A regulatory module controlling stress-induced cell cycle arrest in Arabidopsis
Figures
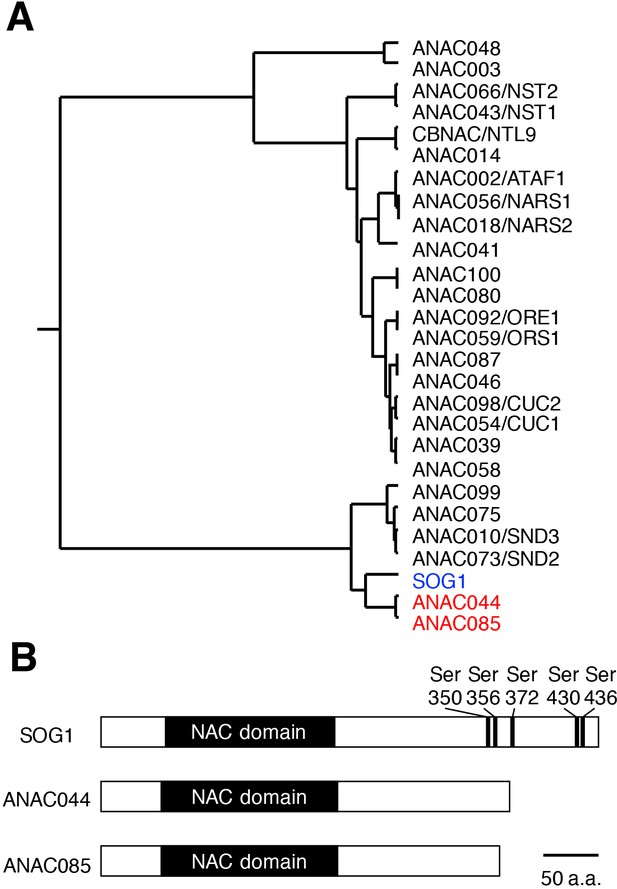
Similarities among SOG1, ANAC044 and ANAC085.
(A) Phylogenetic tree of the NAC transcription factors in Arabidopsis. SOG1 (blue letters) and ANAC044 and ANAC085 (red) are highlighted. (B) Protein structures of SOG1, ANAC044 and ANAC085. Serine (Ser) residues of the SQ motifs in SOG1 are indicated with numbers of the amino acid sequence.
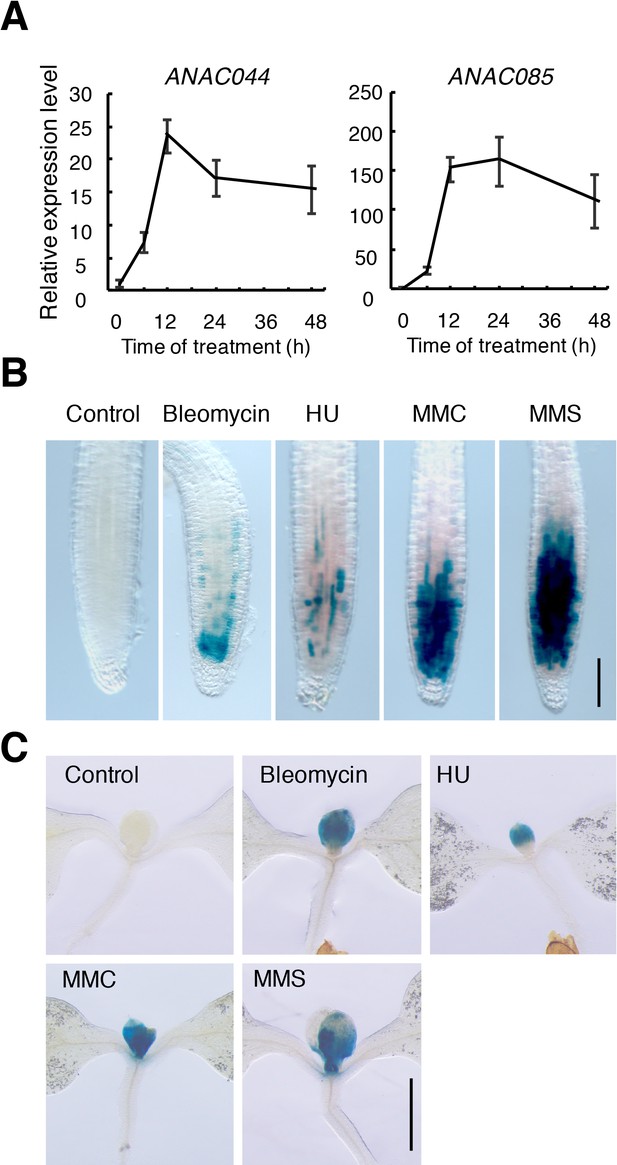
ANAC044 and ANAC085 are induced by DNA damage.
(A) Transcript levels of ANAC044 and ANAC085 after bleomycin treatment. Five-day-old WT seedlings were treated with 0.6 µg/ml bleomycin for 0, 6, 12, 24 or 48 hr. The mRNA levels were normalized to that of ACTIN2 and are indicated as relative values, with that for 0 hr set to 1. Data are presented as mean ±SD (n = 3). (B, C) Five-day-old seedlings harbouring ProANAC044:GUS were transferred to medium supplemented with or without 0.6 µg/ml bleomycin, 1.5 mM hydroxyurea (HU), 3.3 µg/ml mitomycin C (MMC) or 80 ppm methyl methanesulfonate (MMS), and grown for 24 hr. GUS-stained seedlings were observed for the root tip (B) and shoot apex (C). Bars = 100 µm (B) and 1 mm (C).
-
Figure 1—figure supplement 1—source data 1
Source data.
- https://doi.org/10.7554/eLife.43944.005
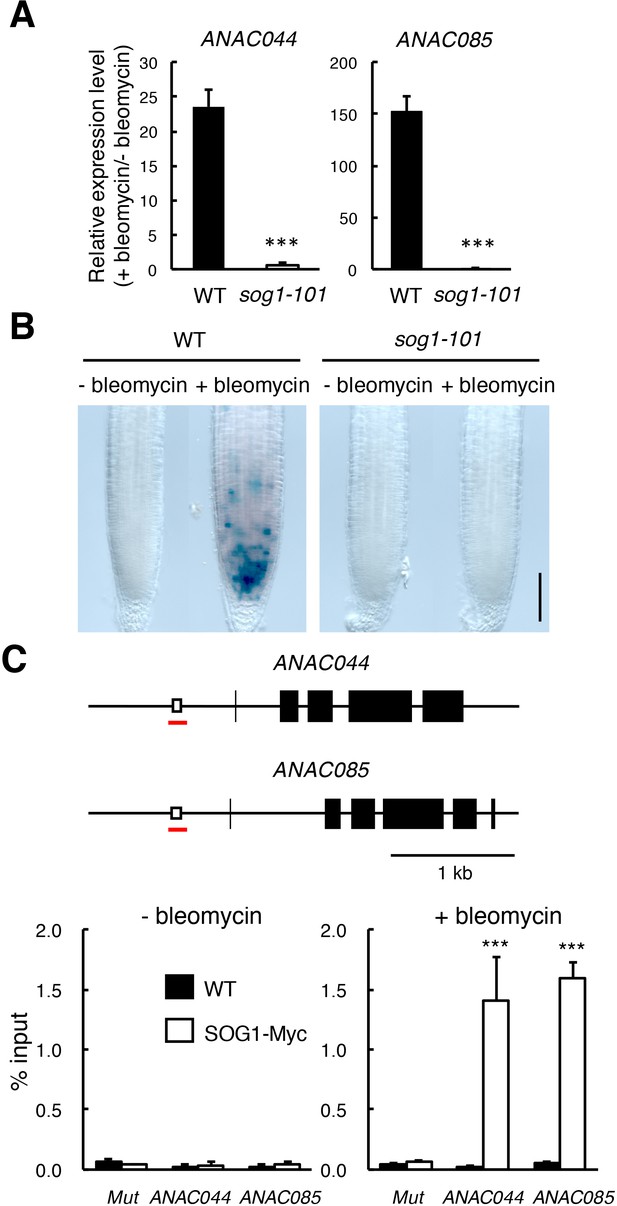
SOG1-mediated induction of ANAC044 and ANAC085.
