CD4-lineage gene sets (A) and CD8-lineage gene sets (B). Box-and-whisker plots depict H3K9ac signal at the TSS at each CD4- or CD8-lineage gene between OT-II and OT-II HDAC3-cKO mice. Snapshots of example ChIP-seq tracks for each gene set are below plots depicting the average signal. P values were calculated using Wilcoxon signed rank test.
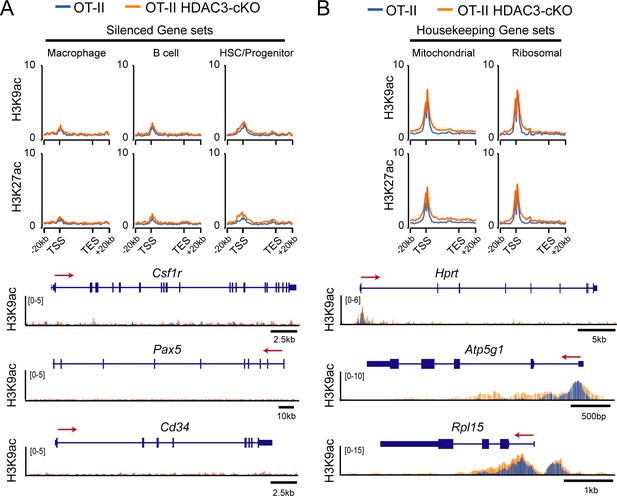
H3K9ac signal and H3K27ac signal at silenced genes and housekeeping genes in thymocytes.
(A–B) Average H3K9ac and H3K27ac signal ±20 kb around the gene body among gene sets that are silenced in thymocytes (A) or housekeeping genes (B). Below are example H3K9ac ChIP-seq snapshots of genes within each of the genesets—Csf1r for macrophage, Pax5 for B cell, Cd34 for HSC/progenitor, Hprt for housekeeping, Atp5g1 for mitochondrial, and Rpl15 for ribosomal. Each image depicts an overlay of ChIP-seq tracks between OT-II (blue) and OT-II HDAC3-cKO (orange) mice. Refer to Figure 3—source data 2 for list of genes in these gene sets (macrophage, B cell, HSC/Progenitor, mitochondrial, ribosomal). TSS represents transcription start site; TES represents transcription end site. Scale bar in kb below ChIP-seq tracks identifies scale of snapshot.
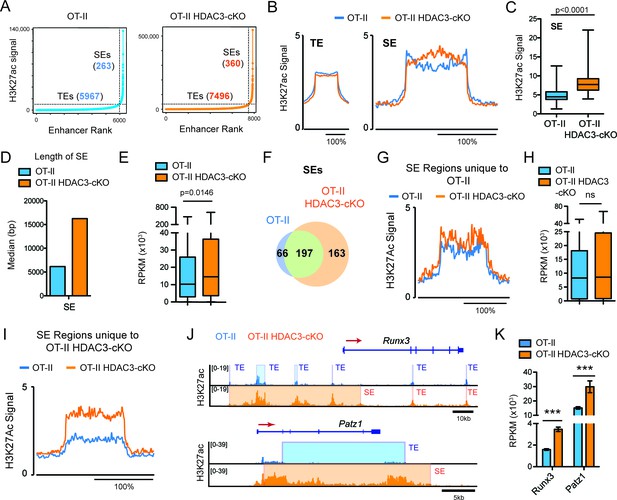
DP thymocytes from HDAC3-cKO mice show a CD8-lineage bias at the chromatin level.
