Transcriptional down-regulation of ccr5 in a subset of HIV+ controllers and their family members
Figures
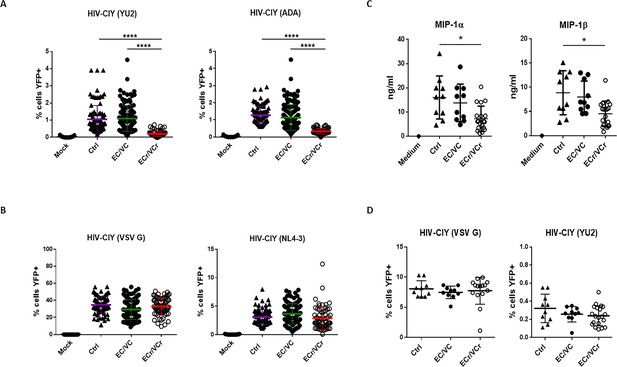
CD4 +T cell resistance to infection in prospective single cycle assay, specific to R5-tropic viruses in a subset of EC/VCs.
(A) Five-fold resistance to R5-tropic viruses in 16% of EC/VC (ECr/VCr) infected using replication defective HIV-cycT1-IRES-eYFP (CIY) with R5-tropic envelopes YU2 and ADA. A > 95% power was determined based on comparisons of means using PASS statistical software between ECr/VCr and all other groups (Ctrl and EC/VC). (B) Equivalent susceptibility to both X4-tropic (NL4-3) and VSV G pseudoviral particles in ECr/VCr. A and B are pooled results from different experiments with samples tested at least in triplicate (Ctrl n = 35, EC/VC n = 38, representative from the initial population (Figure 1—figure supplement 1) and selected based upon specimen availability, and ECr/VCr (n = 21). (C) Comparable levels of chemokines (MIP-1α and MIP-1β) in cell culture supernatants from activated CD4 +T cells, measured by ELISA. (D) CD4 +T cells from Ctrl were exposed to cell culture supernatants from activated T cells of Ctrl and EC/VC with or without the resistance phenotype, in the presence of HIV particles pseudotyped with YU2 or VSV G. C and D are pooled results from different experiments with n = 10 (Ctrl and EC/VCs) and n = 21 (ECr/VCr). Shown are individual values with Means ± Standard Deviation (SD). Data were analyzed by using the Kruskal-Wallis test and Dunn’s multiple-comparison test. *p<0.05; ****p<0.0001.
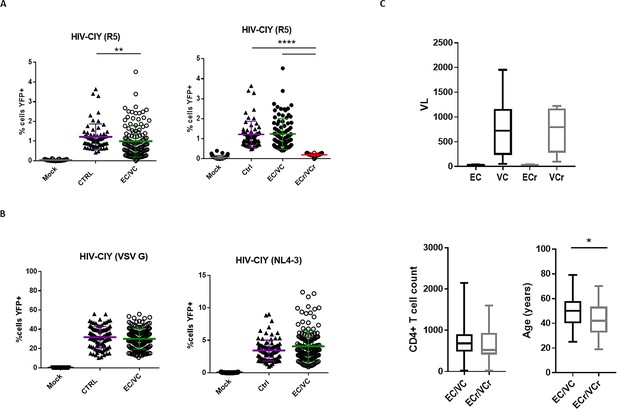
Initial testing showing CD4 +T cell resistance to infection in single-cycle pseudotyping assay, specific to R5-tropic viruses, in a subset of EC/VCs.
