Luciferase-LOV BRET enables versatile and specific transcriptional readout of cellular protein-protein interactions
Figures
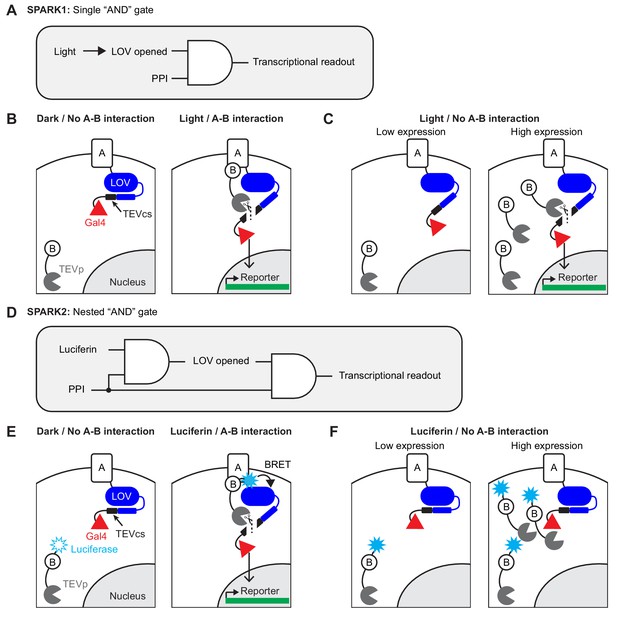
Motivation and design for SPARK2.
(A) Logic diagram for single ‘AND’ gate used in first-generation SPARK1 (Kim et al., 2017). Transcription of the reporter gene requires both light (to open the LOV domain) AND a protein-protein interaction (PPI). (B) Schematic of SPARK1. Protein A is fused to the LOV domain, protease cleavage site (TEVcs), and transcription factor (TF, Gal4). Protein B is fused to a low-affinity variant of the protease TEVp. In the absence of an interaction between proteins A and B, TEVp recognition of and binding to TEVcs is minimal. Furthermore in the absence of light, the LOV domain cages the TEVcs, protecting it from spurious cleavage by the TEVp. If both light and an A-B interaction are present, then TEVp is recruited to the exposed TEVcs, resulting in the release of the TF Gal4 to the nucleus where it can drive expression of an exogenous reporter gene. (C) Schematic of PPI-independent background at high SPARK1 expression levels. If protein B-TEVp levels are sufficiently high, TEVp cleavage of the TEVcs could occur even in the absence of protein A-protein B interaction, when light is present. (D) Logic diagram for nested ‘AND’ gate used in second-generation SPARK2. Both TEVp recruitment to TEVcs and LOV domain opening (via Luciferase-LOV BRET) require a PPI. This double filter for PPI increases the specificity of SPARK2 for the PPI of interest and reduces PPI-independent background. (E) Schematic of SPARK2. A blue light-emitting luciferase is fused to protein B and TEVp. Instead of externally supplied light, the luciferase’s substrate luciferin is supplied as a small-molecule drug. When there is an A-B interaction and luciferin is present, the luciferase activates the LOV domain via BRET, allowing the TEVp to cleave the now-accessible TEVcs. (F) Schematic of the reduction in background signal in the Luciferin/No PPI condition with SPARK2. Even if the luciferase-arrestin-TEVp is expressed at higher levels, TEVp cleavage of the TEVcs does not occur in the Luciferin/no PPI condition, as the LOV domain remains caged.
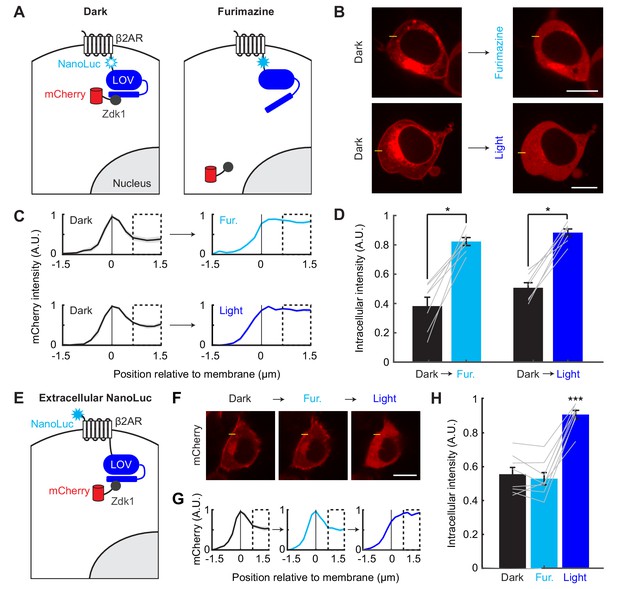
Efficient and proximity-dependent NanoLuc-LOV BRET detected using LOVTRAP.