(A) Transcript levels of ANAC044 and ANAC085 in sog1-101. Five-day-old WT and sog1-101 seedlings were treated with or without 0.6 µg/ml bleomycin for 12 hr. The mRNA levels were normalized to that of ACTIN2 and are indicated as fold induction following bleomycin treatment. Data are presented as mean ± SD (n = 3). Significant differences from WT were determined by Student’s t-test: ***, p<0.001. (B) Promoter activity of ANAC044 in the root tip. Five-day-old seedlings of WT and sog1-101 carrying ProANAC044:GUS were transferred to MS medium with or without 0.6 µg/ml bleomycin and cultured for 24 hr, followed by GUS staining. Bar = 100 µm. (C) (Upper) Schematic representation of the genomic regions of ANAC044 and ANAC085. Open and closed boxes indicate SOG1-binding motifs and exons, respectively. Red lines represent the regions amplified in ChIP-qPCR. (Lower) ChIP-qPCR assay. Two-week-old WT and ProSOG1:SOG1-Myc (SOG1-Myc) seedlings were treated with or without 0.6 µg/ml bleomycin. Chromatin bound to SOG1-Myc was collected by immunoprecipitation with anti-Myc antibodies, and qPCR was conducted to amplify the promoter regions of ANAC044 and ANAC085. Mutator-like transposon (Mut) was used as a negative control. The recovery rate of each DNA fragment was determined against input DNA. Data are presented as mean ± SD (n = 3). Significant differences from WT were determined by Student’s t-test: ***, p<0.001.
-
Figure 1—figure supplement 2—source data 1
Source data.
- https://doi.org/10.7554/eLife.43944.007
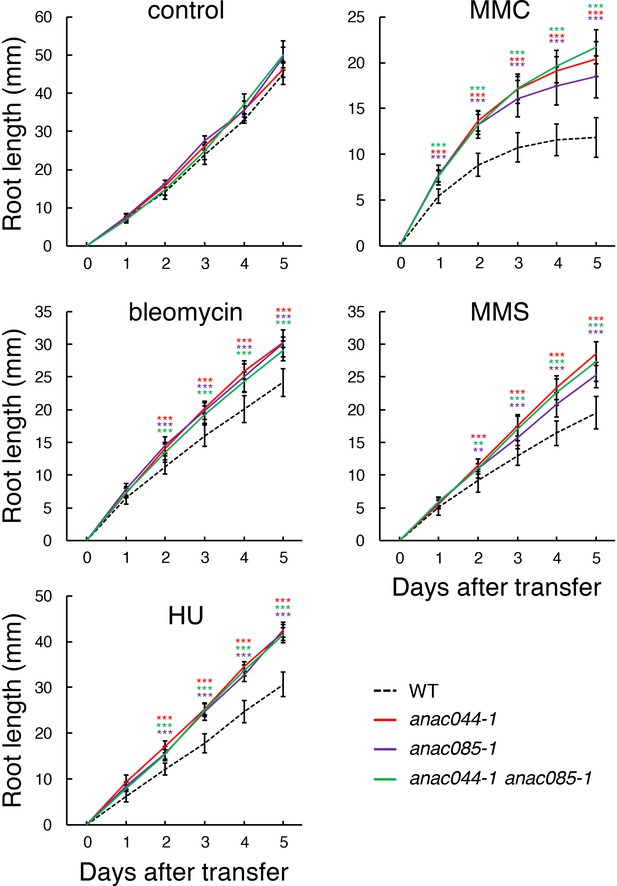
anac044 and anac085 are tolerant to DNA damage.
Five-day-old seedlings of WT, anac044-1, anac085-1 and anac044-1 anac085-1 were transferred to medium with or without 0.6 µg/ml bleomycin, 1.5 mM hydroxyurea (HU), 3.3 µg/ml mitomycin C (MMC) or 80 ppm methyl methanesulfonate (MMS), and root length was measured every 24 hr. Data are presented as mean ± SD (n = 13). Significant differences from WT were determined by Student’s t-test: **, p<0.01; ***, p<0.001.
-
Figure 2—source data 1
Source data.
- https://doi.org/10.7554/eLife.43944.013
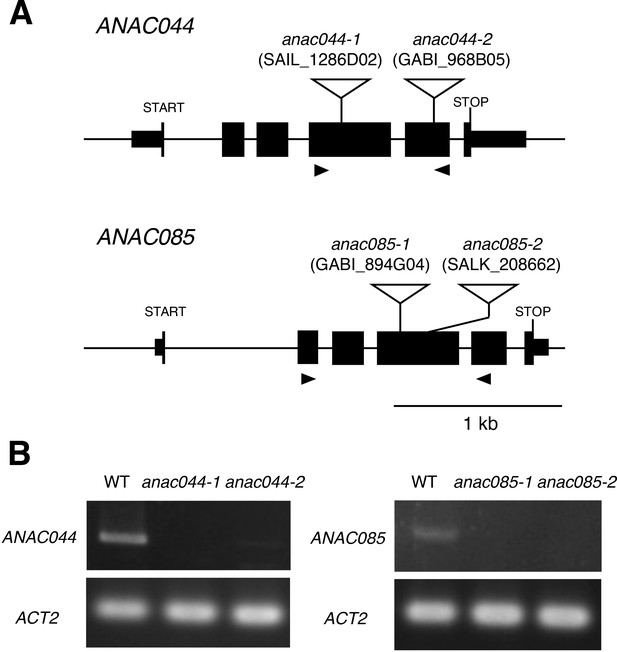
T-DNA insertion mutants of ANAC044 and ANAC085.
(A) Schematic representation of the ANAC044 and ANAC085 genes. T-DNA insertion sites in anac044-1, anac044-2, anac085-1 and anac085-2 are shown. Exons and introns are indicated by black boxes and solid lines, respectively. The positions of primers used for RT-PCR are depicted by black triangles. (B) Semi-quantitative RT-PCR analysis of the anac044 and anac085 mutants. Total RNA was extracted from five-day-old seedlings, and subjected to RT-PCR using primers shown in A. ACTIN2 (ACT2) was used as a control.
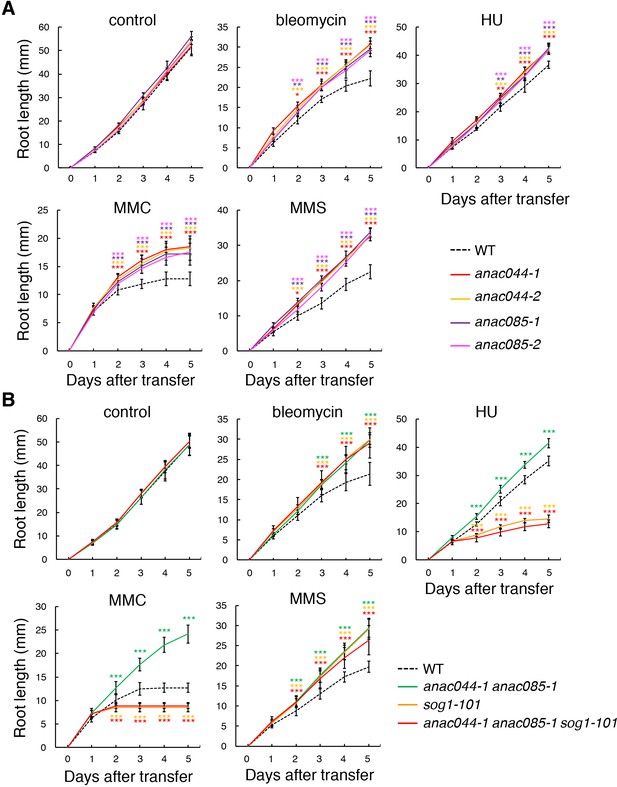
anac044 and anac085 are tolerant to various DNA-damaging agents.
