A hydrophobic gate in the inner pore helix is the major determinant of inactivation in mechanosensitive Piezo channels
Figures
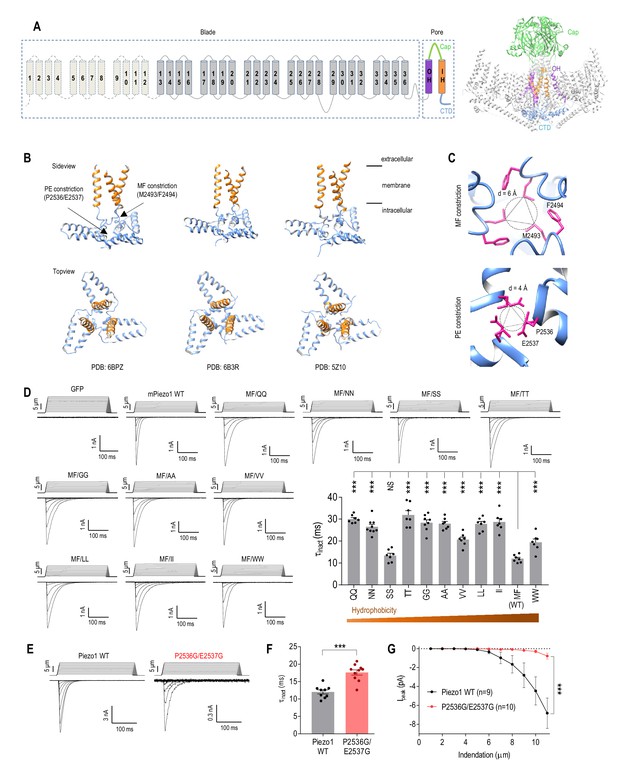
Physical constrictions in the CTD play only moderate roles in Piezo1 inactivation.
(A) Topology diagram and cryo-EM structure of Piezo1 (PDB: 6BPZ). IH, inner pore helix; OH, outer pore helix. (B) Side view and top view of the Piezo1 pore region from three cryo-EM structures showing the location of the MF and PE constrictions. (C) Top view close-up of the MF and PE constrictions in Piezo1 (PDB: 6BPZ). (D) Representative whole-cell MA current traces and quantification of MA current inactivation rate (τinact) in HEK293TΔP1 cells expressing Piezo1 with mutations at the M2493 F2494 (MF) site (n = 7–9 cells). Ehold = −80 mV. ***p<0.001; NS, not significant, p>0.05, one-way ANOVA with Holm-Sidak’s correction. (E and F) Representative whole-cell MA current traces and quantification of MA current inactivation for WT Piezo1 and P2536G/E2537G mutant. **p<0.001, unpaired t-test. (G) Quantification of peak MA current amplitude (Ipeak) at different indentation depths for WT Piezo1 and P2536G/E2537G mutant. ***p<0.001, two-way ANOVA. Data are mean ± SEM.
-
Figure 1—source data 1
Electrophysiological analysis of Piezo1 CTD mutants.
- https://doi.org/10.7554/eLife.44003.005
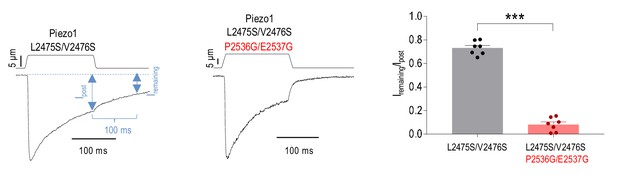
Mutations at the Piezo1 PE site accelerate deactivation of MA current.
(A and B) Representative whole-cell MA current traces recorded in HEK293TΔP1 cells expressing Piezo1 with mutations at the putative inactivation gate (L2475S/V2476S) and PE constriction (P2536G/E2537G). Ehold = −80 mV. Current traces were normalized to peak current amplitude to highlight the differences in the kinetics of persistent current decay. Blue arrows indicate points of measurements of current amplitude at the end of mechanical stimulus (Ipost) and 100 ms after the removal of mechanical stimulus (Iremaining). (C) Quantification of Iremaining/Ipost for the indicated mPiezo1 mutants (***p<0.001, unpaired t-test). Data are mean ± SEM.