(A) Schematic of NanoLuc fused to membrane-localized protein β2AR and asLOV2 in LOVTRAP. When asLOV2 is activated by NanoLuc (or blue light), mCherry-Zdk1 dissociates from the C-terminus of asLOV2. The asLOV2 contains a V416L mutation that slows its off kinetics (return to the dark state conformation), and consequently slows the rate of Zdk1 recapture (V416L reset t1/2=496 s). (B) Example immunofluorescence images of HEK293T cells expressing mCherry-Zdk1 and β2AR-NanoLuc-LOV. Left: mCherry expression is visible along the cell membrane in the dark. Right: following either furimazine for 1 min or blue light for 10 s, mCherry-Zdk1 relocalizes to the cytoplasm. The yellow line indicates pixels quantified for mCherry signal. Scale bars, 11 µm. (C) Mean normalized mCherry fluorescence measured across the cell membrane for the same cells before and after furimazine (top), or before and after blue light (bottom; n = 7 cells for both conditions). Dashed box indicates intracellular region quantified in panel D). Data plotted as mean ±s.e.m. (D) The mean normalized cytosolic mCherry fluorescence increased following either furimazine or light exposure (Dark vs Furimazine: 0.38 ± 0.061 vs 0.82 ± 0.027, n = 7 cells, Wilcoxon’s signed-rank test, *p=0.016. Dark vs Light: 0.51 ± 0.035 vs 0.88 ± 0.025, n = 7 cells, Wilcoxon’s signed-rank test, *p=0.016). The Furimazine/Dark ratio of cytosolic mCherry fluorescence was comparable to the Light/Dark ratio (Furimazine/Dark: 2.75 ± 0.68, Light/Dark: 1.79 ± 0.12, n = 7 cells each group, Wilcoxon’s ranksum test: p=0.84). Data plotted as mean ±s.e.m. (E) Schematic of NanoLuc fused to extracellular N-terminus of β2AR-asLOV2 in LOVTRAP. When NanoLuc is expressed extracellularly, we do not expect to observe NanoLuc-asLOV activation with furimazine. As a result, mCherry-Zdk1 should remain at the membrane. (F) Example immunofluorescence images of HEK293T cells expressing mCherry-Zdk1 and NanoLuc-β2AR-LOV. Left: mCherry expression is visible along the cell membrane in the dark. Middle: following furimazine for 1 min, mCherry-Zdk1 remains along the cell membrane. Right: following 10 s of blue light, mCherry-Zdk1 relocalizes to the cytosol. The yellow line indicates pixels quantified for mCherry signal. Scale bars, 11 µm. (G) Mean normalized mCherry fluorescence measured across the cell membrane for the same cells in the dark, after furimazine, and then after blue light (n = 8 cells). Dashed box indicates intracellular region quantified in panel H). Data plotted as mean ±s.e.m. (H) With the extracellular NanoLuc, the mean normalized cytosolic mCherry fluorescence did not increase following furimazine. However, subsequent blue light exposure resulted in an increase in the mean normalized cytosolic mCherry fluorescence (Dark: 0.56 ± 0.040; Furimazine: 0.53 ± 0.036; Light: 0.91 ± 0.025; n = 8 cells each group, 1-way ANOVA, F(2,21)=37.49, p = 1.18e-7; Tukey’s multiple comparison’s test, ***p < 0.001 Light compared to all other conditions). Data plotted as mean ± s.e.m. See also Figure 2—figure supplement 1.
-
Figure 2—source data 1
Excel spreadsheet containing fluorescence intensity values used to generate panels C-D and G-H.
- https://doi.org/10.7554/eLife.43826.006
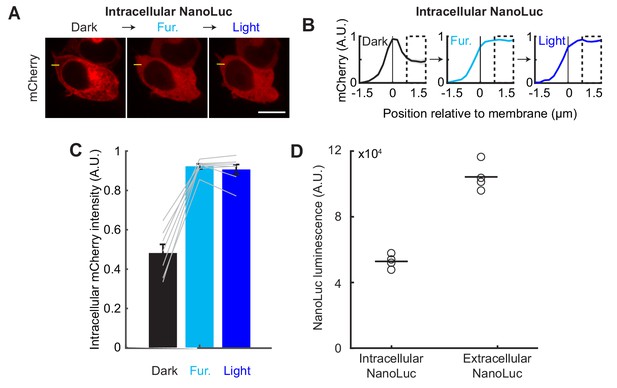
Quantification and controls for LOVTRAP detection of NanoLuc-LOV activation.
