Species specific differences in use of ANP32 proteins by influenza A virus
Figures
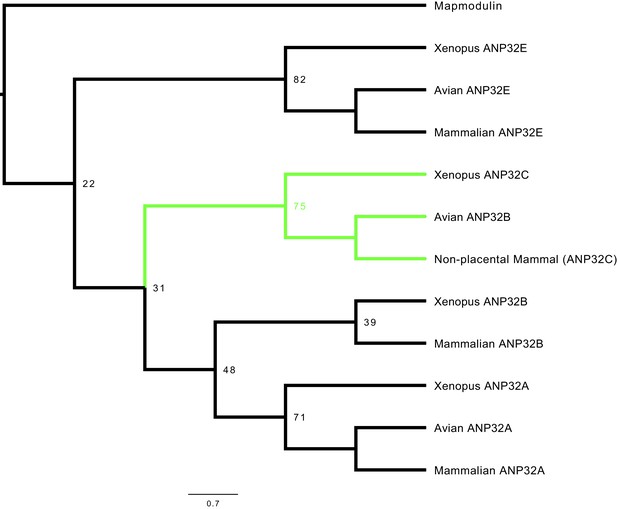
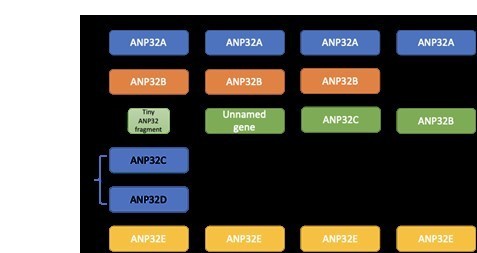
Phylogenetic and sequence analysis reveals avian ANP32B to be a paralog of mammalian ANP32B.
The best maximum-likelihood tree was calculated from a set of ANP32 proteins with mapmodulin from Drosophila melanogaster as an outgroup using RAxML with 100 bootstraps. This figure is a cladogram showing the relationships between mammalian ANP32s, avian ANP32s and ANP32s from Xenopus tropicalis. Selected bootstrap values show the relationship between different ANP32 protein clades. Avian ANP32B clade is shown in green. The full tree is shown in Figure 1—figure supplement 1.

Phylogenetic and sequence analysis reveals avian ANP32B to be a paralog of mammalian ANP32B.
Full tree used to make the cladogram in Figure 1. This tree was made using RAxML with 100 bootstraps and mapmodulin as an outgroup.
Synteny of ANP32 genes.
Chromosome locations of ANP32 family members A, B, C and E from coelacanth (Latimeria chalumnae), Xenopus (Xenopus tropicalis), chicken (Gallus gallus), zebra finch (Taeniopygia guttata), opossum (Monodelphis domestica), mouse (Mus musculus) and Human (Homo sapiens).
Chicken ANP32B is not functional for IAV polymerase.
Cells were transfected with avian H5N1 50–92 polymerase (PB2 627E or 627K) together with NP, firefly minigenome reporter, Renilla expression control, either Empty vector (control) or ANP32 expression plasmid and incubated at 37°C for 24 hr. (a) Minigenome assay in human eHAP1 cells with co-expressed Empty vector, FLAG-tagged chANP32A or chANP32B. (b) Minigenome assay in double knockout (dKO) eHAP1 cells. (c) Western blot analysis of dKO eHAP1 cell minigenome assay confirming expression of PB2 and FLAG-tagged chANP32A and B. (d) Minigenome assay in WT DF-1 cells with either co-expressed Empty vector or chANP32B. (e) Minigenome assay in DF-1 ANP32B knockout (bKO) cells with either co-expressed Empty vector or chANP32B. Data shown are firefly activity normalised to Renilla, plotted as mean ± SEM (n = 3 biological replicates). Two-way ANOVA with Dunnet’s multiple comparisons to Empty vector. ns = not significant, ****p<0.0001.