(A) Left panel shows relative resistance to R5-tropic viruses in EC/VC compared to Ctrl, infected with replication-defective HIV-CIY pseudotyped with R5-tropic envelope ADA. EC/VC (n = 131) and Ctrl (n = 35) tested in duplicate (80% power was determined based on comparisons between groups). Right panel shows resistance to R5-tropic viruses in a subset of EC/VC (ECr/VCr, 21/131 or 16%) compared to Ctrl after separated out from the remaining EC/VC. These 21 ECr/VCr were identified as resistant based upon the % eYFP +cells being lower than that of the Ctrl group and confirmed after additional retesting using two R5 envelopes in triplicate (data shown in Figure 1). (B) Equivalent susceptibility to both VSV G and X4-tropic (NL4-3) pseudoviral particles between groups in all samples analyzed. Data were analyzed by using the Kruskal-Wallis test and Dunn’s multiple-comparison test. **p<0.01; ***p<0.001; ****p<0.0001. (C) Clinical data in EC/VC population. Comparable Viral Load (VL) and CD4 +T cell counts between EC and VC with or without the resistance phenotype to R5-tropic viruses. Age (years) at the time of the diagnosis to HIV between EC/VC (n = 109) and ECr/VCr (n = 21). Data presented as box and whisker plots. Statistical analysis was performed by using the U-Mann Whitney test; *p<0.05.
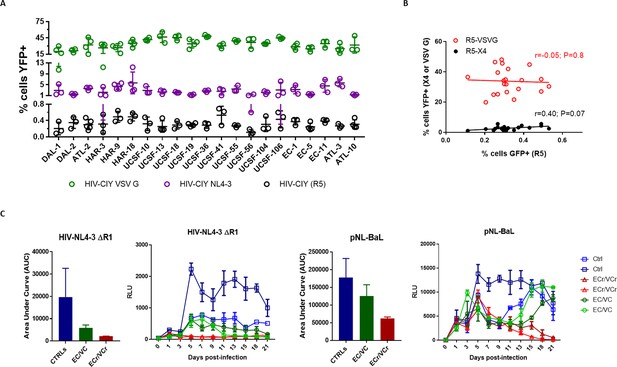
CD4 +T cell resistance to infection in single-cycle pseudotyping assays in 21 ECr/VCr and multi-cycle infection in a subset of ECr/VCr, Ctrl and EC/VC.
(A) Susceptibility to R5-, X4-, and pan-tropic virus in all 21 ECr/VCr, using single cycle infection. (B) Virus susceptibility correlations in all 21 ECr/VCr. (C) Replication kinetics using replication-competent HIV-NL4-3ΔR1 and pNL-BaL. Supernatants from CD4 +T cells infected with HIV-NL4-3ΔR1 (0.01 ml) and pNL-Bal (0.001 ml) were harvested every other day for 21 days and use to infect TZM-bl targets. Shown is a representative experiment out of 2 performed, with n = 2 per group, tested in triplicate and quantified as Relative Light Units (RLU) using luciferase assay with Means ± SD. ECr/VCr T cells were still fully viable at the end of the 3 week period, with no evidence of cytotoxicity. Differences were analyzed using AUC, with Means ± SD.
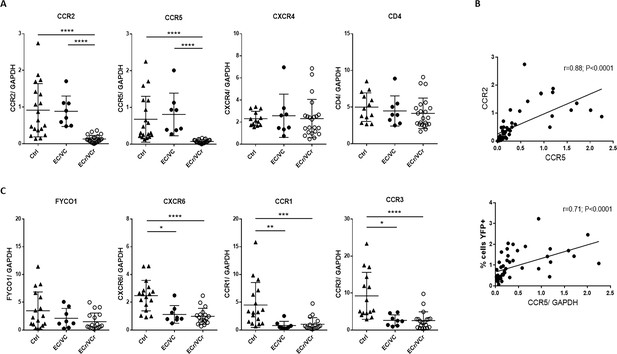
Decreased mRNA levels of several chromosomal three genes in ECr/VCrs.
(A) Decreased ccr2/ccr5 RNA levels in activated CD4 +T cells from EC/VCs with the resistance phenotype, with comparable cxcr4 and cd4 RNA levels in all groups. Shown are individual values with Means ± SD. Pooled results from different experiments are shown with representative samples per group, n = 19 (Ctrl), n = 8 (EC/VC) and n = 21 (ECr/VCr) per group. (B) Positive correlation between ccr2 and ccr5 RNA levels in activated CD4 +T cells. ccr5 RNA levels positively correlated with % of YFP +infected cells by single cycle assay using R5-tropic viruses but not with cd4 or cxcr4 (Figure 2—figure supplement 1A). (C) Decreased RNA levels in multiple chromosomal 3p21 genes in T cells of HIV +infected individuals (Figure 2—figure supplement 1C). Statistical analysis performed using Kruskal-Wallis test and Dunn’s multiple-comparison test. r value calculated using the non-parametric Spearman correlation test. Graphs show individual values with Means ± SD. *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001.