(A) HEK293T cells were transfected as in Figure 2A. Example sequential immunofluorescence images of mCherry-Zdk1 during baseline (Dark), 10 µM furimazine, and blue light for the intracellular β2AR-NanoLuc-asLOV2 construct. Left: mCherry expression is visible along the cell membrane in the dark. Middle: following furimazine for 1 min, mCherry-Zdk1 relocalizes to the cytosol. Right: subsequent blue light does not result in any additional mCherry-Zdk1 release to the cytosol. The yellow line indicates pixels quantified for mCherry signal. Scale bar, 11 µm. (B) Mean normalized mCherry fluorescence measured across the cell membrane for the same cells before and after furimazine and then blue light (n = 7 cells). Dashed box indicates intracellular region quantified in panel C). Data plotted as mean ± s.e.m. (C) The mean normalized cytosolic mCherry fluorescence increased following furimazine exposure. There was no an additional increase in relative mCherry cytosolic fluorescence following subsequent light exposure (Dark: 0.48 ± 0.045; Furimazine: 0.92 ± 0.012; Light: 0.91 ± 0.026; n = 7 cells each group, 1-way ANOVA, F(2,18)=67.42, p = 4.36e-9; Tukey’s multiple comparison’s test for Furimazine vs Light, p = 0.93). (D) Even though the Extracellular NanoLuc construct did not activate LOVTRAP, NanoLuc bioluminescence measured in the Extracellular condition was greater than in the Intracellular condition (Intracellular: 104 × 104 ± 4.3 × 104; Extracellular: 53 × 104 ± 2.1 × 104; n = 4 replicates, Wilcoxon’s ranksum test, p = 0.03).
-
Figure 2—figure supplement 1—source data 1
Excel spreadsheet containing fluorescence intensity and luminescence values used to generate panels B-D.
- https://doi.org/10.7554/eLife.43826.005
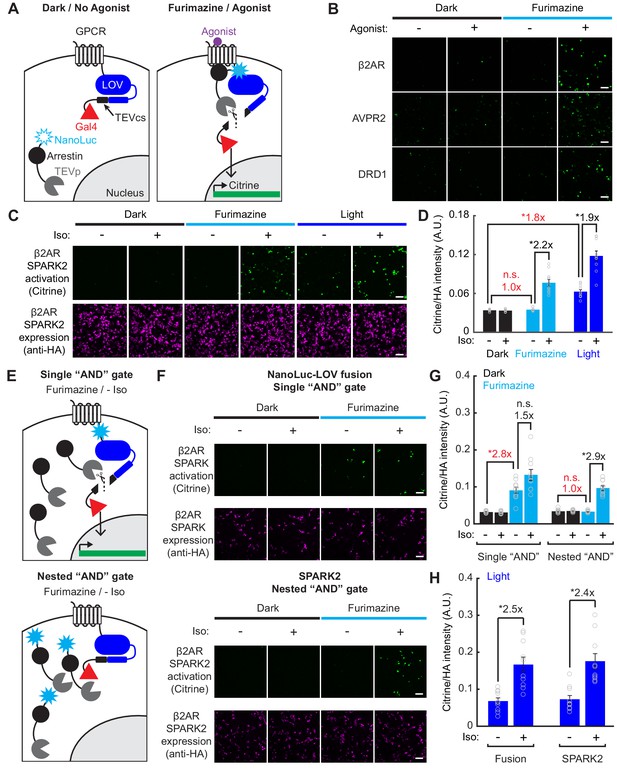
Reduced background during transcriptional read-out of PPIs with a nested ‘AND’ gate in SPARK2.