(A) Typical enhancers (TEs) and super enhancers (SEs) in Immature (DP) cells from OT-II and OT-II HDAC3-cKO mice. (B) Average H3K27ac signal (normalized to input) for TE and SE regions from OT-II and OT-II HDAC3-cKO mice. (C) Box-and-whisker plot of H3K27ac signal for each super enhancer in OT-II and OT-II HDAC3-cKO mice. (D) Median length of super-enhancers between OT-II and OT-II HDAC3-cKO mice. (E) Box-and-whisker plots of mRNA expression (reads per kilobase of exon per million mapped reads, RPKM) from super-enhancer-associated genes from OT-II and OT-II HDAC3-cKO mice. (F) Venn Diagram of shared and unique SEs between OT-II and OT-II HDAC3-cKO mice. (G) H3K27ac signal at super-enhancer regions unique to OT-II mice and the corresponding regions in OT-II HDAC3-cKO mice. (H) Box-and-whisker plots of mRNA expression (RPKM) between OT-II and OT-II HDAC3-cKO mice of genes associated with super-enhancers unique to OT-II mice. (I) H3K27ac signal at super-enhancer regions unique to OT-II HDAC3-cKO mice and the corresponding regions in OT-II mice. (J) Snapshot of H3K27ac ChIP-seq tracks at the Runx3 and Patz1 locus from OT-II (blue) and OT-II HDAC3-cKO (orange) mice. Shaded regions depict TEs and SEs. (K) Gene expression (RNA-seq) of Runx3 and Patz1 in Immature (DP) cells from OT-II and OT-II HDAC3-cKO mice. (B, G, I) The x-axis represents a surrounding area that corresponds to 200% of the center of each region. (***, p < 0.001). See also Figure 4—source data 1.
-
Figure 4—source data 1
Super-enhancer list.
Genomic data corresponding to super-enhancers analyzed in Figure 4.
- https://doi.org/10.7554/eLife.43821.013
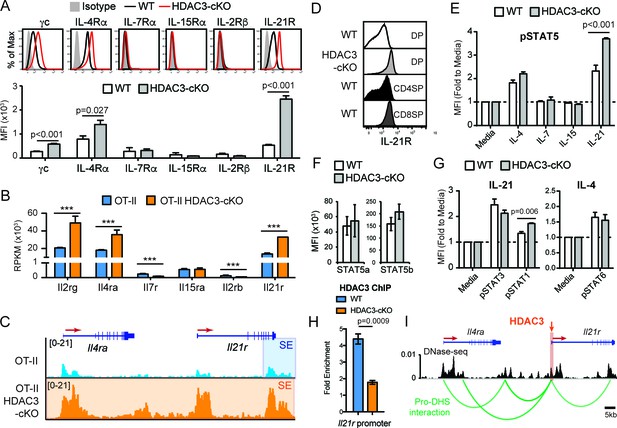
DP thymocytes from HDAC3-cKO mice show increased STAT5 activation in response to IL-21.
(A) Surface expression of cytokine receptor chains γc, IL-4Rα, IL-7Rα, IL-15Rα, IL-2Rβ, and IL-21R on DP thymocytes from WT and HDAC3-cKO mice. Flow cytometric plots have an isotype control to illustrate background level of expression. (B) Gene expression (RNA-seq) of Il2rg, Il4ra, Il7r, Il15ra, Il2rb, and Il21r in Immature (DP) cells from OT-II and OT-II HDAC3-cKO mice. (C) Snapshot of H3K27ac ChIP-seq tracks at the Il4ra and Il21r gene loci in Immature (DP) thymocytes from OT-II and OT-II HDAC3-cKO mice. Shaded regions identify super-enhancers. (D) Protein expression of IL-21R on DP, CD4SP and CD8SP thymocytes from WT mice and DP thymocytes from HDAC3-cKO mice. (E) pSTAT5 levels in DP thymocytes from WT and HDAC3-cKO mice after 10 min ex vivo stimulation with IL-21, IL-15, IL-7, IL4, or media alone. (F) Protein expression of STAT5a and STAT5b in DP thymocytes from WT and HDAC3-cKO mice. (G) pSTAT3 and pSTAT1 levels after a 10 min in vitro IL-21 stimulation, and pSTAT6 levels after IL-4 stimulation of DP thymocytes from WT and HDAC3-cKO mice. (H) Quantitative ChIP (qChIP) of HDAC3 binding at the Il21r promoter in DP thymocytes from WT and HDAC3-cKO mice. Graph depicts fold enrichment over Rpl30 (n = 4 mice/group from four indpendent experiments). (I) DNase-seq and Hi-C arc plots at the Il4ra and Il21r gene loci in DP thymocytes and pooled DN3-to-DP thymocytes, respectively. Shaded region highlights where HDAC3 binds, as shown in Figure 5H. (DHS, DNA hypersensitivity sites). (A, E, G) Bar graph shows mean ± SEM of MFI from 4 to 5 mice from at least three independent experiments. (D, H) Plots are representative of at least three mice from three independent experiments. (F) Bar graph shows mean ± SEM of MFI from three mice from two independent experiments. (***, p < 0.001). See also Figure 5—figure supplements 1–2.