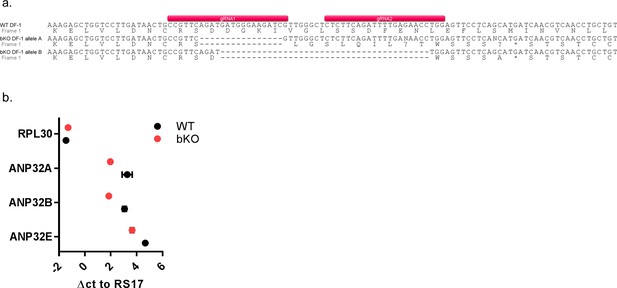
Sequence analysis of ANP32 in genome edited DF-1 chicken cells.
(a) DNA sequence analysis of ANP32B from genomic DNA of DF-1 WT and ΔB clones, showing the target sequence of the gRNA pair used in the CRISPR/Cas9 reaction. Allele A had a 16 bp deletion and allele B a 40 bp deletion, resulting in premature stop codons in the ANP32B sequence. (b) qRT-PCR analysis of mRNA isolated from WT and bKO DF-1 cells. Data are Δct of RPL30, ANP32A, B or E to RS17. Data are Δct of RS17, ANP32A, B or E to RPL30. Annotated alignments generated using Geneious R6 software.
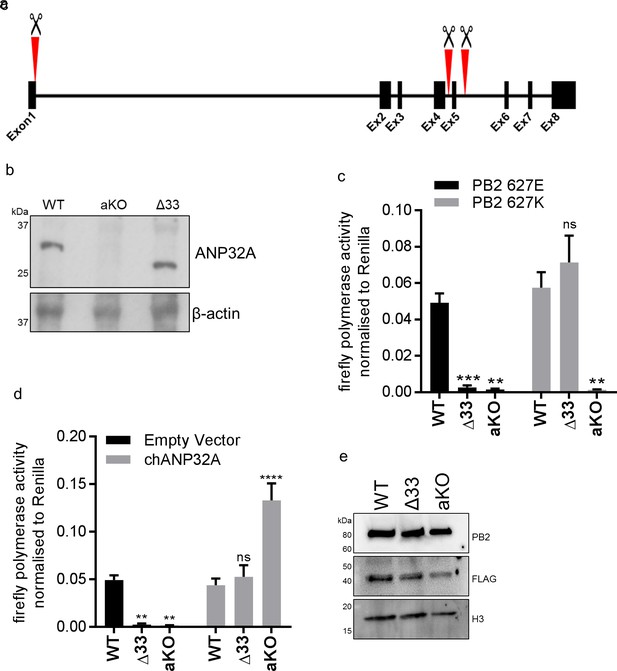
Chicken PGC derived fibroblast cells lacking ANP32A or the 33 amino acid insertion do not support avian IAV polymerase activity.
(a) Schematic of CRISPR/Cas9 RNA guide targets used to generate aKO (exon1) and Δ33 (exon 5) PGC cell lines. (b) Western blot analysis of ANP32A and β-actin expression in WT, KO and Δ33 PGC-derived fibroblast cells. (c) Minigenome assay in WT, Δ33 or aKO PGC derived fibroblast cells with either PB2627E (black) or 627K (grey) polymerase derived from avian H5N1 50–92 virus. (d) Minigenome assay in WT, Δ33 or aKO cells with avian H5N1 50–92 PB2 627E polymerase co-transfected with Empty vector (black) or FLAG-tagged chANP32A (grey). (e) Western blot analysis of PB2, FLAG and Histone 3. Data shown are firefly activity normalised to Renilla, plotted as mean ± SEM (n = 3 biological replicates). Two-way ANOVA with Dunnet’s multiple comparisons to WT. ns = not significant, **p<0.01, ***p<0.001, ****p<0.0001.
Sequence analysis of ANP32 in genome edited PGC chicken cells.
Alignment of DNA sequence from WT, Δ33 and KO PGCs, showing the target sequence of the gRNAs used in the CRISPR/Cas9 reaction. (a) Comparison between WT and KO PGCs showed an 8 bp deletion in exon 1 of ANP32A in KO PGC cells, resulting in a truncated ANP32A protein. (b) Intron and exon five sequence comparison of WT and Δ33 cells revealed a 400 bp deletion resulting in the loss of exon 5. (c) qRT-PCR analysis of mRNA isolated from WT, Δ33 or aKO PGC derived fibroblast cells. Data are Δct of RS17, ANP32A, B or E to RPL30. Annotated alignments generated using Geneious R6 software.