Root growth of WT, anac044-1, anac044-2, anac085-1 and anac085-2 (A), and of WT, anac044-1 anac085-1, sog1-101, and anac044-1 anac085-1 sog1-101 (B). Five-day-old seedlings were transferred to medium with or without 0.6 µg/ml bleomycin, 1.5 mM hydroxyurea (HU), 3.3 µg/ml mitomycin C (MMC) or 80 ppm methyl methanesulfonate (MMS), and root length was measured every 24 hr. Data are presented as mean ± SD (n > 11). Significant differences from WT were determined by Student’s t-test: *, p<0.05; **, p<0.01; ***, p<0.001.
-
Figure 2—figure supplement 2—source data 1
Source data.
- https://doi.org/10.7554/eLife.43944.011
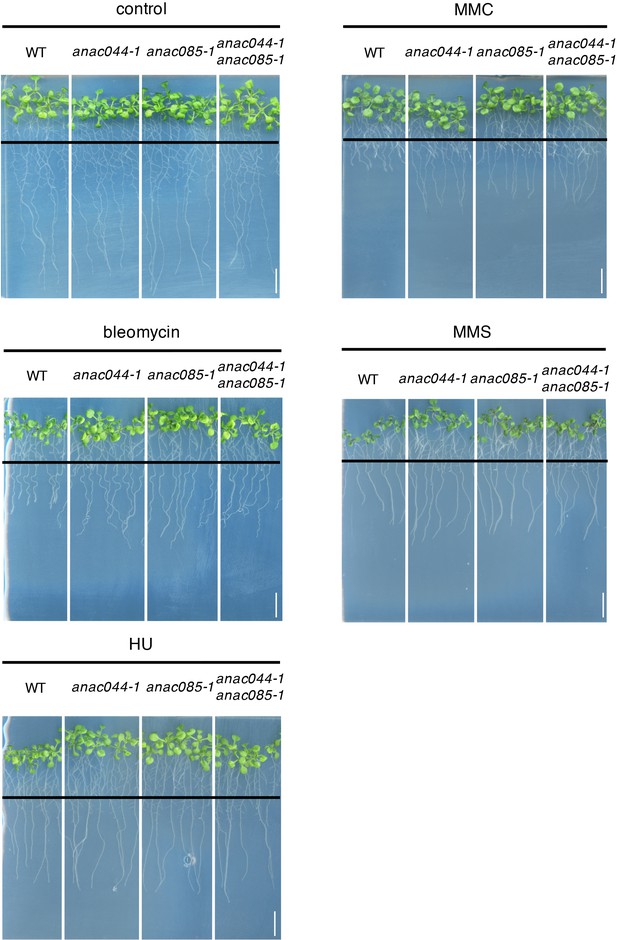
Root growth of anac044-1, anac085-1 and anac044-1 anac085-1 in the presence of DNA-damaging agents.
Five-day-old seedlings were transferred to medium with or without 0.6 µg/ml bleomycin, 1.5 mM hydroxyurea (HU), 3.3 µg/ml mitomycin C (MMC) or 80 ppm methyl methanesulfonate (MMS), and grown for a further five days. Black lines indicate the positions of root tips when seedlings were transferred onto each medium. Bar = 1 cm.
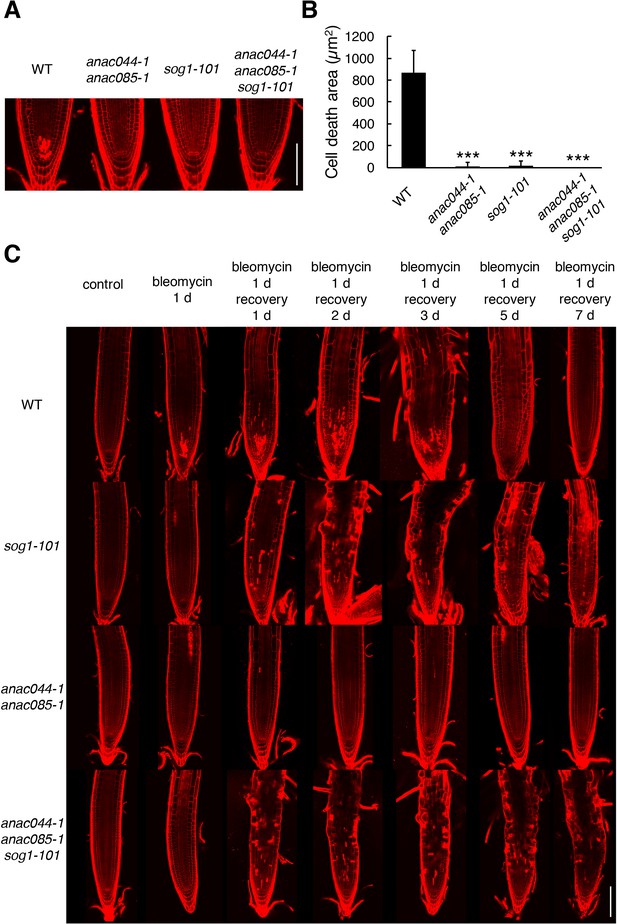
ANAC044 and ANAC085 are involved in DNA damage-induced stem cell death.
(A) Root tips of WT, anac044-1 anac085-1, sog1-101, and anac044-1 anac085-1 sog1-101. Five-day-old seedlings were transferred to medium containing 0.6 µg/ml bleomycin and grown for 24 hr, followed by propidium iodide (PI) staining. Bar = 100 µm. (B) Cell death area in the root tip. The total area of cells stained with PI (A) was measured by ImageJ software. Data are presented as mean ± SD (n > 9). Significant differences from WT were determined by Student’s t-test: ***, p<0.001. (C) Root tips of sog1-101, anac044-1 anac085-1 and anac044-1 anac085-1 sog1-101 after recovery from bleomycin-containing medium. Five-day-old seedlings were transferred to medium supplemented with 0.6 µg/ml bleomycin, and grown for 24 hr. The seedlings were then transferred back to medium without bleomycin, and grown for the indicated number of days. Root tips were observed after PI staining. Bar = 100 µm.
-
Figure 3—source data 1
Source data.
- https://doi.org/10.7554/eLife.43944.015
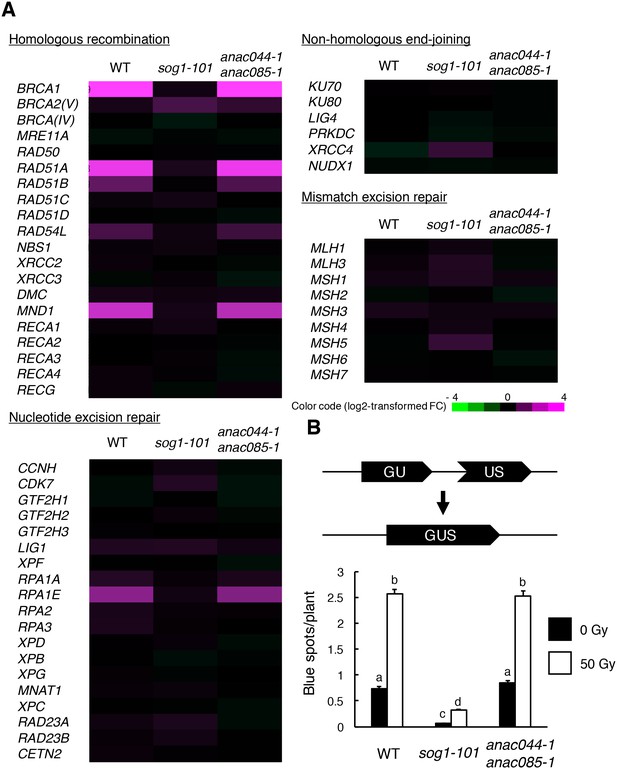
ANAC044 and ANAC085 are not required for HR-mediated DNA repair.