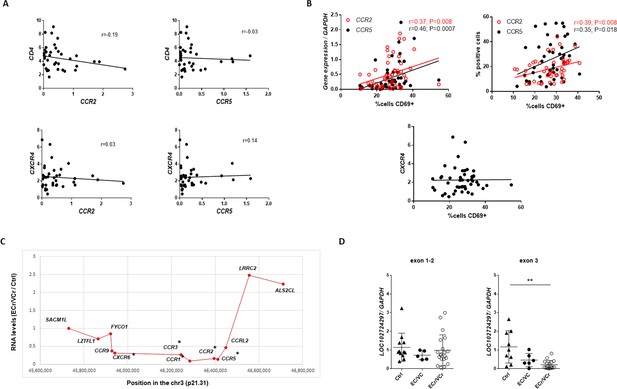
Correlations and fold-change in RNA levels in ECr/VCr.
(A) Correlation between ccr2 and ccr5 with cd4 or cxcr4 RNA levels in activated CD4 +T cells. (B) Positive correlation observed between ccr2/ccr5 RNA and cell surface protein levels and CD69 +cell surface expression, with no correlation versus cxcr4 RNA levels. Values obtained using the non-parametric Spearman correlation test. **p<0.01 (CCR2) and ***p<0.001 or *p<0.05 (CCR5). (C) Fold-change in ECr CD4 +T cell RNA levels, normalized to that of Ctrl, of genes in ~1 Mb region of chr 3p21. Each gene is plotted as the midpoint of the transcriptional unit or TU. RTP3, TDGF1, SLC6A20 and XCR1 are not included because RPKM values were too low for statistical analysis. * indicates statistically significant genes. (D) loc102724291 RNA levels in activated CD4 +T cells from ECr/VCr compared EC/VC and Ctrl, using two different qPCR primer pairs: (i) exon 1–2, centromeric to ccr5, (ii) exon 3, within intron 2 of ccr5. Shown are individual values with Means ± SD. Statistical analysis was performed by using Kruskal-Wallis with Dunn’s multiple-comparison test. **p<0.01.
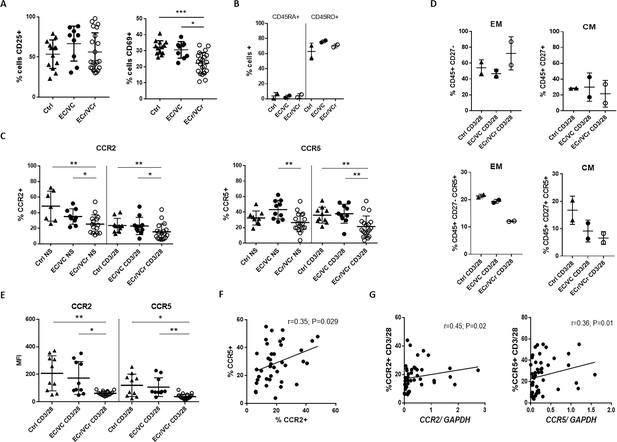
Lower proliferative responses and CCR2 and CCR5 cell surface levels in activated CD4 +T cells from ECr/VCrs.