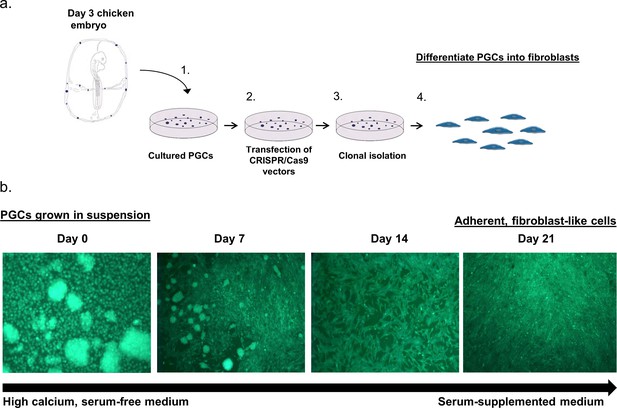
In vitro reprogramming of chicken PGCs into adherent fibroblast-like cells.
(a) Diagram describing the method to differentiate PGCs from day three chicken embryos. (b) Fluorescent images of live cells demonstrating the differentiation of GFP+ PGCs into fibroblast-like cells.
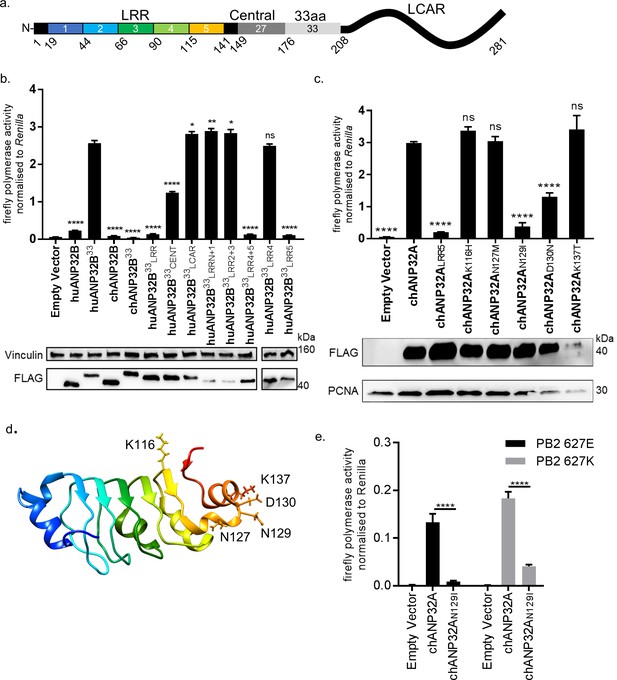
Lack of functional support for IAV polymerase by chicken ANP32B maps to differences in LRR5 domain.
(a) Schematic of chicken ANP32A protein highlighting the different domains and LRR sequences (LRR 1–5). (b) Human 293 T cells were transfected with avian H5N1 50–92 polymerase (PB2 627E) together with NP, pHOM1-firefly minigenome reporter, Renilla expression control, either Empty vector or FLAG-tagged ANP32 expression plasmid and incubated at 37°C for 24 hr. Western blot analysis shown below (FLAG and Vinculin). (c) Minigenome assay in 293 T cells (PB2 627E) with FLAG-tagged WT or mutant chANP32A expression plasmids with associated western blot (FLAG and PCNA). (d) huANP32A crystal structure (PDB 4 × 05) with residues K116, N127, N129, D130 and K137 highlighted using UCSF Chimaera (Pettersen et al., 2004). (e) Minigenome assay of avian H5N1 50–92 polymerase with either PB2 627E or 627K in PGC-derived fibroblast aKO cells, together with co-expressed Empty vector, chANP32A or chANP32AN129I. Data shown are firefly activity normalised to Renilla, plotted as mean ± SEM (n = 3 biological replicates). One-way ANOVA with Tukey’s comparison to chANP32A (b and c) or two-way ANOVA with Dunnet’s multiple comparisons to chANP32A (e). ns = not significant, *p<0.05, **p<0.01, ****p<0.0001.