(A) Transcriptional response of DNA repair-related genes to bleomycin. Five-day-old seedlings of WT, sog1-101 and anac044-1 anac085-1 were treated with or without 0.6 µg/ml bleomycin for 10 hr. Total RNA was extracted from root tips and subjected to microarray analysis. Purple and green colours indicate up- and down-regulation, respectively, of genes by bleomycin treatment. (B) HR assay. The GUS reporter constructs before and after HR are shown (upper panel). Two-week-old plants of WT, sog1-101 and anac044-1 anac085-1 carrying the GUS reporter construct were irradiated with or without gamma rays (50 Gy), and grown for 3 days. Numbers of blue spots on leaves were counted. Data are presented as mean ± SD (n = 50). Different letters indicate significant differences between samples (Student’s t-test, p<0.05).
-
Figure 4—source data 1
Source data.
- https://doi.org/10.7554/eLife.43944.017
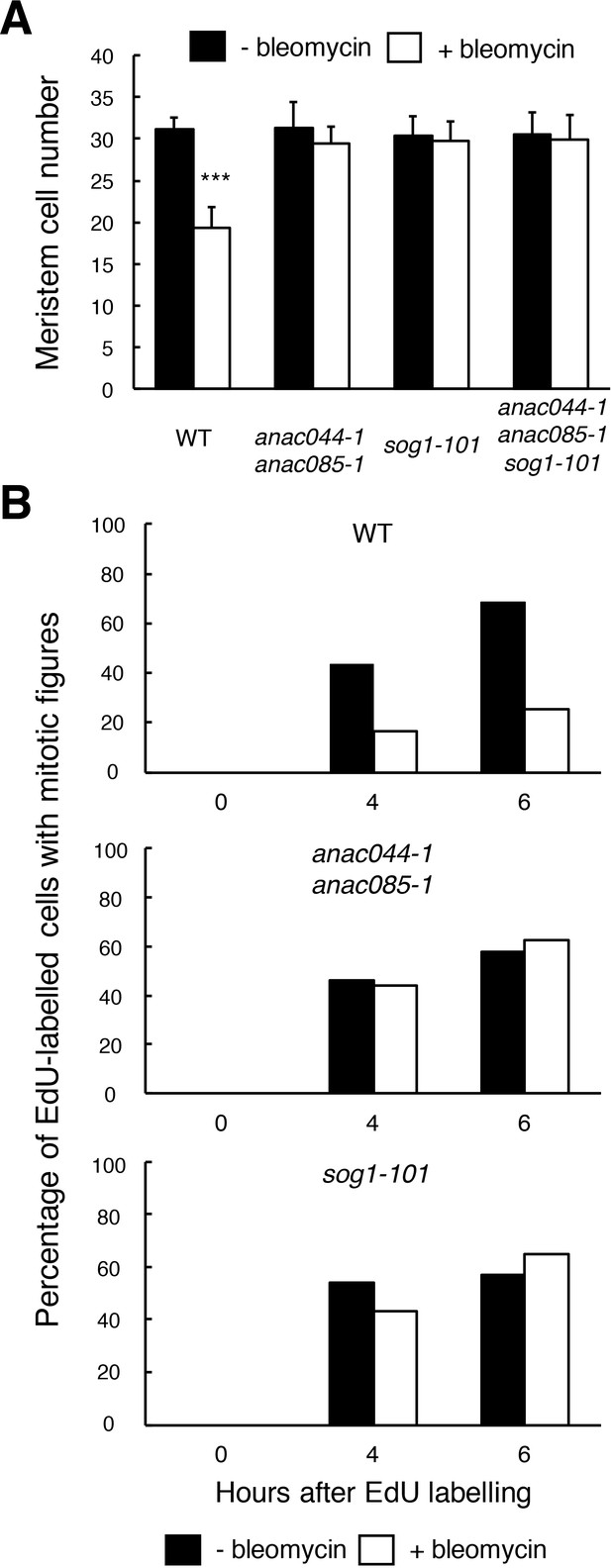
ANAC044 and ANAC085 are required for DNA damage-induced cell cycle arrest.
(A) Cell number in the meristematic zone of WT, anac044-1 anac085-1, sog1-101 and anac044-1 anac085-1 sog1-101. Five-day-old seedlings were transferred to MS medium supplemented with or without 0.6 µg/ml bleomycin, and the number of cortex cells between the QC and the first elongated cell was counted after 24 hr. Data are presented as mean ± SD (n = 20). Significant differences from the non-treated control were determined by Student’s t-test: ***, p<0.001. (B) Cell cycle progression through the G2/M phase. Five-day-old seedlings of WT, anac044-1 anac085-1 and sog1-101 were transferred to MS medium supplemented with or without 0.6 µg/ml bleomycin for 12 hr, and pulse-labelled with EdU for 15 min. Seedlings were then transferred back to MS medium with or without bleomycin, and collected at the indicated time points. Root meristematic cells were double-stained with EdU and DAPI, and cells with mitotic figures were counted. Data are presented as the percentage of EdU-labelled cells among those with mitotic figures (n > 20).
-
Figure 5—source data 1
Source data.
- https://doi.org/10.7554/eLife.43944.019
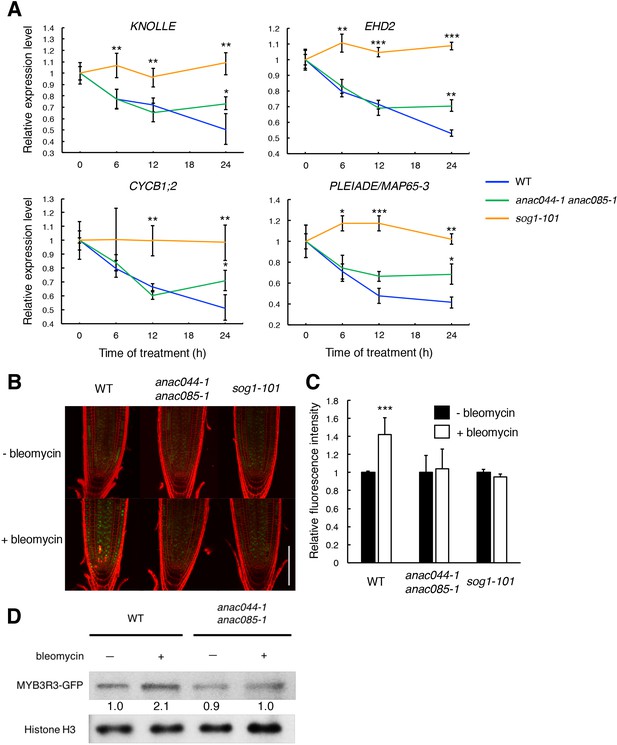
ANAC044 and ANAC085 promote Rep-MYB accumulation in response to DNA damage.