(A) Reduced CD69, but not CD25 levels in activated CD4 +T cells from ECr/VCrs. Graph shows representative data N = 13 (Ctrl), n = 9 (EC/VC) and n = 21 (ECr/VCr). (B) Comparable frequencies of naïve CD45RA + and memory CD45RO + T cells after anti-CD3/CD28 activation between groups (n = 2 per group). (C) CCR5 and CCR2 cell surface levels measured by flow cytometry are reduced in freshly thawed (NS, non-stimulated) and activated CD4 +T cells (anti-CD3/28) from ECr/VCr. (D) Percentages of CCR5 +in effector memory (EM) and central memory (CM) compartments of activated CD4 +T cells (n = 2 per group). (E) Reduced CCR2 and CCR5 cell surface levels, expressed as MFI, in activated (anti-CD3/28) CD4 +T cells from ECr/VCr. Data in D-E shown pooled results from different experiments with n = 10 (Ctrl and EC/VC) and n = 19 (ECr/VCr). (F) Positive correlation between CCR2 and CCR5 cell surface levels. (G) Positive correlation observed between ccr2/ccr5 RNA levels and cell surface expression. Values obtained using the non-parametric Spearman correlation test. *p<0.05.
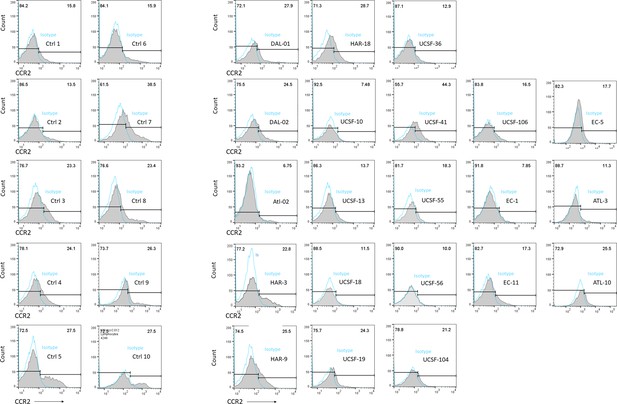
Flow cytometric histograms showing CCR2 +T cells in all 21 ECr/VCr, compared to 10 Ctrl, with isotype control staining in blue (open histograms).
https://doi.org/10.7554/eLife.44360.008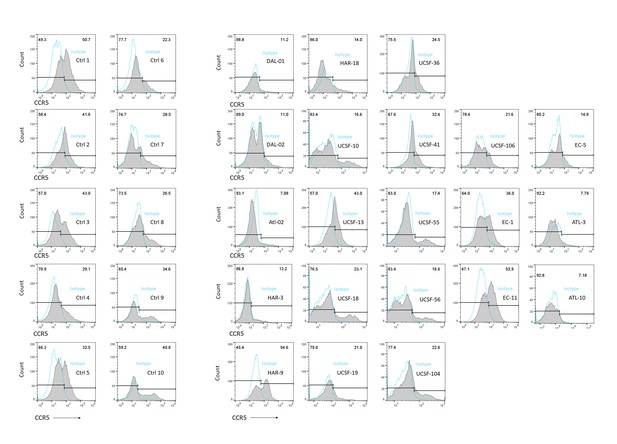
Flow cytometric histograms showing CCR5 +T cells in all 21 ECr/VCr, compared to 10 Ctrl, with isotype control staining in blue (open histograms).
https://doi.org/10.7554/eLife.44360.009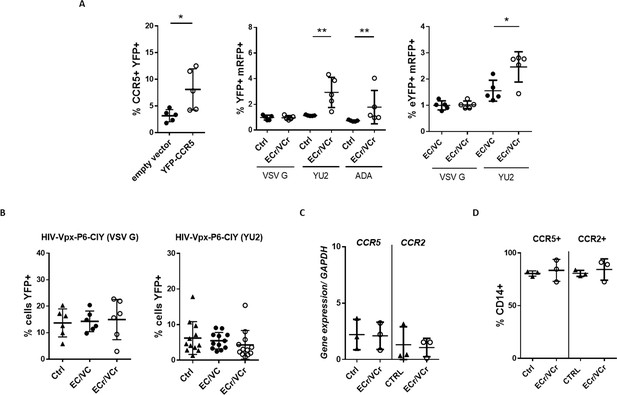
Resistance to R5-tropic viruses is due to down-regulation of CCR5 in ECr/VCr.