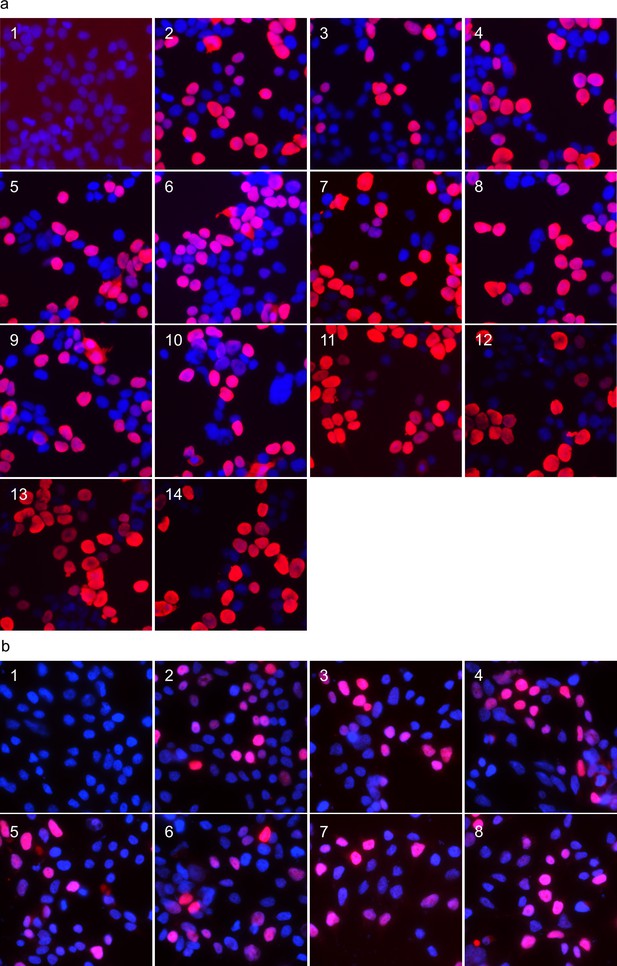
Nuclear localisation of exogenously expressed ANP32 proteins in 293 T cells.
Immunofluorescent images of 293 T cells expressing ANP32 constructs fixed and stained with DAPI to highlight the nucleus. (a) FLAG-tagged ANP32A constructs were imaged by probing with mouse α-FLAG antibody and detected by α-mouse AlexaFluor-568: Empty vector(1), huANP32B(2), huANP32B33(3), chANP32B(4), chANP32B33(5), huANP32B33LRR(6), huANP32B33CENT(7), huANP32B33LCAR(8), huANP32B33N+LRR1(9), huANP32B33LRR2+3 (10), huANP32B33LRR4+5 (11), chANP32A(12), chANP32Ascr149-175(13), chANP32Ascr176-208(14). (b) ANP32A with mCherry fused to the C-terminus: Empty Vector(1), chANP32A(2), chANP32B(3), chANP32AK116H(4), chANP32AN127M(5), chANP32AN129I(6), chANP32AD130N(7), chANP32AK137T(8). All images were prepared using ImageJ (Rueden et al., 2017) and Microsoft PowerPoint.
Avian ANP32B proteins share the I129 and N130 residues in LRR5.
Alignment comparing sequence of LRR5 from chicken ANP32A and ANP32B sequences from Homo sapiens and 22 avian species (residues 115 to 141). Protein sequences downloaded from NCBI and aligned using Geneious R6 software.
Sequence of amino acids 149–175 of the central domain of chANP32A are required to support activity of both avian and human-adapted IAV polymerase.