(A) Transcript levels of KNOLLE, CYCB1;2, EHD2 and PLEIADE/MAP65-3 after bleomycin treatment. Five-day-old seedlings of WT, anac044-1 anac085-1 and sog1-101 were transferred to MS medium containing 0.6 µg/ml bleomycin, and grown for 0, 6, 12 and 24 hr. Total RNA was extracted from root tips. mRNA levels were normalized to that of ACTIN2, and are indicated as relative values, with that for 0 hr set to 1. Data are presented as mean ± SD (n = 3). Significant differences from WT were determined by Student’s t-test: *, p<0.05; **, p<0.01; ***, p<0.001. (B–D) MYB3R3 accumulation in the root tip. Five-day-old seedlings of WT, anac044-1 anac085-1 and sog1-101 harbouring ProMYB3R3:MYB3R3-GFP were transferred to MS medium supplemented with or without 0.6 µg/ml bleomycin, and grown for 24 hr. GFP fluorescence was observed after counterstaining with PI (B). Bar = 100 µm. Intensities of GFP fluorescence in the root tip are shown as relative values, with that of non-treated control set to 1 (C). Data are presented as mean ± SD (n = 5). Significant differences from the non-treated control were determined by Student’s t-test: ***, p<0.001. Twenty micrograms of total protein extracted from root tips were subjected to immunoblotting using antibodies against GFP or histone H3 (D). Relative levels of MYB3R3-GFP are expressed as fold change, normalized with respect to the corresponding band of histone H3.
-
Figure 6—source data 1
Source data.
- https://doi.org/10.7554/eLife.43944.025
-
Figure 6—source data 2
Uncut blot.
- https://doi.org/10.7554/eLife.43944.026
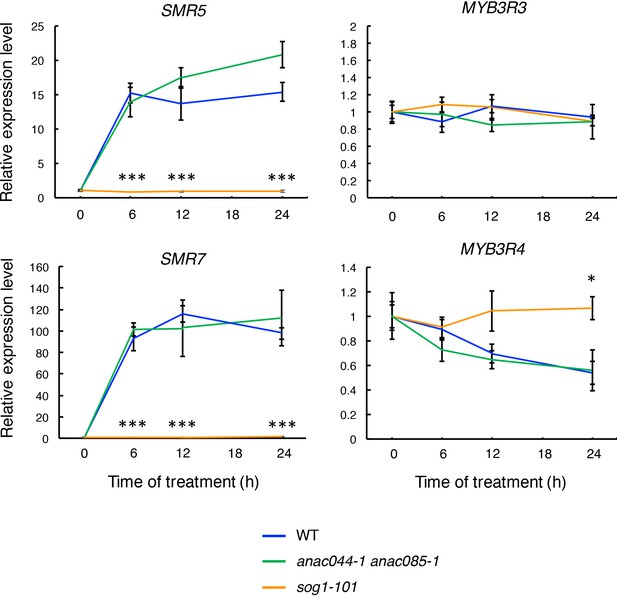
mRNA levels of SMR5, SMR7, MYB3R3 and MYB3R4 after bleomycin treatment.
Five-day-old seedlings of WT, anac044-1 anac085-1 and sog1-101 were transferred to medium containing 0.6 µg/ml bleomycin, and grown for 0, 6, 12 and 24 hr. Total RNA was extracted from root tips, and subjected to qRT-PCR analysis. mRNA levels were normalized to that of ACTIN2, and are indicated as relative values, with that for 0 hr set to 1. Data are presented as mean ± SD (n = 3). Significant differences from WT were determined by Student’s t-test: *, p<0.05; ***, p<0.001.
-
Figure 6—figure supplement 1—source data 1
Source data.
- https://doi.org/10.7554/eLife.43944.022
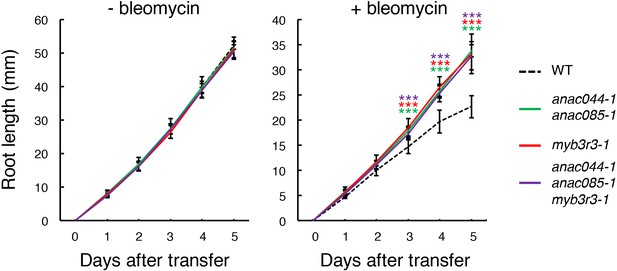
Root growth of anac044-1 anac085-1, myb3r3-1 and anac044-1 anac085-1 myb3r3-1 in the presence of bleomycin.
Five-day-old seedlings were transferred to medium supplemented with or without 0.6 µg/ml bleomycin, and root length was measured every 24 hr. Data are presented as mean ± SD (n > 12). Significant differences from WT were determined by Student’s t-test: ***, p<0.001.
-
Figure 6—figure supplement 2—source data 1
Source data.
- https://doi.org/10.7554/eLife.43944.024
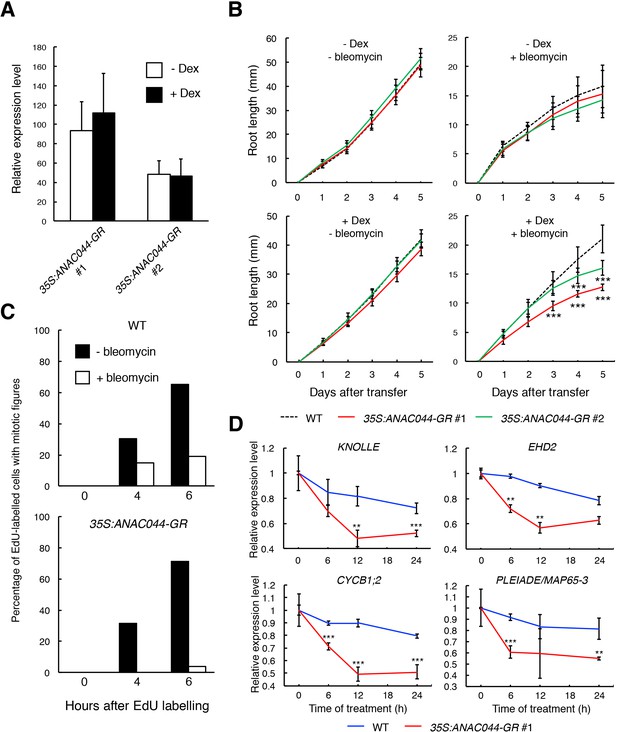
ANAC044-overexpressing plants exhibit hypersensitivity to DNA damage.
(A) Transcript levels of ANAC044-GR. Five-day-old seedlings of two independent lines of 35S:ANAC044-GR (#1 and #2) were treated with or without 10 µM Dex for 24 hr. mRNA levels were quantified by qRT-PCR using ANAC044 primers and normalized to that of ACTIN2, and are indicated as relative values, with that of endogenous ANAC044 in WT set to 1. Data are presented as mean ± SD (n = 3). (B) Root growth of 35S:ANAC044-GR seedlings. Five-day-old seedlings were transferred to MS medium supplemented with or without 10 µM Dex and/or 0.6 µg/ml bleomycin, and root length was measured every 24 hr. Data are presented as mean ± SD (n > 12). Significant differences from WT were determined by Student’s t-test: ***, p<0.001. (C) Cell cycle progression through the G2/M phase. Five-day-old seedlings of WT and 35S:ANAC044-GR #1 were grown in 10 µM Dex-containing medium with or without 0.6 µg/ml bleomycin for 12 hr. After pulse-labelling with EdU for 15 min, seedlings were transferred back to Dex-containing medium with or without bleomycin, and collected after 0, 4 and 6 hr. Root meristematic cells were double-stained with EdU and DAPI, and cells with mitotic figures were counted. Data are presented as the percentage of EdU-labelled cells among those with mitotic figures (n > 20). (D) Transcript levels of G2/M-specific genes. Five-day-old seedlings of WT and 35S:ANAC044-GR #1 were transferred to MS medium containing 10 µM Dex and 0.6 µg/ml bleomycin, and grown for 0, 6, 12 and 24 hr. Total RNA was extracted from root tips and subjected to qRT-PCR. mRNA levels were normalized to that of ACTIN2, and are indicated as relative values, with that for 0 hr set to 1. Data are presented as mean ± SD (n = 3). Significant differences from WT were determined by Student’s t-test: **, p<0.01; ***, p<0.001.
-
Figure 7—source data 1
Source data.