(A) Overexpression of CCR5 in CD4 +T cells using a lentiviral vector (YFP-CCR5). Increased susceptibility to R5-tropic virus after overexpression of CCR5 in EC/VCs with R5 resistance, as measured by YFP+/mRFP +double positive cells (n = 5 per group). (B) Comparable susceptibility to infection specific to R5-tropic virus in MDMs from ECr/VCr, EC/VCs and Ctrl (n = 6 per group; samples tested in duplicate for YU2). (C–D) Similar ccr2/ccr5 mRNA (C) and cell surface protein levels (D) in MDMs from EC/VCs (n = 3 per group). Shown in all cases are individual values with Means ± SD, analyzed using U-Mann Whitney test. *p<0.05; **p<0.01.
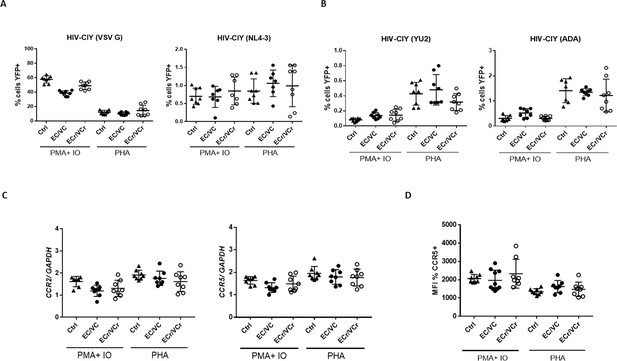
Resistance specific to R5-tropic virus is dependent upon T cell activation method.
(A) Comparable CD4 +T cell susceptibility to X4- and VSV G or (B) R5- pseudotyped particles in all groups after PMA plus ionomycin or PHA stimulation. Decreased susceptibility to R5-tropic infection in ECr/VCr compared to Ctrl and remaining EC/VCs after PHA stimulation was not significant. Shown are Means ± SD. (C) Comparable ccr2 and ccr5 mRNA expression levels between experimental groups after PMA plus ionomycin or PHA treatment. (D) Comparable frequency of CCR5 +cells between samples after PMA plus ionomycin or PHA stimulation in activated cells, analyzed as the MFI (n = 8 per experimental group).
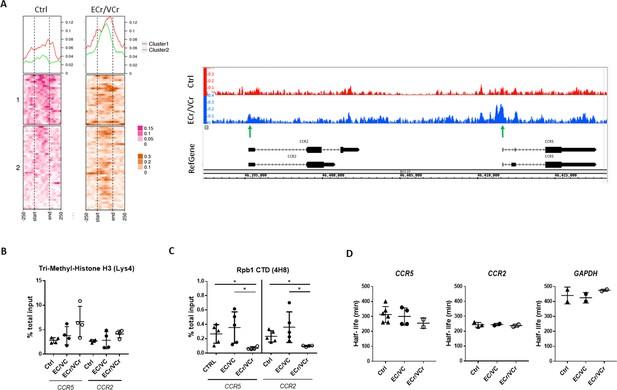
Increased chromatin accessibility and lower active transcription in activated CD4 +T cells from ECr/VCr.
(A) Left panel: ATAC-Seq coverage profiles of region of chr 3p21 (45,920,704-46,497,303) of ECr/VCr CD4 +T cells, compared to those of Ctrl (n = 4 replicates per group). Heat map showing gene TSS aligned, with a window of −250 bp to +250 bp, calculated as a normalized coverage around each TSS. Matrix was divided it into two clusters, based upon Ctrl data. At top is average coverage profile for each of the clusters (cluster one in red and cluster two in green). Right panel: ATAC-Seq peaks of chr 3p21 (46,392,331-46,418,348) of ECr/VCr vs Ctrl visualized using Integrated Genome Browser (IGB), see also Figure 6—figure supplement 1. Green arrows highlight increased peaks near the TSS of both genes, ccr2/ccr5, in ECr/VCr relative to Ctrl. (B–C) ChIP-qPCR, using either Tri-Methyl-Histone H3 (Lys4) (B) or Rpb1 (C) antibodies, with ccr2 and ccr5 DNA quantified by qPCR. Data normalized by the % total input DNA. Shown are Means ± SD (n = 4 and n = 5 per group in B and C, respectively), with statistical analysis performed using Kruskal-Wallis with Dunn’s multiple-comparison test. *p<0.05. (D) Quantitation of mRNA half-lives of indicated genes in activated CD4 +T cells, using Act D as a transcription inhibitor. T cells were incubated with Act D and harvested (from time 0 to 8 hr). RNA was extracted, and RNA levels quantified by RT-qPCR and half-life calculated using GraphPad PRISM software.