(A) Schematic of NanoLuc fused to arrestin-TEVp, co-expressed with GPCR-LOV-TEVcs-Gal4 and UAS-Citrine. In the dark and in the absence of the GPCR’s agonist, the TEVcs is caged by the LOV domain and the NanoLuc-arrestin-TEVp is not in proximity to the TEVcs. In the presence of both furimazine and agonist, NanoLuc-arrestin-TEVp is recruited to the GPCR, allowing NanoLuc to uncage eLOV and the TEVp to cleave the TEVcs. The released Gal4 then drives transcription of a UAS reporter gene. (B) Example immunofluorescence images of UAS-Citrine expression in HEK293T cells ~ 8 hr following 10 µM furimazine and agonist (20 min 100 µM isoetharine for β2AR, 15 min 10 µM vasopressin for AVPR2, 2 hr 10 µM dopamine for DRD1). Control conditions without agonist and/or without furimazine had low Citrine expression. Scale bars, 100 µm. (C) HEK293T cells were transfected with β2AR SPARK2 components as in panel A). SPARK2 activation (Citrine reporter) and SPARK2 expression (HA tag on NanoLuc-arrestin-TEVp) were imaged ~8 hr after 15 min of exposure to 10 µM furimazine or light and 10 µM isoetharine. (D) All HA-positive cells were quantified by their Citrine to HA fluorescence intensity ratio. Background fold changes in Furimazine/-Iso and Light/-Iso signal are displayed in red, while ±Ligand signal ratios are displayed in black. There was no additional background signal in the Furimazine/-Iso condition compared to the Dark/-Iso condition (1.0-fold), whereas the Light/-Iso condition had a significant 1.8-fold increase in background compared to the Dark/-Iso condition. There was a 2.2-fold ± Iso signal ratio with furimazine, and a 1.9-fold ± Iso signal ratio with light (n = 10 FOVs each group, two-way ANOVA, interaction F(2,54)=25.49, p = 1.61e-8; Tukey’s multiple comparison’s test, *p < 0.001). Data plotted as mean ± s.e.m. (E) Top: Schematic illustrating potential for background TEVcs cleavage in the absence of a PPI using a luciferase-mediated single ‘AND’ gate (NanoLuc directly fused to N-terminus of LOV domain). Bottom: Schematic illustrating negligible background TEVcs cleavage in the absence of a PPI using the luciferase-mediated nested ‘AND’ gate in SPARK2 (NanoLuc fused to arrestin-TEVp). (F) HEK293T cells were transfected either with a modified β2AR-NanoLuc-LOV direct fusion SPARK construct and Arrestin-TEVp (top) or β2AR SPARK2 components (bottom). SPARK activation (Citrine reporter) and SPARK expression (HA tag on Arrestin-TEVp or NanoLuc-arrestin-TEVp) were imaged ~8 hr after continuous exposure to 10 µM furimazine and 10 µM isoetharine. Scale bars, 100 µm. (G) Data were quantified as in panel D). In the modified fusion SPARK (single ‘AND’ gate), there was not a robust ±Iso signal ratio with furimazine (1.5-fold, not significant, n.s.), and a 2.8-fold background increase due to furimazine alone (n = 10 FOVs each group, two-way ANOVA, interaction F(2,54)=14.11, p = 1.18e-5; Tukey’s multiple comparison’s test, *p < 0.001). In SPARK2 (nested ‘AND’ gate), there was a 2.9-fold ± Iso signal ratio with furimazine, and no significant background due to furimazine alone (1.0-fold, n.s.; n = 10 FOVs each group, two-way ANOVA, interaction F(2,54)=9.82, p = 2.0e-4; Tukey’s multiple comparison’s test, *p < 0.001). Data plotted as mean ± s.e.m. (H) HEK293T cells transfected as in panel F) were exposed to blue light for 5 min, with or without 10 µM isoetharine. The fusion and SPARK2 conditions resulted in similar ±Iso signal ratios with light, 2.5- and 2.4-fold, respectively (Data analyzed in two-way ANOVA with data from panel (G). Fusion: n = 10 FOVs each group, two-way ANOVA, interaction F(2,54)=14.11, p = 1.18e-5; SPARK2: n=10 FOVs each group, two-way ANOVA, interaction F(2,54) = 9.82, p = 2.0e-4; Tukey’s multiple comparison’s test, *p < 0.01). Data plotted as mean ± s.e.m. See also Figure 3—figure supplement 1.
-
Figure 3—source data 1
Excel spreadsheet containing fluorescence ratio intensity values used to generate panels D, G, and H.
- https://doi.org/10.7554/eLife.43826.010
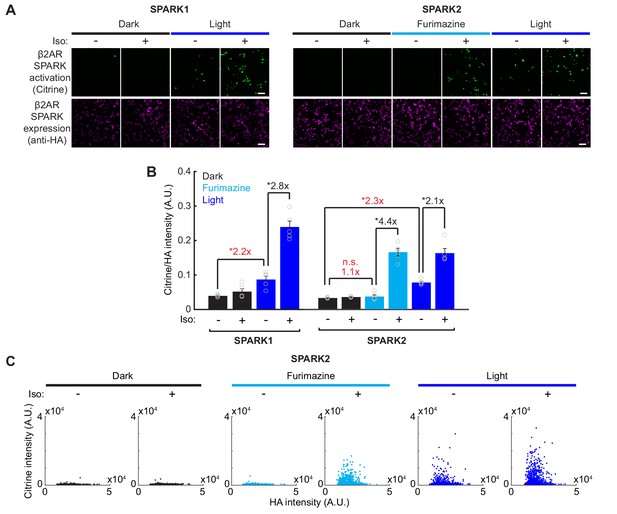
Direct comparison of SPARK1 versus SPARK2 and raw fluorescence intensities for SPARK2 characterization.