- https://doi.org/10.7554/eLife.43944.029
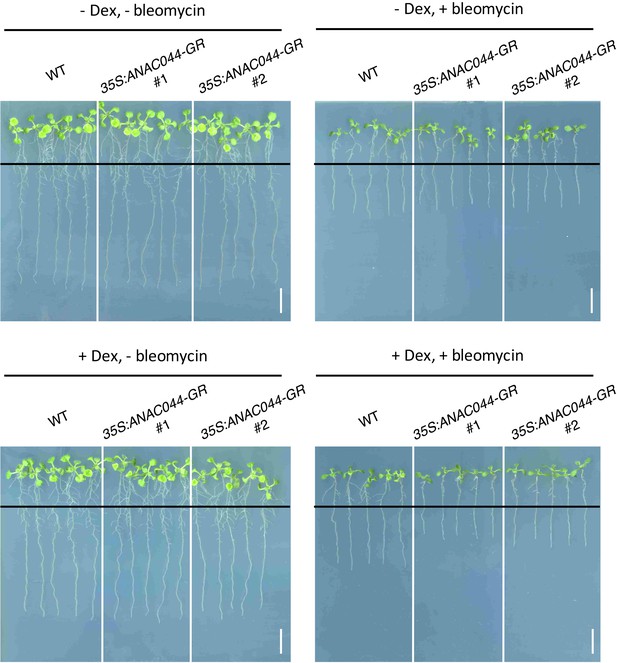
Root growth of 35S:ANAC044-GR seedlings.
Five-day-old seedlings were transferred to medium with or without 10 µM Dex and/or 0.6 µg/ml bleomycin, and grown for a further five days. Black lines indicate the positions of root tips when seedlings were transferred onto each medium. Bar = 1 cm.
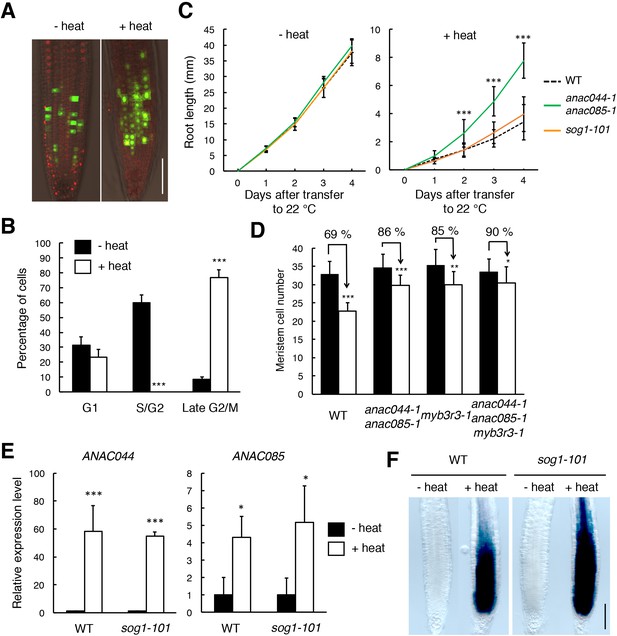
ANAC044 and ANAC085 are involved in the heat stress response in roots.
(A, B) Heat stress-induced cell cycle arrest in the root tip. Five-day-old seedlings carrying the cell cycle marker system Cytrap were incubated at 22°C (- heat) or 37°C (+heat) for 24 hr. Signals of ProHTR2:CDT1a (C3)-RFP (S/G2 marker; red) and ProCYCB1;1:CYCB1;1-GFP (late G2/M marker; green) were observed (A). Bar = 100 µm. Cells with RFP or GFP signals were estimated to be in S/G2 or late G2/M, respectively, and those with no fluorescence were counted as G1-phase cells (B). Data are presented as mean ± SD (n = 10). Significant differences from the control (22°C) were determined by Student’s t-test: ***, p<0.001. (C) Root growth of WT, anac044-1 anac085-1 and sog1-101 under heat stress. Five-day-old seedlings were incubated at 22°C (- heat) or 37°C (+heat) for 24 hr, and transferred to 22°C to measure root length every 24 hr. Data are presented as mean ± SD (n > 13). Significant differences from WT were determined by Student’s t-test: ***, p<0.001. (D) Cortical cell number in the root meristem after heat stress. Five-day-old seedlings of WT, anac044-1 anac085-1, myb3r3-1 and anac044-1 anac085-1 myb3r3-1 were incubated at 22°C (black bars) or 37°C (white bars) for 6 hr. Data are presented as mean ± SD (n > 15). Significant differences from the control (22°C) were determined by Student’s t-test: *, p<0.05; **, p<0.01; ***, p<0.001. (E) Transcript levels of ANAC044 and ANAC085 under heat stress. Five-day-old seedlings of WT and sog1-101 were incubated at 22°C (- heat) or 37°C (+heat) for 24 hr, and total RNA was extracted. mRNA levels were normalized to that of ACTIN2, and are indicated as relative values, with that for the control (22°C) set to 1. Data are presented as mean ± SD (n = 3). Significant differences from the control (22°C) were determined by Student’s t-test: *, p<0.05, ***, p<0.001. (F) GUS staining of WT and sog1-101 roots harbouring ProANAC044:GUS. Five-day-old seedlings were incubated at 22°C (- heat) or 37°C (+heat) for 24 hr, followed by GUS staining. Bar = 100 µm.
-
Figure 8—source data 1
Source data.
- https://doi.org/10.7554/eLife.43944.034
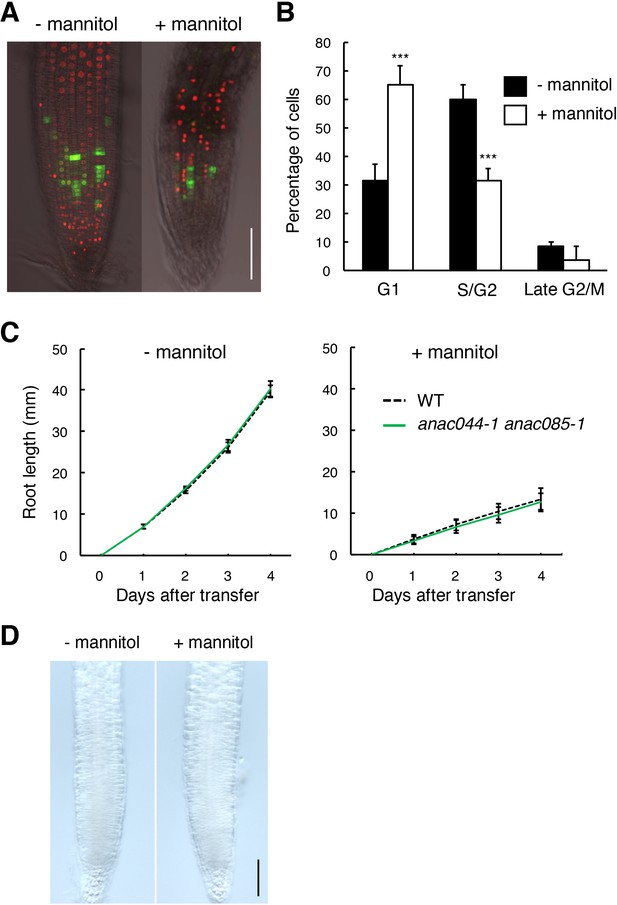
ANAC044 and ANAC085 are not involved in the osmotic stress response.
(A, B) Omotic stress-induced cell cycle arrest in the root tip. Five-day-old seedlings carrying the cell cycle marker system Cytrap were transferred to medium supplemented with or without 400 mM mannitol, and grown for 24 hr. Signals of ProHTR2:CDT1a (C3)-RFP (S/G2 marker; red) and ProCYCB1;1:CYCB1;1-GFP (late G2/M marker; green) were observed (A). Bar = 100 µm. Cells with RFP or GFP signals were estimated to be in S/G2 or late G2/M, respectively, and those with no fluorescence were counted as G1-phase cells (B). Data are presented as mean ± SD (n = 10). Significant differences from the control without mannitol treatment were determined by Student’s t-test: ***, p<0.001. (C) Root growth of WT and anac044-1 anac085-1 under osmotic stress. Five-day-old seedlings were transferred to medium supplemented with or without 400 mM mannitol, and root length was measured every 24 hr. Data are presented as mean ± SD (n > 15). (D) GUS staining of roots harbouring ProANAC044:GUS. Five-day-old seedlings were transferred to medium supplemented with or without 400 mM mannitol, and grown for 24 hr, followed by GUS staining. Bar = 100 µm.