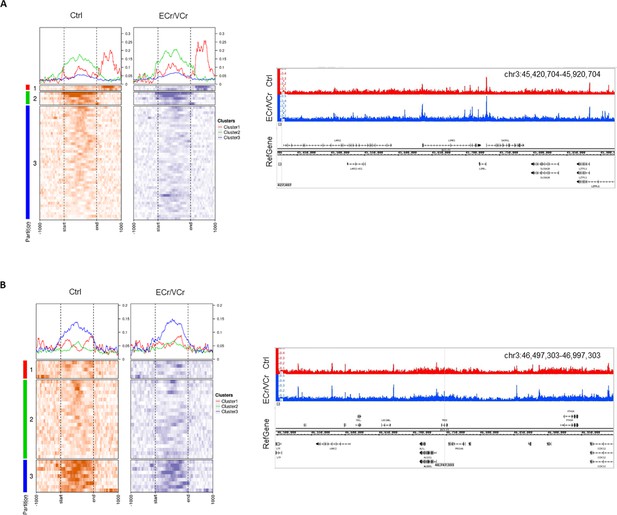
ATAC-Seq identifies comparable peaks between samples in other regions around chr 3p21.
(A) ATAC-Seq heat map representation upstream and (B) downstream of ccr5, showing all gene TSS aligned, with a window from −1 kb to +1 kb. The matrix was divided it into three clusters, generated based upon the Ctrl sample data (n = 4 replicates per group). At the top of each heat map is shown the average peak profile for the three clusters. Panels show the actual ATAC-Seq peaks across each region, visualized with the IGB.
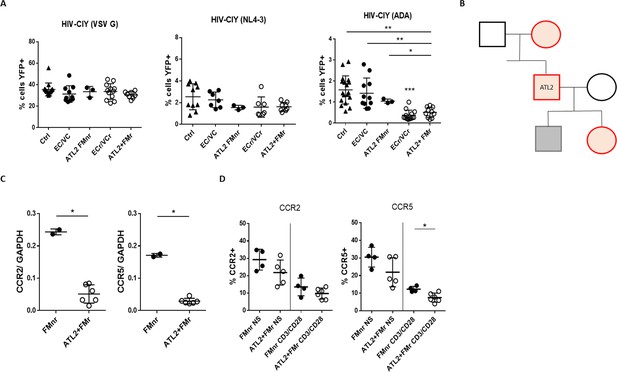
Pedigree analysis of an Index VC with R5 resistance phenotype.