-
Figure 1—figure supplement 1—source data 1
Electrophysiological analysis of Piezo1 PE site mutants.
- https://doi.org/10.7554/eLife.44003.004
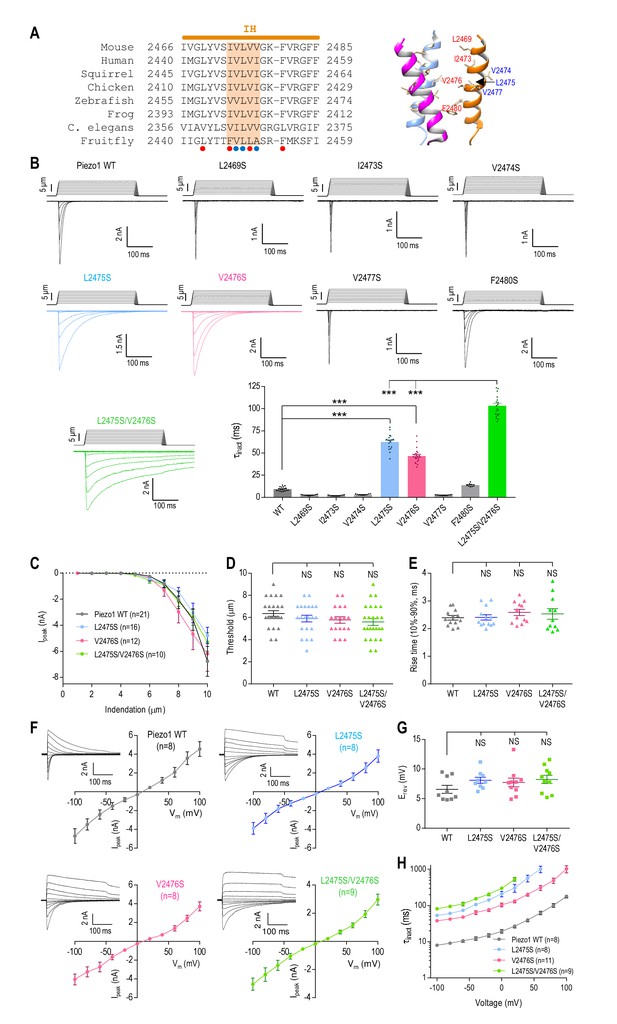
The pore-lining inner helix plays a major role in Piezo1 inactivation.
(A) Left panel, amino acid sequence alignment of the Piezo1 inner helix (IH) from different species. A cluster of five conserved hydrophobic residues in the middle are highlighted. Red and blue dots indicate hydrophobic residues facing and pointing away from the pore, respectively. Right panel, cryo-EM structure of the Piezo1 inner helix (PDB: 6BPZ) showing the hydrophobic residues in the left panel. (B) Representative whole-cell MA current traces and quantification of MA current inactivation rate (τinact) in HEK293TΔP1 cells expressing Piezo1 with mutations in the hydrophobic cluster in the inner helix (n = 8–22 cells). Ehold = −80 mV. ***p<0.001; NS, not significant, p>0.05, one-way ANOVA with Dunnet’s correction. (C–E) Quantification of peak MA current amplitude (Ipeak) at different indentation depths (C), apparent indentation threshold of MA current activation (D) and MA current rise time (E) for WT and mutant Piezo1. NS, not significant, p>0.05, one-way ANOVA with Dunnet’s correction. (F) Peak MA current-voltage relationship in response to mechanical indentation at 9 μm for WT Piezo1 or indicated mutants. Insets show representative traces of whole-cell MA currents evoked at Ehold ranging from −100 mV to +100 mV, in 20 mV increments. (G) Quantification of the reversal potential (Erev) from current-voltage plots in (F). NS, not significant, p>0.05, one-way ANOVA with Dunnet’s correction. (H) Quantification of MA current inactivation rate for WT or mutant Piezo1 at different voltages. Data are mean ± SEM.
-
Figure 2—source data 1
Electrophysiological analysis of Piezo1 IH mutants.