(a) Schematic of chANP32A showing the sequence of amino acids in the central domain (149–175 or 33 amino acid insertion (176-208) and the randomly scrambled sequence in red. (b) Minigenome assay of avian H5N1 50–92 polymerase with either PB2 627E or 627K in PGC-derived fibroblast aKO cells with co-expressed Empty plasmid or FLAG-tagged WT chANP32A, chANP32Ascr149-175 or chANP32Ascr176-208 expression plasmids. (c) Western blot analysis of PB2 (627E), lamin B1 and FLAG. Data shown are firefly activity normalised to Renilla, plotted as mean ± SEM (n = 3 biological replicates). Two-way ANOVA with Dunnet’s multiple comparisons to chANP32A. ns = not significant, ****p<0.0001.
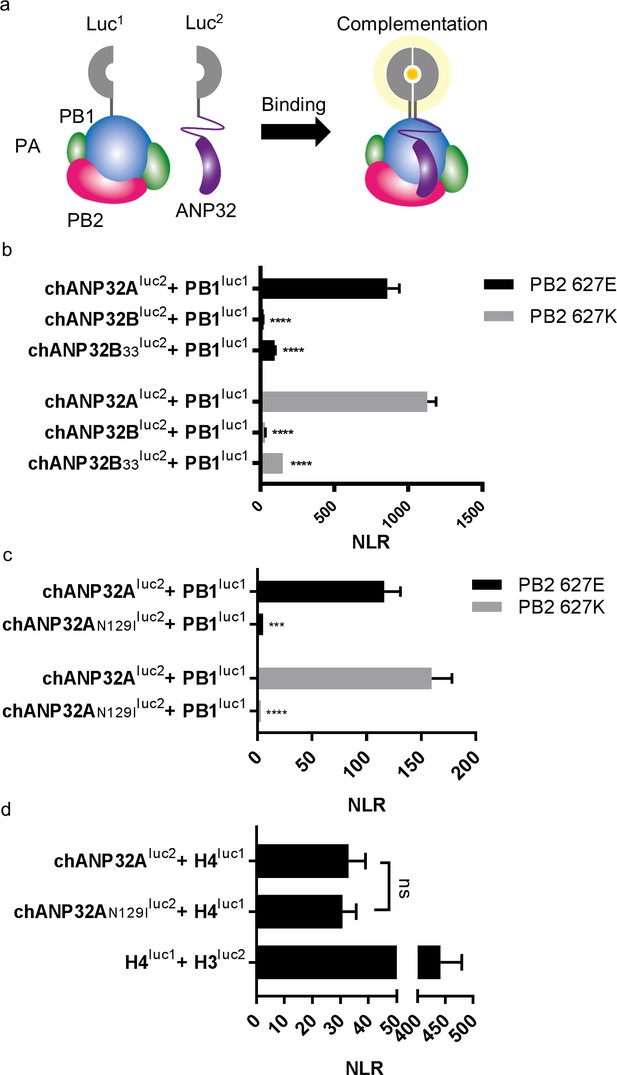
A single amino acid change (N129I) derived from chANP32B disrupts chANP32A support of influenza polymerase activity by abrogating binding to IAV polymerase.
(a) Diagram of the split Gaussia luciferase system, demonstrating how ANP32 fused to luciferase fragment luc2 may bind to polymerase containing PB1 fused to luciferase fragment luc1 and complement full luciferase, which then reacts with substrate to generate a measurable bioluminescent signal. (b) Human 293 T cells were transfected with PB1 fused to luc1 (PB1luc1), PB2 (627E or K), PA and either chANP32A, chANP32B or chANP32B33 fused to luc2 (control wells were transfected with all components but with unfused PB1 and luc1 or chANP32 and luc2). (c) As (b) but with either chANP32Aluc2 or chANP32AN129Iluc2. (d) 293 T cells transfected with either chANP32Aluc2 or chANP32AN129Iluc2 and histone four fused to luc1 (or with unfused controls) or with H4luc1 and histone three fused to luc2. All data are Normalised Luciferase Ratio (n = 3 biological replicates) (Figure 6—figure supplement 1). One-way ANOVA (d) or two-way ANOVA with Dunnet’s multiple comparisons to chANP32A (b and c). ns = not significant, ****p<0.0001.