(A) Resistance specific to R5-tropic virus, with equivalent susceptibility to X4- and VSV G, in activated CD4 +T cells from 2 of 3 analyzed ATL2 VC family members. Shown are pooled results from different experiments, with samples tested at least in triplicate. Statistical differences between ECr/VCr and other groups (Ctrl, EC/VC, and ATL2 FMnr) are also shown (**). (B) Pedigree analysis of ATL2 EC. Red are individuals with the R5 resistance phenotype (ATL2 FMr); grey represents full susceptibility to infection (ATL2 FMnr); black not available for testing. (C) Decreased ccr2/ccr5 RNA levels in activated CD4 +T cells from family members with R5 resistance. Samples were tested in duplicate. (D) Decreased CCR2 and CCR5 surface expression in resting (NS) and activated CD4 +T cells in family members with the resistance phenotype. Samples were tested at least in duplicate; shown are individual values with Mean ±SD. Statistical analysis was performed by using the U-Mann Whitney test or Kruskal-Wallis with Dunn’s multiple-comparison test. *p<0.05; **p<0.01. FMr: family member with R5 resistance. FMnr: family member without R5 resistance.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | anti-CD3 mouse Monoclonal Antibody (OKT3), PerCP-Cyanine5.5 | eBioscience | Cat # 45-0037-42; RRID: AB_10548513 | Dilution (1:100) |
| Antibody | anti-CD4 mouse Monoclonal Antibody (RPA-T4), APC | eBioscience | Cat # 17-0049-42; RRID: AB_1272048 | Dilution (1:100) |
| Antibody | anti-CD14 mouse Monoclonal Antibody (61D3), FITC | eBioscience | Cat # 11-0149-42; RRID: AB_10597597 | Dilution (1:100) |
| Antibody | anti-CD8a mouse Monoclonal Antibody (HIT8a), PE | eBioscience | Cat # 12-0089-42; RRID: AB_10804039 | Dilution (1:100) |
| Antibody | CD3 mouse Monoclonal Antibody (OKT3), Functional Grade | eBioscience | Cat # 16-0037-81; RRID: AB_468854 | 10 µg/ml |
| Antibody | CD28 mouse Monoclonal Antibody (CD28.2), Functional Grade | eBioscience | Cat # 16-0289-81; RRID: AB_468926 | 4 µg/ml |
| Antibody | CD25 mouse Monoclonal Antibody (BC96), PE | eBioscience | Cat # 12-0259-42; RRID: AB_1659682 | Dilution (1:200) |
| Antibody | CD69 mouse Monoclonal Antibody (FN50), FITC | eBioscience | Cat # 11-0699-42; RRID: AB_10853975 | Dilution (1:200) |
| Antibody | CD45RA mouse Monoclonal Antibody (HI100), FITC | eBioscience | Cat # 11-0458-42; RRID: AB_11219672 | Dilution (1:100) |
| Antibody | CD45RO, mouse Monoclonal PE-Cyanine5, clone: UCHL1 | eBioscience | Cat # 15597726; Gene ID: 5788 | Dilution (1:100) |
| Antibody | PE anti-human CD195 (CCR5) rat Monoclonal Antibody | Biolegend | Cat # 313707; RRID: AB_345307 | Dilution (1:100) |
| Antibody | APC anti-human CD192 (CCR2) mouse Monoclonal Antibody | Biolegend | Cat # 357207; AB_2562238 | Dilution (1:100) |
| Antibody | anti-Rpb1 CTD mouse Monoclonal | Cell Signaling | Cat # 2629; 4H8 | ChIP (1:50) |
| Antibody | Tri-Methyl-Histone H3-Lysine 4 (H3Lys4) rabbit Monoclonal | Cell Signaling | Cat # 9727 | ChIP (1:50) |
| Peptide, recombinant protein | Recombinant Human IL-2 | E. coli-derived human IL-2 protein | R and D: P60568 | |
| Recombinant DNA reagent | HIV-cycT1-IRES-YFP (HIV-CIY) | this paper | Sutton lab | plasmid |
| Recombinant DNA reagent | pSM-ADA Env | this paper | Sutton lab | plasmid |
| Recombinant DNA reagent | pSRα-YU2 Env | this paper | Heinrich Gottlinger, UMass Medical Cener | plasmid |
| Recombinant DNA reagent | pSRα-NL4-3 Env | this paper | Heinrich Gottlinger, UMass Medical Cener | plasmid |