(A) HEK293T cells were transfected with either β2AR SPARK1 (arrestin-TEVp) or SPARK2 (NanoLuc-arrestin-TEVp) and UAS-Citrine. The same transmembrane construct was used in both conditions (β2AR-eLOV-TEVcs-Gal4). SPARK1/2 activation (Citrine reporter) and SPARK1/2 expression (HA tag on arrestin-TEVp or NanoLuc-arrestin-TEVp) were imaged ~8 hr after 15 min of exposure to 10 µM furimazine or blue light, and 10 µM isoetharine. Scale bars, 100 µm. (B) All HA-positive cells were quantified by their Citrine to HA fluorescence intensity ratio. Background fold changes in Furimazine/-Iso and Light/-Iso signal are displayed in red, while ±Ligand signal ratios are displayed in black. With SPARK1, there was a 2.8-fold ± Iso signal ratio with light, but a significant 2.2-fold background increase due to light alone (n=5 FOVs each group, two-way ANOVA, interaction F(1,16)=41.85, p = 7.74e-6; Tukey's multiple comparison's test, *p < 0.05). With SPARK2, there was a 4.4-fold ± Iso signal ratio with furimazine, and no significant background due to furimazine alone (1.1-fold, n.s.). As expected, SPARK2 with light-gating had a lower 2.1-fold ± Iso signal ratio, and a significant 2.3-fold background increase due to light alone (n = 5 FOVs each group, two-way ANOVA, interaction F(2,24)=36.74, p = 4.96e-8; Tukey’s multiple comparison’s test, *p < 0.01). Data plotted as mean ± s.e.m. (C) All individual cells analyzed in the SPARK2 experiment in Figure 3C–D plotted according to their mean HA and Citrine intensities.
-
Figure 3—figure supplement 1—source data 1
Excel spreadsheet containing fluorescence intensity ratio values used to generate panel B.
- https://doi.org/10.7554/eLife.43826.009
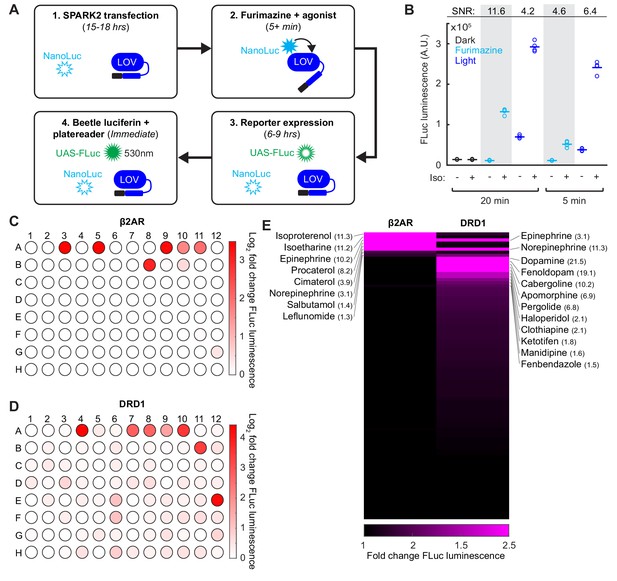
SPARK2 is compatible with an orthogonal luciferase reporter to enable high-throughput drug-screens for GPCR activation.
(A) Schematic of the general timeline for SPARK2 assays using an orthogonal luciferase reporter such as FLuc. For simplicity, only the luciferases and LOV domain are illustrated (unfilled luciferases indicate no luminescence; filled luciferases indicate luciferin substrate-mediated luminescence). (B) HEK293T were transfected with β2AR SPARK2 components and UAS-FLuc.~8 hr after SPARK2 labeling with 10 µM furimazine/blue light and 10 µM isoetharine, the UAS-FLuc luminescence recorded using a platereader (n = 4 replicates each condition). The highest ±Iso signal ratio was achieved using 20 min exposure to furimazine (11.6-fold). 5 min of blue light resulted in a ± Iso signal ratio of 6.4-fold, whereas 20 min of blue light or 5 min of furamizine resulted in similar ±Iso signal ratios of 4.2- and 4.6-fold, respectively. (C–D) HEK293T cells were transfected with either β2AR or DRD1 SPARK2 components and UAS-FLuc. Cells were exposed to 10 µM furimazine and 5 µM each of 92 different drugs for 1 hr (BioMol FDA-approved compound library). In addition, we performed two DMSO vehicle measurements (A1–A2), and two positive control spike-ins (isoetharine in A3 and dopamine in A4). ~8 hr later, FLuc luminescence was measured on a platereader. The Log2 fold change in FLuc luminescence is plotted corresponding to each compound’s position in the 96-well plate. Data plotted as the average of two biological replicates. Compound list and positions in Figure 4—source data 2. (E) Sorted heatmap of fold changes in FLuc luminescence for β2AR and DRD1 SPARK2 for all 94 compounds (excluding DMSO replicates). Example compound hits for each receptor are labeled. Heatmap thresholded from 1 to 2.5 for visualization, mean fold-changes for selected hits listed in parentheses. Fold-changes for all compounds listed in Figure 4—source data 2.