- https://doi.org/10.7554/eLife.44003.009
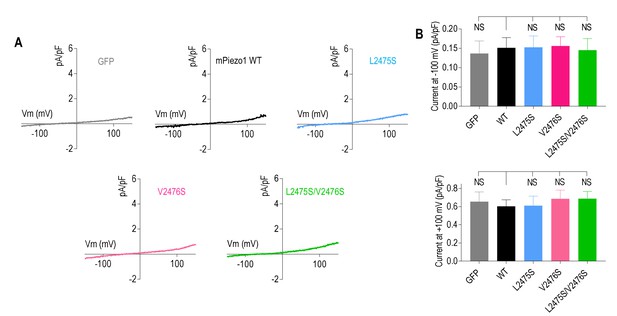
Mutations that prolong inactivation in Piezo1 do not affect basal current.
(A) Representative current-voltage curves measured from HEK293TΔP1 cells expressing WT or mutant Piezo1, in response to a voltage ramp from Ehold = −80 mV in the absence of mechanical stimulation. (B) Quantification of current densities at −100 mV or +100 mV for control (GFP, n = 6), WT Piezo1 (n = 7), L2475S (n = 8), V2476S (n = 7), or L2475S/V2476S (n = 9) (NS, not significant, one-way ANOVA analysis with Dunnett’s correction). Data are mean ± SEM.
-
Figure 2—figure supplement 1—source data 1
Quantification of basal current in Piezo1 mutants.
- https://doi.org/10.7554/eLife.44003.008
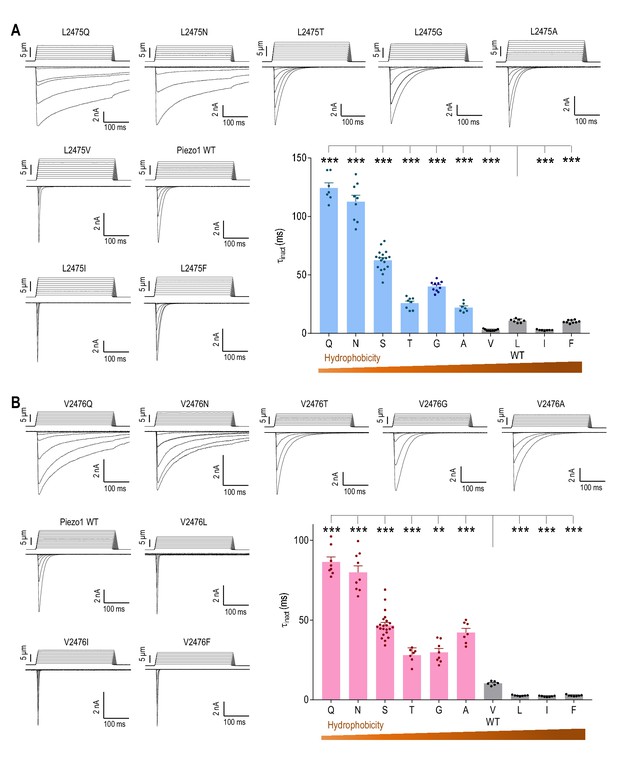
The hydrophobicity of L2475 and V2476 determines the rate of Piezo1 inactivation.
(A and B) Representative whole-cell MA current traces and quantification of MA current inactivation rate (τinact) in HEK293TΔP1 cells expressing Piezo1 with indicated mutations of variable hydrophobicity at L2475 (A, n = 7–18 cells) and V2476 (B, n = 6–22 cells). Ehold = −80 mV. **p<0.01, ***p<0.001, one-way ANOVA with Holm-Sidak’s correction. Data are mean ± SEM.
-
Figure 3—source data 1
Electrophysiological analysis of Piezo1 L2475 and V2476 mutants.