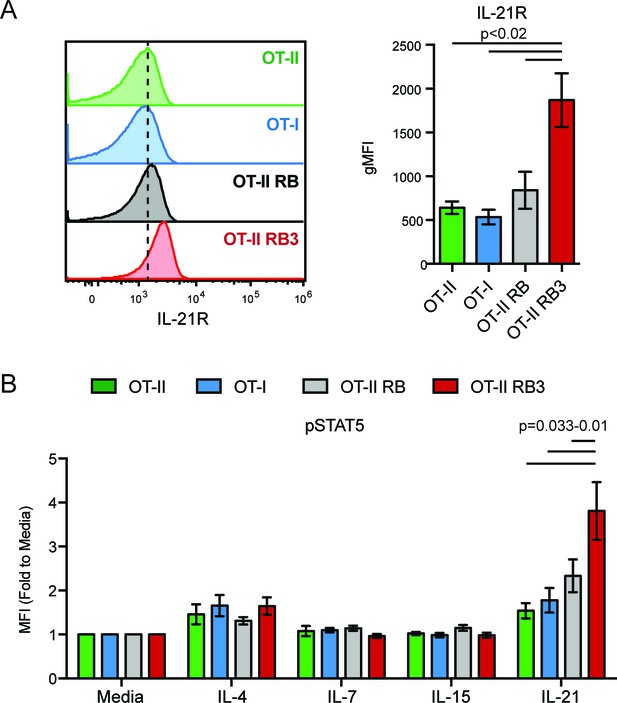
IL-21R expression and signaling in OT-II RB3 mice.
(A) IL-21R expression in DP thymocytes from OT-II, OT-I, OT-II RB, and OT-II RB3 mice. Bar graph is mean ± SEM of MFI. N = 3–7 mice/group. (B) p-STAT5 expression in DP thymocytes from OT-II, OT-I, OT-II RB, and OT-II RB3 mice that were stimulated with the indicated cytokines for 10mins. Experimental conditions are the same as performed in Figure 5E. Bar graph shows mean ± SEM of p-STAT5 MFI fold to unstimulated conditions. N = 3–7 mice/group.
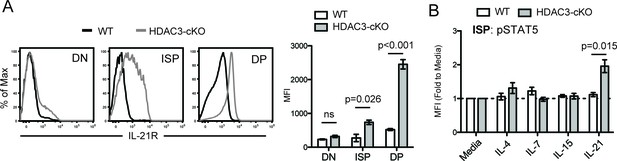
IL-21R expression and signaling in CD2-icre HDAC3-cKO mice.
(A) IL-21R expression in DN (CD4-CD8-), immature SP (ISP; CD4-CD8+TCRβ-), and DP (CD4+CD8+) thymocytes from WT and HDAC3-cKO mice. Bar graph is mean ±SEM of MFI. N = 3 mice/group. (B) p-STAT5 expression in ISPs from WT and CD2-icre HDAC3-cKO mice that were stimulated with the indicated cytokines for 10mins. Experimental conditions are the same as performed in Figure 5E. Bar graph shows mean ±SEM of p-STAT5 MFI fold to unstimulated conditions. N = 4–5 mice/group.
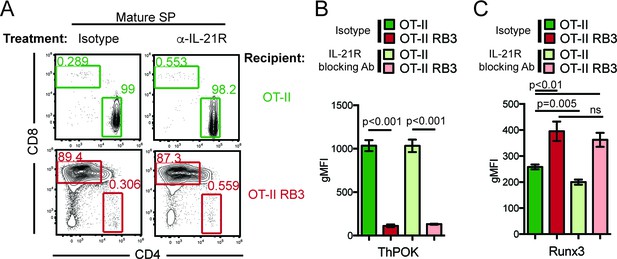
Blocking IL-21R does not restore CD4-lineage commitment in OT-II RB3 mice.
(A) OT-II and OT-II RB3 mice were injected with isotype or anti-IL-21R blocking antibodies retro-orbitally (i.v.) for two weeks every 3–4 days. Flow plots depict CD4/CD8 profile in mature SP thymocytes (Vβ5+Vα2+H2K+CD24lo). Data is representative of 3–4 mice per group from two independent experiments. (B–C) ThPOK (B) and Runx3 (C) expression in Vβ5hiH2K- thymocytes from OT-II and OT-II RB3 mice treated with isotype or anti-IL-21R blocking antibody, as performed in A. Data show mean ± SEM of geometric MFI from 3 to 4 mice from two independent experiments. See also Figure 6—figure supplement 1.