-
Figure 4—source data 1
Excel spreadsheet containing luminescence values used to generate panel B.
- https://doi.org/10.7554/eLife.43826.014
-
Figure 4—source data 2
Excel spreadsheet containing SPARK2 drug screen compounds and mean fold-changes (n = 2 biological replicates) to generate panels C-E.
- https://doi.org/10.7554/eLife.43826.015
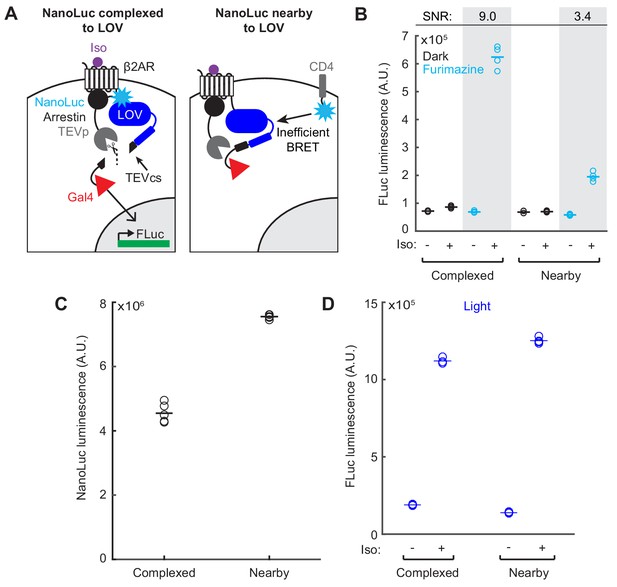
Quantification and controls for proximity-dependence of SPARK2.
(A) Schematic for testing whether NanoLuc activation of eLOV is proximity-dependent. NanoLuc was fused either to arrestin-TEVp (left, Complexed), or expressed at the membrane as CD4-NanoLuc (right, Nearby). (B) HEK293T cells were transfected with constructs in Panel A), with either the Complexed or Nearby (membrane-bound NanoLuc) configurations of β2AR SPARK2. The UAS-FLuc reporter luminescence was measured ~8 hr after 20 min of exposure to 10 µM furimazine and 10 µM isoetharine (n = 4 replicates each condition). The Complexed condition resulted in a ± Iso signal ratio of 9.0-fold, and the Nearby condition resulted in a ± Iso signal ratio of 3.4-fold. (C) Even though the Nearby NanoLuc construct did not activate LOV as efficiently as the Complexed NanoLuc construct, NanoLuc bioluminescence measured in the Nearby condition was greater than in the Complexed condition (Complexed: 4.55e6 ±1.46e5, Nearby: 7.55e6 ±3.47e4; n = 5 technical replicates each condition; Student’s t-test, t8 = −20.01, p=4.05e-8). (D) FLuc reporter luminescence measurements following 10 min of blue light with and without 10 µM isoetharine were similar between the Complexed and Nearby conditions (n = 4 replicates each condition).
-
Figure 4—figure supplement 1—source data 1
Excel spreadsheet containing luminescence values used to generate panels B-D.
- https://doi.org/10.7554/eLife.43826.013
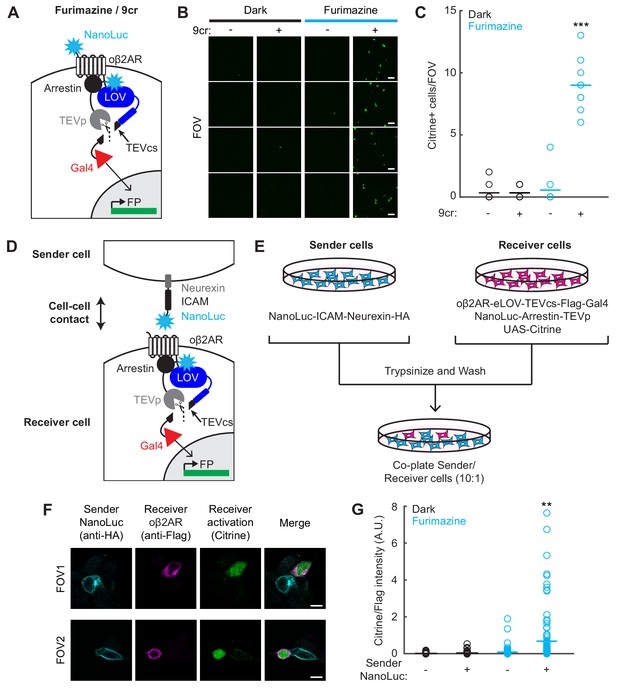
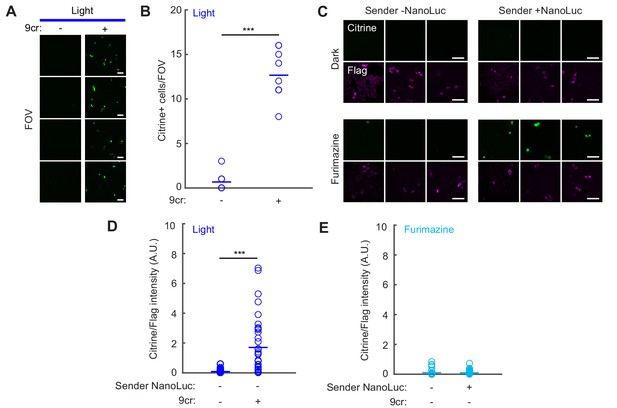
Detection of cell-cell contacts using NanoLuc-oβ2AR activation with SPARK2.