- https://doi.org/10.7554/eLife.44003.011
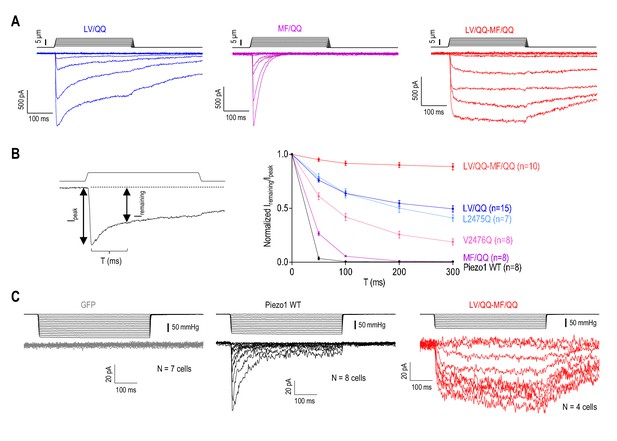
Mutation of the inner helix and MF constriction eliminates Piezo1 inactivation.
(A) Representative whole-cell MA current traces from HEK293TΔP1 cells expressing Piezo1 with glutamine mutations in the putative hydrophobic gate (L2475/V2476, LV), or the MF constriction (M2493/F2494, MF). Ehold = −80 mV. (B) Left panel, an example trace of Piezo1 MA current illustrating the measurement of the ratio of remaining MA current amplitude (Iremaining) to peak (Ipeak) at different time points during current decay. Right panel, quantification of Iremaining/Ipeak for WT or mutant Piezo1. Data are mean ± SEM. (C) Representative cell-attached MA current traces induced by high-speed pressure clamp via application of a negative pipette pressure in HEK293TΔP1 cells expressing GFP (negative control), WT or mutant Piezo1. Ehold = −80 mV.
-
Figure 4—source data 1
Quantification of current decay in Piezo1 mutants.
- https://doi.org/10.7554/eLife.44003.013
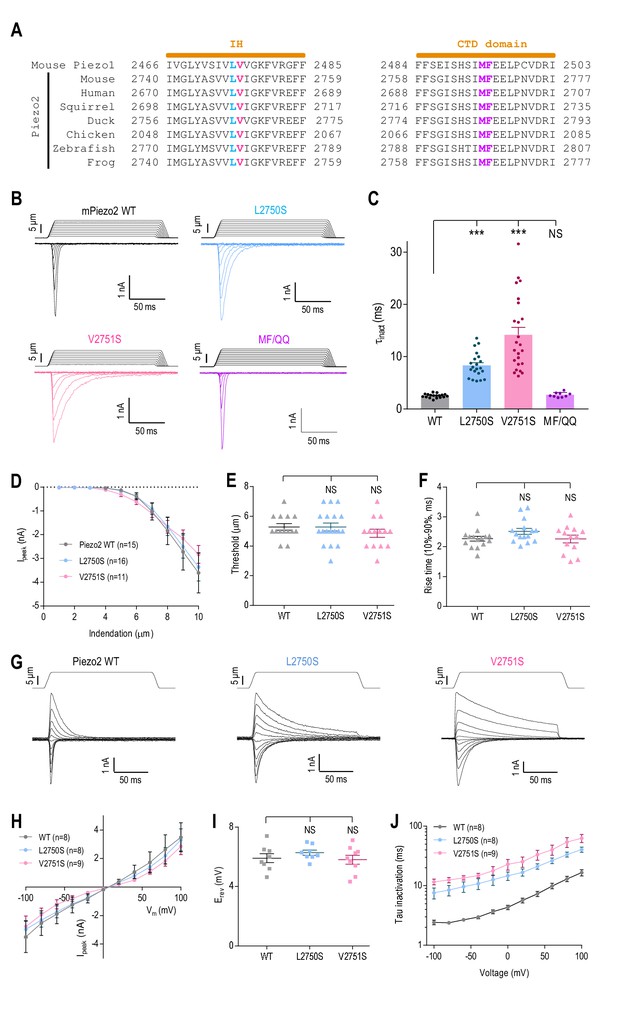
The putative inner helix inactivation gate is functionally conserved in Piezo2.