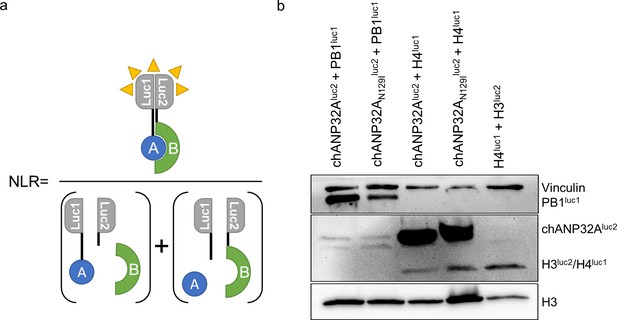
Western blot analysis of split luciferase constructs.
(a) Normalised luciferase Ratio was calculated by the equation described in this diagram, whereby the bioluminescence measured by interacting partners A and B fused to luc1 and luc2 are divided by the sum of the bioluminescence of the unfused controls. (b) Western blot analysis of luc-fused constructs (α-Vinculin, PB1, Gaussia luciferase and histone 3).
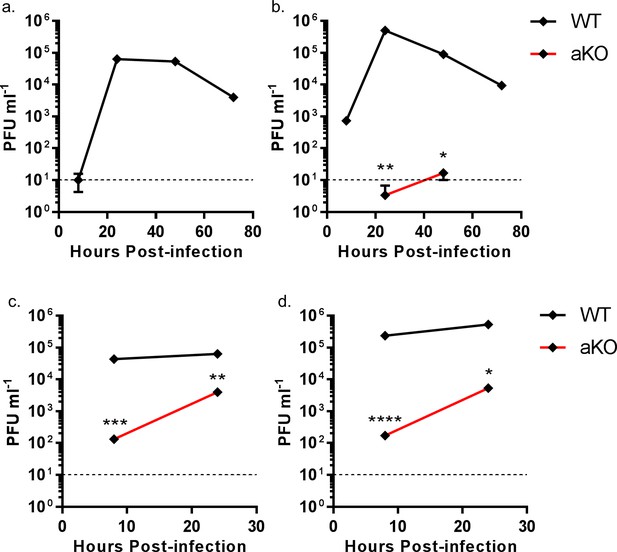
Viral replication is abrogated in chicken PGC fibroblast cells lacking ANP32A.
WT (black lines) or aKO (red lines) PGC-derived fibroblast cells were infected with recombinant viruses (containing PR8 HA, NA and M genes and internal genes from either H5N1 50–92 or H7N9 Anhui), at an MOI of either 0.0001 (a,b) or 1.0 (c,d), incubated at 37°C in the presence of trypsin, cell supernatants harvested at described time-points and PFU ml−1 measured by plaque assay on MDCK cells. (a) H5N1 50–92 (MOI 0.0001). b. H7N9 Anhui (MOI 0.0001). c. H5N1 50-−92 (MOI 1.0). d. H7N9 Anhui (MOI 1.0). vRNAs from supernatants 24 hr post infection MOI 1.0 (c and d) were extracted, PCR amplified and sequenced by Sanger sequencing. Limit of detection by plaque assay shown by dotted line (10PFU ml−1). n = 3 biological replicates. Multiple t-tests with Holm-Sidak comparison. ns = not significant, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (G. gallus) | GFP+/Hyline cross | Roslin Institute | Fertile heterozygous eggs for PGC derivations (Pettersen et al., 2004) | |
| Primary cells (G. gallus) | Primordial Germ Cells (PGCs) | G. gallus GFP+/Hyline cross, Roslin Institute (This study) | ||
| Cell line (G. gallus) | DF-1 fibroblasts | American Type Culture Collection | CRL-12203; RRID:CVCL_0570 | |
| Cell line (C. sapiens) | MDCK | American Type Culture Collection | CCL- 34; RRID:CVCL_0422 | |
| Cell line (H. sapiens) | 293T | American Type Culture Collection | CRL-3216; RRID:CVCL_0063 | |
| Cell line (H. sapiens) | eHAP1 | Horizon Discovery | C669 | |
| Antibody | rabbit polyclonal α-ANP32A | Sigma-Aldrich | AV40203; RRID:AB_1844874 | Dilution 1:500-1:1000 |
| Antibody | mouse monoclonal α-β-actin | Sigma-Aldrich | A2228; RRID:AB_476697 | Dilution 1:1000 |
| Antibody | mouse monoclonal α-FLAG | Sigma-Aldrich | F1804; RRID:AB_262044 | Dilution 1:1000 (WB), 1:300 (IF) |
| Antibody | mouse monoclonal α-Lamin B1 | Merck | MAB5492; RRID:AB_2085944 | Dilution 1:1000 |
| Antibody | mouse monoclonal α-PCNA | Santa Cruz | sc-25280, RRID:AB_628109 | Dilution 1:1000 |