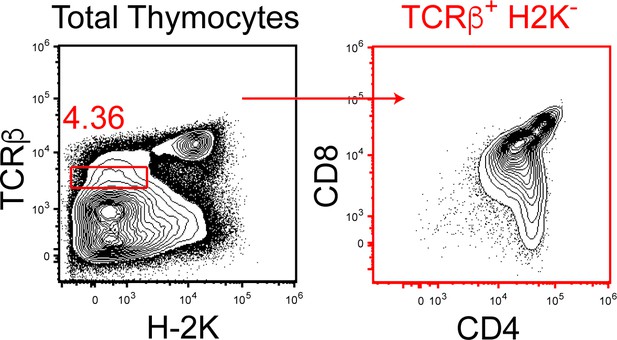
Gating strategy for Figure 6B–C.
To examine thymocytes early in CD4/CD8-lineage choice, thymocytes were gated as TCRβ+H-2K-. As shown here, TCRβ+H-2K- thymocytes are CD4+CD8lo, also known as intermediate thymocytes that undergo CD4/CD8-lineage choice (Singer et al., 2008).
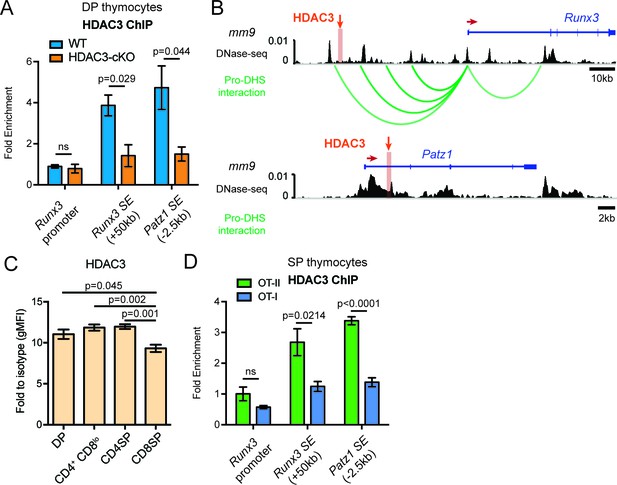
HDAC3 binds to Runx3 and Patz1 regulatory regions in DP and CD4SP thymocytes.
(A) HDAC3 qChIP in DP thymocytes from WT and HDAC3-cKO mice. Plots show mean ± SEM of fold enrichment over Rpl30 (n = 3 mice/group from three independent experiments). (B) DNase-seq and Hi-C arc plots at the Runx3 and Patz1 gene loci in DP thymocytes and pooled DN3-to-DP thymocytes, respectively. Shaded region highlights where HDAC3 binds, as shown in (A). (DHS, DNA hypersensitivity sites). (C) HDAC3 protein expression measured by flow cytometry in DP, CD4+CD8lo, CD4SP, and CD8SP thymocytes from WT mice. Plots show mean ± SEM of fold enrichment over isotype (n = 4 mice/group from three independent experiments). (D) HDAC3 qChIP in SP thymocytes from OT-II and OT-I mice. Plots show mean ± SEM of fold enrichment over Rpl30 (n = 4 mice/group from two independent experiments). See also Figure 7—figure supplements 1–2.
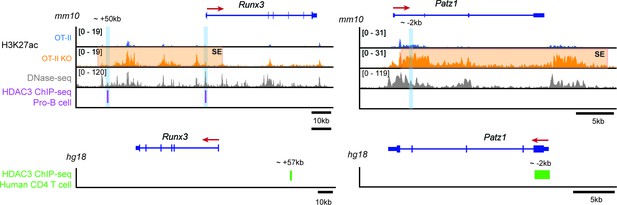
HDAC3 ChIP-seq in pro-B cells and human CD4 T cells.