-
Figure 8—figure supplement 1—source data 1
Source data.
- https://doi.org/10.7554/eLife.43944.032
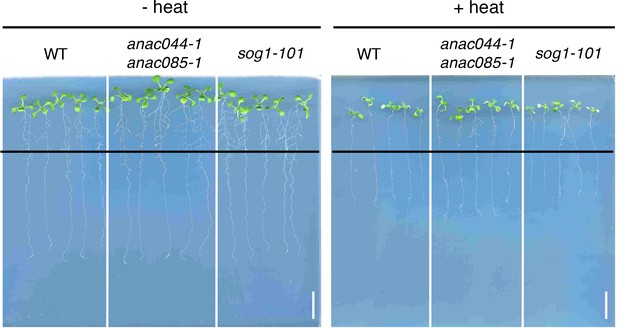
Root growth of anac044-1 anac085-1 and sog1-101 after heat stress.
Five-day-old seedlings were incubated at 22°C (- heat) or 37°C (+heat) for 24 hr, and grown at 22°C for a further four days. Black lines indicate the positions of root tips when seedlings were incubated after 24 hr treatment at 22°C or 37°C. Bar = 1 cm.
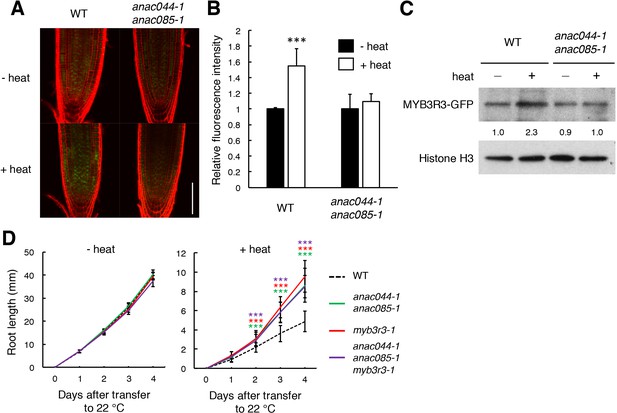
Heat stress induces ANAC044/085-mediated Rep-MYB accumulation in the root tip.
(A–C) Rep-MYB accumulation under heat stress. Five-day-old seedlings of WT and anac044-1 anac085-1 harbouring ProMYB3R3:MYB3R3-GFP were incubated at 22°C (- heat) or 37°C (+heat) for 12 hr. GFP fluorescence was observed after counterstaining with PI (A). Bar = 100 µm. Fluorescence intensities are indicated as relative values, with that of the control (22°C) set to 1 (B). Data are presented as mean ± SD (n > 5). Significant differences from the control (22°C) were determined by Student’s t-test: ***, p<0.001. Twenty micrograms of total protein were extracted from root tips and subjected to immunoblotting using antibodies against GFP or histone H3 (C). Relative levels of MYB3R3-GFP are expressed as fold change, normalized with respect to the corresponding band of histone H3. (D) Root growth of WT, anac044-1 anac085-1, myb3r3-1 and anac044-1 anac085-1 myb3r3-1 under heat stress. Five-day-old seedlings were incubated at 22°C (- heat) or 37°C (+heat) for 24 hr, and transferred to 22°C to measure root length every 24 hr. Data are presented as mean ± SD (n > 12). Significant differences from WT were determined by Student’s t-test: ***, p<0.001.
-
Figure 9—source data 1
Source data.
- https://doi.org/10.7554/eLife.43944.039
-
Figure 9—source data 2
Uncut blot.
- https://doi.org/10.7554/eLife.43944.040
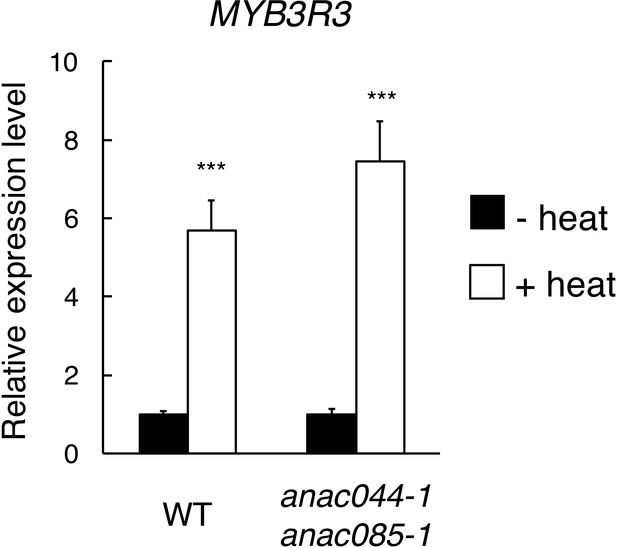
MYB3R3 expression under heat stress.
Five-day-old seedlings of WT and anac044-1 anac085-1 were incubated at 22°C (- heat) or 37°C (+heat) for 24 hr, and total RNA was extracted for qRT-PCR. mRNA levels were normalized to that of ACTIN2, and are indicated as relative values, with that for the control (22°C) set to 1. Data are presented as mean ± SD (n = 3). Significant differences from the control (22°C) were determined by Student’s t-test: ***, p<0.001.
-
Figure 9—figure supplement 1—source data 1
Source data.
- https://doi.org/10.7554/eLife.43944.037
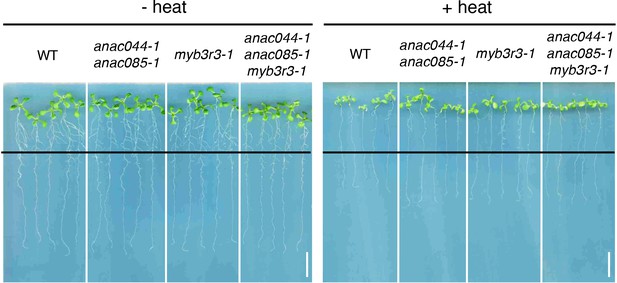
Root growth of anac044-1 anac085-1, myb3r3-1 and anac044-1 anac085-1 myb3r3-1 after heat stress.
Five-day-old seedlings were incubated at 22°C (- heat) or 37°C (+heat) for 24 hr, and grown at 22°C for a further four days. Black lines indicate the positions of root tips when seedlings were incubated after 24 hr treatment at 22°C or 37°C. Bar = 1 cm.
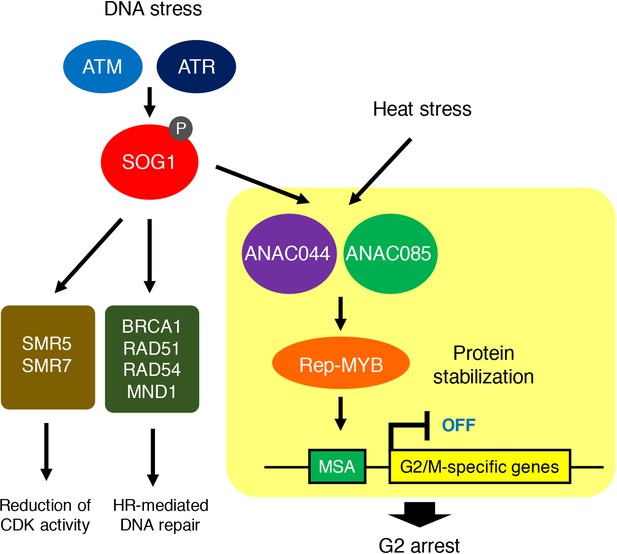
ANAC044/ANAC085-mediated pathway involved in stress-induced G2 arrest.