| Antibody | rabbit polyclonal α-Histone 3 | Abcam | AB1791; RRID:AB_302613 | Dilution 1:2000 |
| Antibody | rabbit monoclonal α-vinculin | Abcam | AB129002; RRID:AB_11144129 | Dilution 1:1000 |
| Antibody | rabbit polyclonal α-Gaussia Luc | NEB | E80235 | Dilution 1:2000 |
| Antibody | rabbit polyclonal α-PB1 | Invitrogen | PA5-34914; RRID:AB_2552264 | Dilution 1:2000 |
| Antibody | rabbit polyclonal α-PB2 | GeneTex | GTX125926; RRID:AB_11162999 | Dilution 1:2000 |
| Antibody | goat polyclonal anti-rabbit HRP | CST | 7074 | Dilution 1:2000 |
| Antibody | Horse polyclonal anti-mouse HRP | CST | 7076 | Dilution 1:2000 |
| Antibody | Sheep polyclonal α-rabbit HRP | Merck | AP510P | Dilution 1:20000 |
| Antibody | goat polyclonal α-mouse HRP | AbD Serotec | STAR117P; RRID:AB_323839 | Dilution 1:10000 |
| Antibody | goat polyconal α-mouse AlexaFluor-568 | Invitrogen | A11031; RRID:AB_144696 | Dilution 1:1000 |
| Recombinant DNA reagent | pGEM-T Easy vector | Promega | A1360 | |
| Recombinant DNA reagent | pSpCas9(BB)−2A-Puro PX459 V2.0 vector | Gift from Dr. Feng Zhang | RRID:Addgene_62988 | |
| Recombinant DNA reagent | pSpCas9n(BB)−2A-GFP PX461 vecotr | addgene | Plasmid 48140; RRID:Addgene_48140 | |
| Recombinant DNA reagent | pCAGGS vector | Belgium Co-ordinated Collections of Microorganisms (BCCM), University of Ghent, Belgium | ||
| Recombinant DNA reagent | H5N1 A/turkey/England/50-92/1991 polI plasmids | APHA, Weybridge, UK | ||
| Recombinant DNA reagent | H7N9 Anhui/1/2013 pHW2000 plasmids | Pirbright Institute, UK | ||
| Recombinant DNA reagent | H1N1 A/PR/8/34 (PR8) polI or pHW2000 plasmids | (Neumann et al., 1999) | ||
| Commercial assay or kit | RNeasy mini kit | Qiagen | 74106 | |
| Commercial assay or kit | Quick Start Bradford Protein Assay Kit | Biorad | 5000202 | |
| Commercial assay or kit | Dual-luciferase Reporter assay system | Promega | E1910 | |
| Commercial assay or kit | Renilla luciferase kit | Promega | E2810 | |
| Commercial assay or kit | QIAamp Viral RNA Mini Kit | Qiagen | 52906 | |
| Chemical compound, drug | Lipofectamine 3000 | Invitrogen | L3000008 | |
| Chemical compound, drug | Lipofectamine 2000 | Invitrogen | 11668019 | |
| Software, algorithm | Image J | ImageJ (http://imagej.nih.gov/ij/) | ||
| Software, algorithm | GraphPad Prism | GraphPad Prism (https://graphpad.com) | version 6 | |
| Software, algorithm | Geneious | Geneious https://www.geneious.com | R6 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.45066.018