| Recombinant DNA reagent | pME-VSV G | this paper | Sutton lab | plasmid |
| Recombinant DNA reagent | pCCL3L1 | Origene | NM_021006.4, NP_066286 | plasmid |
| Recombinant DNA reagent | pCCL4 | this paper | generated by PCR using pcDNA3/1 + CAT plasmid; Sutton lab | plasmid |
| Recombinant DNA reagent | Vpx-myc-his | Ned Landau laboratory, NYU Medical Center | plasmid | |
| Recombinant DNA reagent | pMDL-Chp6 | Ned Landau laboratory, NYU Medical Center | plasmid | |
| Cell line (H. Sapiens) | HEK 293T | ATCC | Cat# CRL-3216, RRID:CVCL_0063 | |
| Cell line (H. Sapiens) | GHOST.Hi5 | NIH AIDS Reagent Program | NIH-ARP Cat# 3944–343, RRID:CVCL_1E17 | |
| Cell line (H. Sapiens) | GHOST.CXCR4 | NIH AIDS Reagent Program | NIH-ARP Cat# 3685–448, RRID:CVCL_S492 | |
| Cell line (H. Sapiens) | TZM-bl cells | NIH AIDS Reagent Program | NIH-ARP Cat# 8129–442, RRID:CVCL_B478 | |
| Commercial assay or kit | RNeasy Mini Kit | Qiagen | ID: 74104 | |
| Commercial assay or kit | Mouse MIP-1 alpha (CCL3) ELISA | Invitrogen | LS885601322 | |
| Commercial assay or kit | Human CCL4 (MIP-1 beta) ELISA | Invitrogen | Invitrogen 88703476 | |
| Commercial assay or kit | High-Capacity cDNA Reverse Transcription Ki | ThermoFisher | ID: 4368814 | |
| Commercial assay or kit | DNeasy blood and tissue kit | Qiagen | Cat No./ID: 69504 | |
| Commercial assay or kit | SimpleChIP enzymatic ChIP kit agarose beads | Cell Signaling | Cat #9002 | |
| Commercial assay or kit | MinElute Reaction Cleanup kit | Qiagen | Cat No./ID: 28204 | |
| Commercial assay or kit | Transposase mixture | Illumina | Nextera DNA library prep kit; FC-131–1024 | |
| Chemical compound, drug | Phorbol 12-myristate 13-acetate | Sigma | PubChem CID: 27924 | |
| Chemical compound, drug | Ionomycin calcium salt | Sigma | I3909 | |
| Chemical compound, drug | Actinomycin D | Sigma. From Streptomyces sp | Cat # A1410 | |
| Chemical compound, drug | Digitonin | Promega | G944A | |
| Other | Power SYBR Green PCR Master Mix | ThermoFisher | Cat # 4367659 | Commercial reagent |
| Other | NEBnext PCR master mix | New England BioLabs | Cat # M0541S | Commercial reagent |
| Software, algorithm | CummeRbund | R package version 2.24.0 | DOI: 10.18129/B9.bioc.cummeRbund | |
| Software, algorithm | Illumina's CASAVA 1.8.2 | Illumina | Ref. 15011197 | |
| Software, algorithm | GraphPad Prism | GraphPad Prism (https://graphpad.com) | RRID:SCR_015807 | |
| Software, algorithm | FlowJo | https://www.flowjo.com/solutions/flowjo | RRID:SCR_008520 |
Additional files
-
Supplementary file 1
Clinical characteristics of EC/VC cohort.
Legend: F: female; M: male; F*: transgender male to female; No: number; Yr: year; ABC: Abacavir; ATV: Atazanavir; AZT: Zidovudine; COB: Cobicistat; DRV: Darunavir; DTG: Dolutegravir; EFV: Efavirenz; EGV: Elvitegravir; FTC: Emtricitabine; IFNA: Interferon alpha; LPV/r: Lopinavir/Ritonevir; MVC: Maraviroc; NVP: Nevirapine; PRED: Prednisolone; RGV: Raltegravir; RTV: Ritonavir; STRIBLD (EGV, COB, FTC, TDF); TAF: Tenofovir alafenamide; TDF: Tenofovir; 3TC: Lamivudine; IVDU: intravenous drug use; Homo: homosexual contact; Hetero: heterosexual contact; HCV: hepatitis C virus
- https://doi.org/10.7554/eLife.44360.015
-
Supplementary file 2
Significant genes obtained by RNA-seq analysis in CD4 +T cells from ATL2 family members.
- https://doi.org/10.7554/eLife.44360.016
-
Transparent reporting form
- https://doi.org/10.7554/eLife.44360.017





