Response to comment on 'Valid molecular dynamics simulations of human hemoglobin require a surprisingly large box size'
Figures
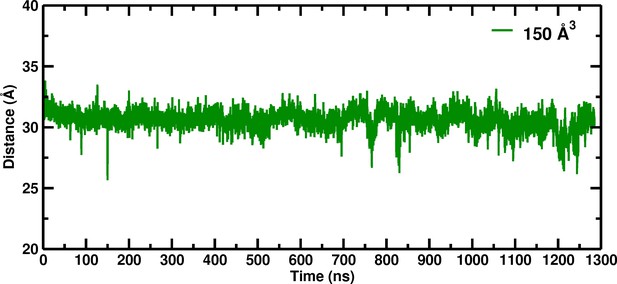
Temporal change of the Cα–Cα distance between His146B2 and His146B2 for the 150 Å box during 1280 nanoseconds of molecular dynamics simulation.
https://doi.org/10.7554/eLife.45318.002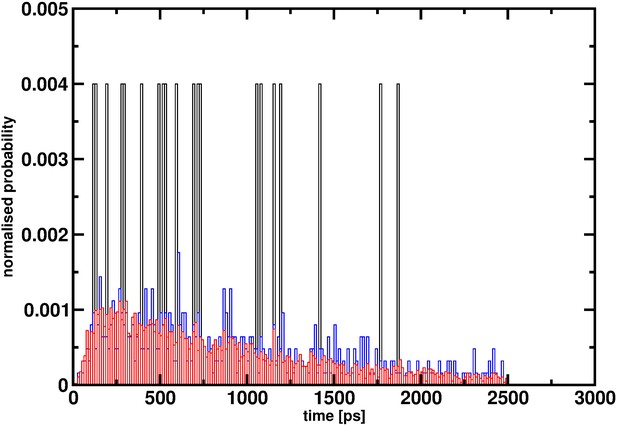
Reaction time distribution p(τ) for the decomposition HSO3Cl -> SO3+HCl from 20 (black), 500 (red) and 5000 (blue) reactive trajectories.
With 20 trajectories the distribution is far from converged and even with 500 trajectories convergence appears to be incomplete. It is only for 5000 independent events that p(τ) approaches a smooth distribution.
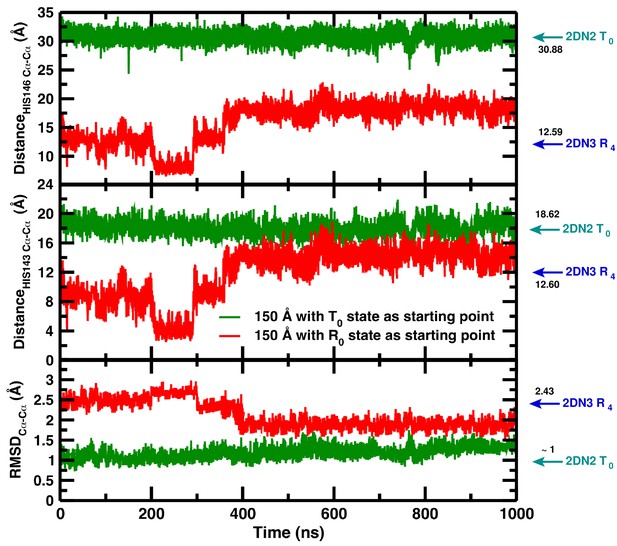
Temporal change in the 150 Å box of (from top to bottom) the Cα–Cα distance between His146B1 and His146B2, the Cα–Cα distance between His143B1 and His143B2, and the Cα RMSD relative to the 2DN2 X-ray structure.
The green and red lines report the time series reported in our article (simulation starting from the T(0) state) and the time series of a simulation starting from the decayed T(0) state (i.e., an R(0) state structure) from R state in the 120 Å box, respectively. Cyan and blue arrows indicate the values of the corresponding observables found for the deoxy T(0)(2DN2) and oxy R(4)(2DN3) states, respectively.