(A) Schematic of an extracellular NanoLuc fused to oβ2AR-eLOV-TEVcs-Gal4, co-expressed with NanoLuc-arrestin-TEVp and UAS-Citrine. (B) Citrine expression in HEK293T cells infected with lentiviruses expressing components in A) and exposed to 10 µM furimazine and 50 µM 9-cis-retinal (9cr) for 30 min. In control conditions, 9cr was omitted and/or furimazine was omitted. Scale bar, 100 µm. (C) There were significantly more Citrine-positive cells per field of view ~24 hr following furimazine and 9cr exposure compared to in control conditions (Dark/−9cr: 0.33 ± 0.24; Dark/+9cr: 0.33 ± 0.17; Fur/−9cr: 0.56 ± 0.44; Fur/+9cr: 9.0 ± 0.71; n = 9 fields of view each condition; two-way ANOVA, interaction F(1,32)=91.32, p = 6.80e-11; Tukey’s multiple comparison’s test ***p < 0.001 compared to all other conditions). (D) Schematic for detecting cell-cell contacts in which a Sender cell expresses NanoLuc-ICAM-Neurexin, and a Receiver cell expresses oβ2AR-eLOV-TEVcs-Gal4, NanoLuc-Arrestin-TEVp, and UAS-Citrine. (E) Experimental paradigm for co-plating Sender and Receiver cells in a 10:1 ratio. (F) Example immunofluorescence images of Citrine-positive Receiver cells that are adjacent to HA-positive Sender cells expressing NanoLuc. The Flag stains against Receiver cells expressing the oβ2AR SPARK2 components. 87% of all detected Citrine-positive Receiver cells were adjacent to an HA-positive Sender cell. Scale bars, 10 µm. (G) Quantification of Citrine/Flag fluorescence intensity ratios for all Flag-positive cells. The Citrine/Flag intensity ratio was significantly higher following furimazine exposure when Sender NanoLuc was expressed compared to in control conditions where NanoLuc was not expressed in the Sender cells (Dark/Sender -NanoLuc: 0.016 ± 0.0037, n = 92 cells; Dark/Sender +NanoLuc: 0.039 ± 0.0074, n = 109 cells; Fur/Sender -NanoLuc: 0.082 ± 0.025, n = 95 cells; Fur/Sender +NanoLuc: 0.68 ± 0.14, n = 104 cells; two-way ANOVA, interaction F(1,396)=15.6, p = 9.28e-5; Tukey’s multiple comparison’s test, **p < 0.001 compared to all other conditions). See also Figure 5—figure supplement 1.
-
Figure 5—source data 1
Excel spreadsheet containing cell count and fluorescence intensity ratio values used to generate panels C and G.
- https://doi.org/10.7554/eLife.43826.019
Quantification and controls for detection of cell-cell contacts using NanoLuc-oβ2AR activation with SPARK2.