ChIP-seq snapshots at Runx3 and Patz1 of publicly available HDAC3 ChIP-seq datasets in pro-B cells (GEO: GSM2096648, genome alignment mm10) and human CD4 T cells (GEO: GSM393952, genome alignment hg18). Alongside the HDAC3 ChIP-seq, DNase-seq (GEO: GSM2195840, genome alignment mm10) and H3K27ac ChIP seq from Immature OT-II and OT-II HDAC3-cKO thymocytes (as shown in Figure 4) were provided to visualize where HDAC3 binds in relation to regulatory elements and changes in histone acetylation between WT and HDAC3-cKO mice, respectively. Orange shaded regions demarcate super enhancers (SE) and blue shaded regions highlight where primers were made for qPCR. In pro-B cells, HDAC3 binds 50 kb upstream of Runx3, at the Runx3 promoter, and approximately 2 kb downstream of the Patz1 promoter. Likewise, HDAC3 binds approximately 57 kb upstream of Runx3 and 2 kb downstream of the Patz1 promoter in human CD4 T cells. These three regions were used to make primers for HDAC3 qChIP performed in Figure 7.
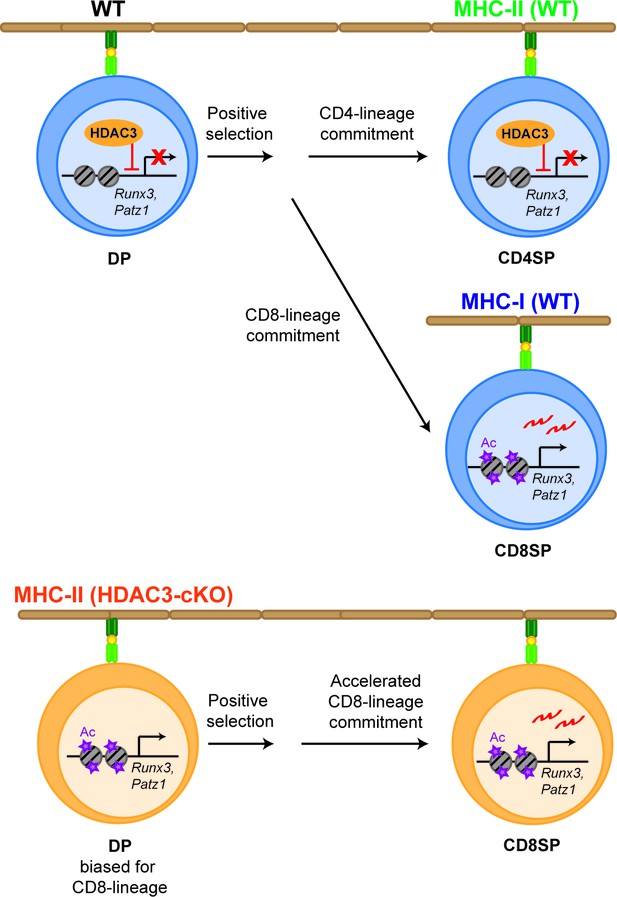
Model.
HDAC3 is required to restrain CD8-lineage genes for CD4-lineage choice. In WT thymocytes (upper panel), HDAC3 associates with Runx3 and Patz1 in DP thymocytes to restrain CD8-lineage gene expression. After positive selection, HDAC3 stays bound to these regions in CD4SP thymocytes for CD4-lineage commitment, while in CD8SP thymocytes HDAC3 no longer binds to these regions for expression of CD8-lineage genes and CD8-lineage commitment. However, deletion of HDAC3 (lower panel) results in an increase in histone acetylation at CD8-lineage genes (Runx3, Patz1) and priming DP thymocytes for the CD8-lineage. As a result, Runx3 is pre-maturely expressing during CD4/CD8-lineage choice and cells commit to the CD8-lineage, which is accelerated.
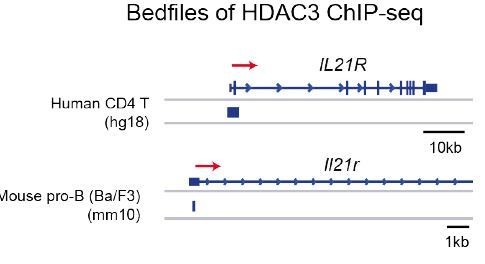
HDAC3 binding at the Il21r promoter in human CD4 T cells and mouse pro-B cells.
Accession numbers for publicly accessible HDAC3 ChIP-seq: human CD4 T cells (GSM393952) and mouse pro-B cells (GSM2096648).