Both DNA damage and heat stress induce ANAC044 and ANAC085, which then promote Rep-MYB accumulation and repress a set of G2/M-specific genes, thereby leading to G2 arrest.
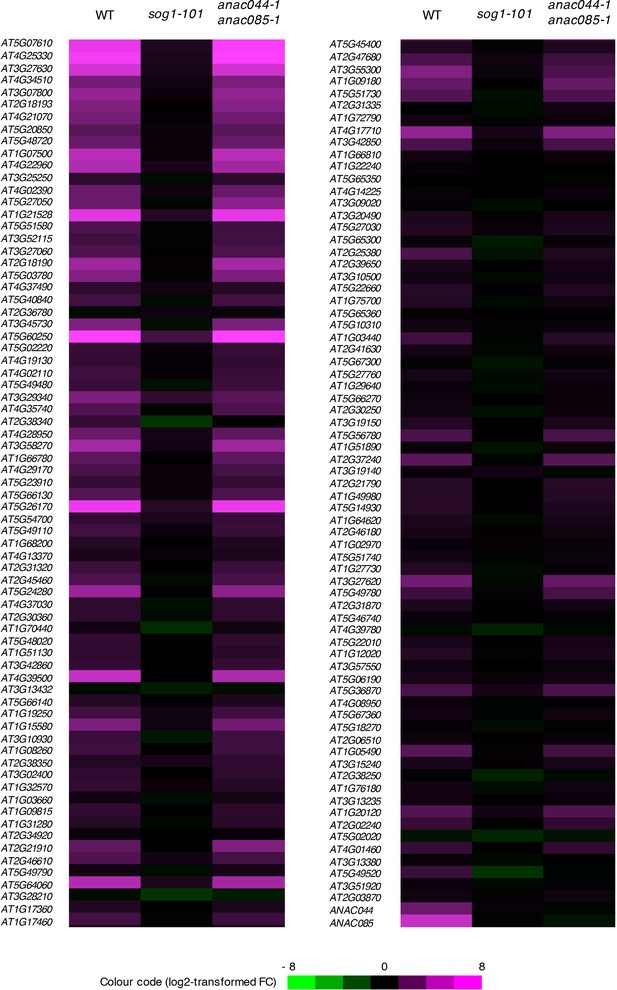
Expression of SOG1 target genes in anac044 anac085.
Five-day-old seedlings of WT, sog1-101 and anac044-1 anac085-1 were treated with or without 0.6 µg/ml bleomycin for 10 hr, and subjected to microarray analysis. Purple and green colours indicate up- and down-regulation, respectively, of 146 SOG1 target genes by bleomycin treatment.
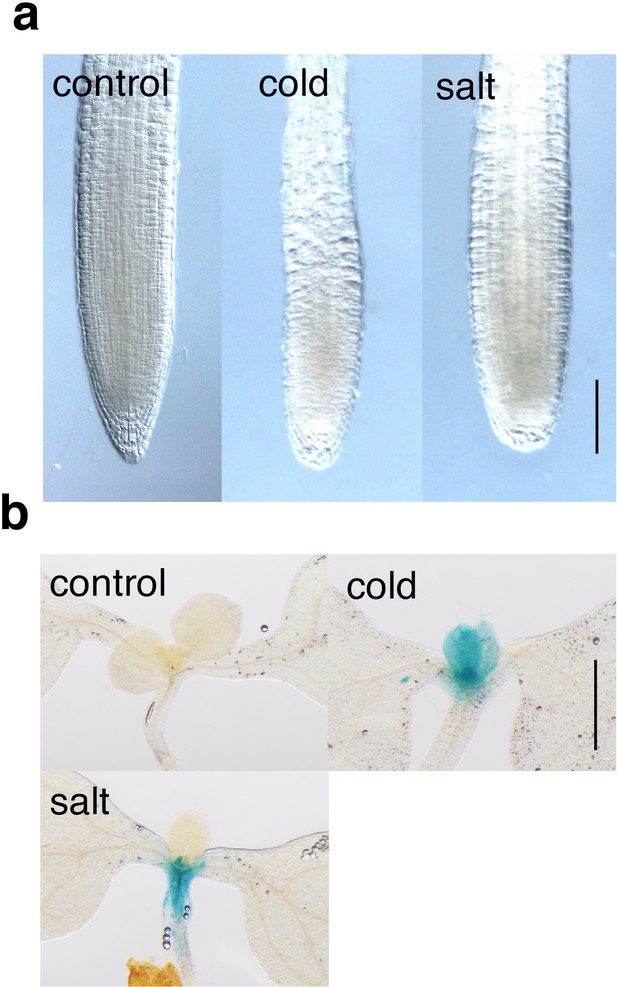
ANAC044 expression under cold and salt stresses.
Five-day-old seedlings harbouring ProANAC044:GUS were incubated at 4°C (cold) or grown in medium containing 200 mM NaCl (salt) for 24 hr. After GUS staining, root tips (A) and shoot apices (B) were observed. Bars = 100 µm.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Arabidopsis thaliana) | anac044-1 | ABRC stock center | SAIL_1286D02 | homozygous mutant plant line |
| Genetic reagent (Arabidopsis thaliana) | anac044-2 | ABRC stock center | GABI_968B05 | homozygous mutant plant line |
| Genetic reagent (Arabidopsis thaliana) | anac085-1 | ABRC stock center | GABI_894G04 | homozygous mutant plant line |
| Genetic reagent (Arabidopsis thaliana) | anac085-2 | ABRC stock center | SALK_208662 | homozygous mutant plant line |
| Genetic reagent (Arabidopsis thaliana) | anac044-1 anac085-1 | this paper stock center | homozygous mutant plant line | |
| Genetic reagent (Arabidopsis thaliana) | anac044-1 anac085-1 sog1-101 | this paper stock center | homozygous mutant plant line | |
| Genetic reagent (Arabidopsis thaliana) | sog1-101 | Ogita et al., 2018 | homozygous mutant plant line | |
| Genetic reagent (Arabidopsis thaliana) | ProMYB3R3:MYB3R3-GFP | Chen et al., 2017 | transgenic plant line | |
| Genetic reagent (Arabidopsis thaliana) | 35S:ANAC044-GR | this paper | transgenic plant line | |
| Genetic reagent (Arabidopsis thaliana) | Cytrap | Yin et al., 2014 | transgenic plant line | |
| Genetic reagent (Arabidopsis thaliana) | myb3r3-1 | Kobayashi et al., 2015 | homozygous mutant plant line | |
| Genetic reagent (Arabidopsis thaliana) | anac044-1 anac085-1 myb3r3-1 | this paper | homozygous mutant plant line | |
| Genetic reagent (Arabidopsis thaliana) | ProANAC044:GUS | this paper | transgenic plant line | |
| Genetic reagent (Arabidopsis thaliana) | ProSOG1:SOG1-Myc | Yoshiyama et al., 2009 | transgenic plant line | |
| Antibody | rabbit polyclonal αGFP | Thermo Fisher Scientific | A-6455 | 1:3000 for immunoblotting |
| Antibody | rabbit polyclonal αHistone H3 | Abcam | ab1791 | 1:2000 for immunoblotting |
| Antibody | Monoclonal antibody | Millipore | clone 4A6 | 1:500 for chromatin immunoprecipitation |
Additional files
-
Supplementary file 1
Primers used for cloning, qRT-PCR, ChIP-qPCR and semi-quantitative RT-PCR.
- https://doi.org/10.7554/eLife.43944.044
-
Transparent reporting form
- https://doi.org/10.7554/eLife.43944.045