(A) HEK293T cells were infected with lentiviruses as in Figure 5A. Immunofluorescence images of Citrine expression were taken ~24 hr following 10 min of blue light to activate NanoLuc-oβ2AR-eLOV-TEVcs-Gal4 and NanoLuc-Arrestin-TEVp, with and without 50 µM 9-cis-retinal. Scale bar, 100 µm. (B) Data quantified from images in panel A). There are significantly more Citrine-positive cells per field of view when blue light is delivered with 9cr versus without 9cr (Light/−9cr: 0.67 ± 0.33; Light/+9cr: 12.67 ± 0.91; n = 9 fields of view each condition; Wilcoxon’s ranksum test ***p=4.11e-5). (C) HEK293T cells were transfected as in Figure 5D–F. Example immunofluorescence images of Citrine expression and Flag antibody staining (detecting oβ2AR fusion construct) were taken ~8 hr following 10 min exposure to 50 µM 9cr with or without 10 µM furimazine. Left: Sender cells were untransfected. Right: Sender cells were transfected with Nrxn-ICAM-NanoLuc. Three example fields of view from different biological replicates are shown. Scale bars, 100 µm. (D) The mean Citrine/Flag fluorescence intensity ratio in Receiver cells was higher following 10 min of blue light with 9cr versus without 9cr (Light/+9cr: 1.70 ± 0.33, n = 35 cells; Light/−9cr: 0.084 ± 0.028, n = 26 cells; Wilcoxon’s ranksum test, ***p = 2.34e-7). NanoLuc was not expressed in the Sender cells. (E) The mean Citrine/Flag fluorescence intensity ratio was negligible in Receiver cells in conditions when there was no 9cr present during the 10 min of furimazine (Fur/−9cr/Sender -NanoLuc: 0.10 ± 0.036, n = 35 cells; Fur/−9cr/Sender + NanoLuc: 0.099 ± 0.024, n = 37 cells).
-
Figure 5—figure supplement 1—source data 1
Excel spreadsheet containing cell counts and fluorescence intensity ratio values used to generate panels B and D-E.
- https://doi.org/10.7554/eLife.43826.018
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (HEK293T) | HEK293T | ATCC | Tested negative for mycoplasma. | |
| Antibody | Rabbit anti-HA | Cell Signaling Technology | C29F4 | 1:100 |
| Antibody | Goat anti-Rabbit AlexaFluor 647 | Life Technologies | A27040 | 1:1000 |
| Antibody | Mouse anti-Flag | Sigma | F3165 | 1:300 |
| Antibody | Goat anti-Mouse AlexaFluor 568 | Life Technologies | A11004 | 1:1000 |
| Recombinant DNA reagent | See Table 1 for all plasmids used or generated in this study. | |||
| Commercial assay or kit | Nano-Glo Live Cell Assay System | Promega | 1:100 substrate | |
| Commercial assay or kit | Bright-Glo Luciferase Assay System | Promega | 1:1 substrate | |
| Software, algorithm | Fiji | Schindelin et al., 2012 | ||
| Software, algorithm | CellSegm toolbox | Hodneland et al., 2013 | ||
| Software, algorithm | MATLAB R2017a | Mathworks |
Plasmids used in this study.
https://doi.org/10.7554/eLife.43826.020| Name | Description | Vector-Promoter | Addgene |
|---|---|---|---|
| P1 | HA-β2AR-NNES-NanoLuc-15aa linker-AsLOV2(V416L) | pAAV-CMV | 125224 |
| P2 | gLuc sp-NanoLuc-HA-β2AR-NNES-AsLOV2(V416L) | pAAV-CMV | 125225 |
| P3 | mCherry-GSGS linker-Zdk1 | pAAV-CMV | 125226 |
| P4 | DRD1-NNES-eLOV-TEVcs-Flag-Gal4-V5 | pAAV-CMV | 125227 |
| P5 | AVPR2-NNES-eLOV-TEVcs-Flag-Gal4-V5 | pAAV-CMV | 104844 |
| P6 | β2AR-NNES-eLOV-TEVcs-Flag-Gal4-V5 | pAAV-CMV | 104841 |
| P7 | NanoLuc-15aa linker-βarrestin2-HA-GS linker-TEVp | pAAV-CMV | 125228 |
| P8 | UAS-Citrine | pAAV-UAS | 104839 |
| P9 | βarrestin2-HA-GS linker-TEVp | pAAV-CMV | 104845 |
| P10 | UAS-FLuc | pAAV-UAS | 104840 |
| P11 | IgK sp-HA-CD4-10aa linker-CIBN-NNES-NanoLuc | pAAV-CMV | 125229 |
| P12 | gLuc sp-NanoLuc-HA-oβ2AR-TS-eLOV-TEVcs-Flag-Gal4 | pLX208-CMV | 125230 |
| P13 | NanoLuc-15aa linker-βarrestin2-HA-GS linker-TEVp | pLX208-CMV | 125231 |
| P14 | UAS-Citrine | pFPGW-UAS | 125232 |
| P15 | gLuc sp-NanoLuc-15aa linker-ICAM-Nrxn3b-HA | pLX208-CMV | 125233 |
| P16 | gLuc sp-GS linker-HiBit-Flag-oβ2AR-TS-NNES-eLOV-TEVcs-Flag-Gal4-V5 | pAAV-CMV | 125234 |
| P17 | NanoLuc-15aa linker-βarrestin2-no HA-GS linker-TEVp | pAAV-CMV | 125235 |
| P18 | β2AR-NNES-NanoLuc-eLOV-TEVcs-Flag-Gal4-V5 | pAAV-CMV | 125236 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.43826.021