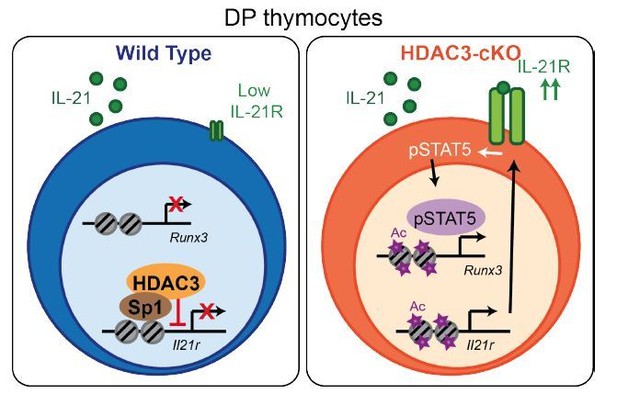
Model.
HDAC3 functions to block expression of IL-21R in DP thymocytes to prevent IL-21R expression, STAT5 activation, and expression of CD8-lineage promoting genes, such as Runx3
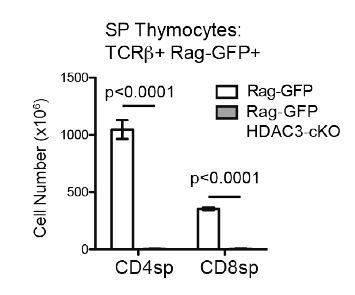
Very few SP thymocytes are generated in CD2-icre HDAC3-cKO mice.
Cell count of CD4SP and CD8SP thymocytes from Rag-GFP and Rag-GFP CD2-icre HDAC3-cKO mice. SP thymocytes were gated on Rag-GFP+ to remove recirculating (GFP-) T cells in the thymus. Graph shows mean +/- SEM of 5 mice/group.
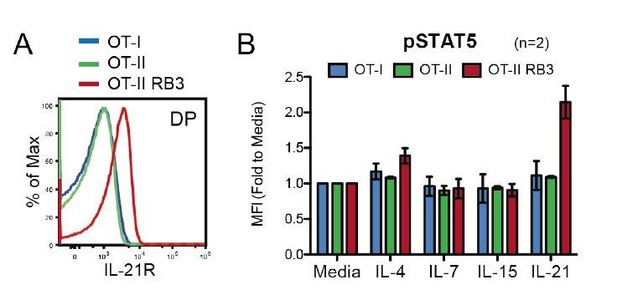
OT-II RB3 mice show increased IL-21R expression and IL-21-induced STAT5 activation.
(A) IL-21R expression on DP thymocytes from OT-I, OT-II, and OT-II RB3 mice. (B) p-STAT5 expression in DP cells from OT-I, OT-II, and OT-II RB3 mice that were stimulated with the indicated cytokines for 10mins. Experimental conditions are the same as performed in Figure 5E. Bar graph shows mean +/- SEM. N=2 mice/group.
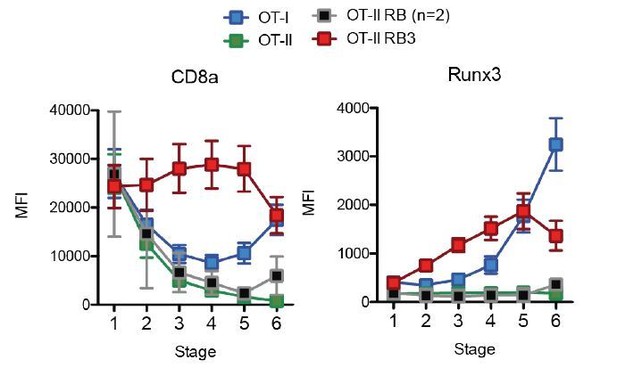
Acceleration of CD8-lineage commitment in HDAC3-deficient thymocytes.
Expression of Runx3 and CD8α during stages of lineage commitment (Stages 1-6), from OT-I, OT-II, and OT-II RB3 straight bone marrow chimeras. CD69-versus-CCR7 plots were gated from DN-removed, CD45.2+ cells. Plots show mean ± SEM of MFI from 4-5 mice per group from three independent experiments, except N=2 mice for OT-II RB.
Additional files
-
Supplementary file 1
Primers used for ChIP-seq and qChIP.
- https://doi.org/10.7554/eLife.43821.022
-
Transparent reporting form
- https://doi.org/10.7554/eLife.43821.023