(A) Amino acid sequence alignments of the IH and part of CTD between mouse Piezo1 and Piezo2 orthologues from indicated species. The conserved L2475 and V2476 residues in the IH are highlighted in blue and red; M2493 and F2494 in the CTD are highlighted purple. (B and C) Representative whole-cell MA current traces of WT and mutant Piezo2 (B), and quantification of MA current inactivation constant (τinact) in HEK293TΔP1 cells (C, n = 9–24 cells). Ehold = −80 mV. Data are mean ± SEM. **p<0.001; NS, not significant, one-way ANOVA with Dunnett’s correction. (D–F) Quantification of peak MA current amplitude (Ipeak) at different indentation depths (D), apparent indentation threshold of MA current activation (E) and MA current rise time (F) for WT and mutant Piezo2 in HEK293TΔP1 cells. Ehold = −80 mV. NS, not significant, p>0.05, one-way ANOVA with Dunnet’s correction. (G and H) Representative current traces (G) and quantification of peak MA current-voltage relationship (H) in response to mechanical indentation at 9 μm for WT or mutant Piezo2, evoked at Ehold ranging from −100 mV to +100 mV, in 20 mV increments. (I) Quantification of the reversal potential (Erev) from current-voltage plots in (H). NS, not significant, p>0.05, one-way ANOVA with Dunnet’s correction. (J) Quantification of MA current inactivation rate for WT or mutant Piezo2 in response to a 9 µm indentation at different voltages. Data are mean ±SEM.
-
Figure 5—source data 1
Electrophysiological analysis of Piezo2 mutants.
- https://doi.org/10.7554/eLife.44003.015
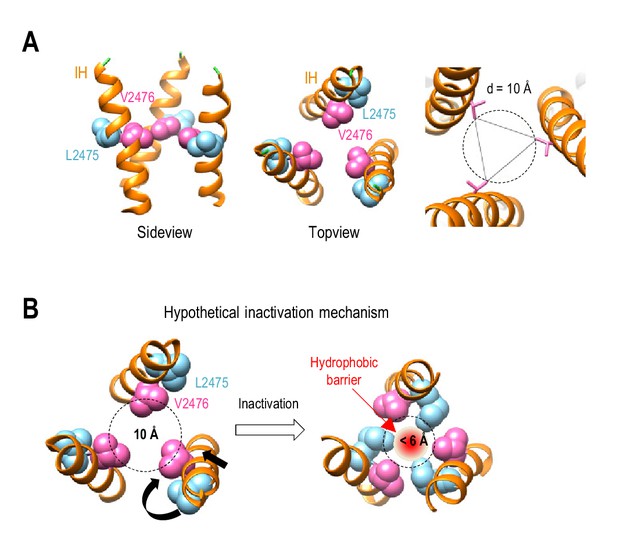
Hypothetical inactivation mechanism of Piezo1.
(A) Left and middle panels, the side view and top view of a portion of Piezo1 inner helix (PDB: 6BPZ) showing the orientations of L2475 and V2476 residues with respect to the ion permeation pore. Right panel, pore diameter at V2476. (B) A hypothetical mechanistic model for Piezo1 inactivation at the hydrophobic gate in the inner helix. Inactivation is proposed to involve a combined twisting and constricting motion of the inner helix (black arrows), allowing both V2476 and L2475 residues to face the pore to form a hydrophobic barrier.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (H. sapiens) | HEK293TPIEZO1-/- (HEK293TΔP1) | Dr. Ardem Patapoutian (Scripps Research Institute) (Lukacs et al., 2015) | ||
| Recombinant DNA reagent | Mouse-Piezo2-Sport6 | Dr. Ardem Patapoutian (Scripps Research Institute) (Coste et al., 2010) | ||
| Recombinant DNA reagent | Mouse-Piezo1-pMO | (Anderson et al., 2018) | ||
| Recombinant DNA reagent | Mouse-Piezo1 -IRES-EGFP | Dr. Ardem Patapoutian (Scripps Research Institute) (Coste et al., 2010) Addgene #80925 | ||
| Software, algorithm | GraphPad Prism | GraphPad Prism (https://graphpad.com) | RRID:SCR_000306 | Version 7 |
| Software, algorithm | pCLAMP | Molecular Devices (https://www.moleculardevices.com/) | RRID:SCR_011323 | Version 10 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.44003.017