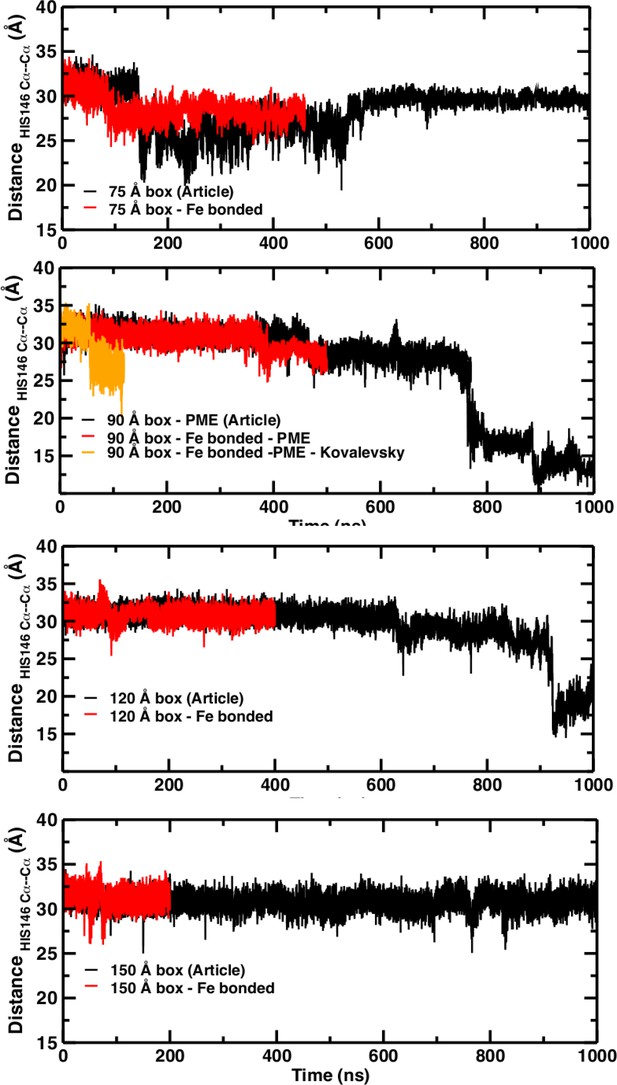
Temporal change of the Cα–Cα distance between His146B1 and His146B2 for the 75 Å box (A), 90 Å box (B), 120 Å box (C), and 150 Å box (D).
In (A), (C), and (D) the black line reports the time series published and the red line shows the new time series obtained with the new simulation with the Fe–N bond. In (B) the black trace is from El Hage et al. (2018) the red trace for the bonded Fe–His simulations, the green line from using finite cutoffs for evaluating the electrostatics (cutoff 14 Å), and the orange line from using Kovalevsky protonation states for all histidines.
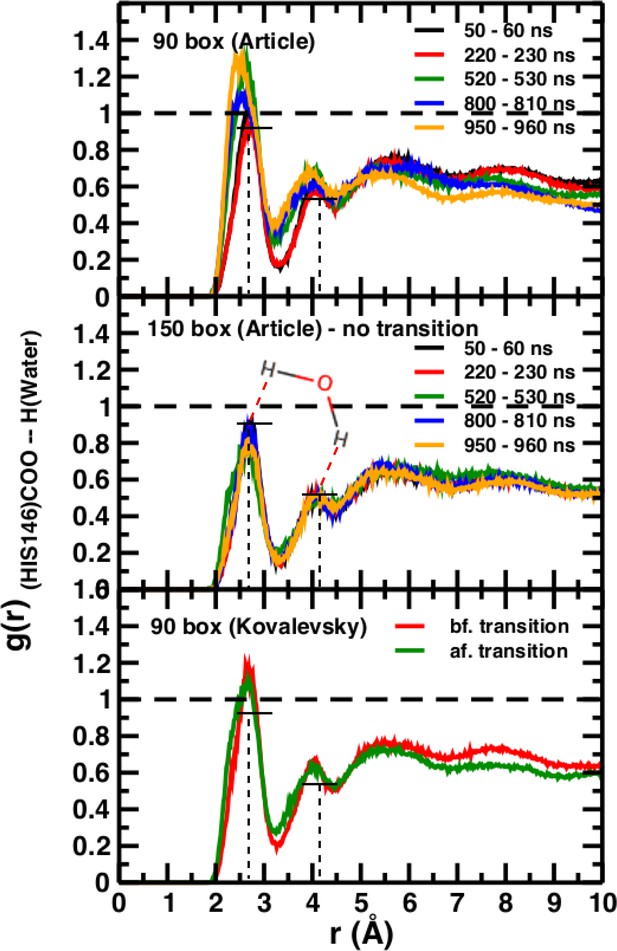
Time-averaged radial distribution functions g(r) between the C-terminal (COO) of His146 and water H for box sizes 90 Å (top) and 150 Å (middle) data from Figure 6 of El Hage et al. (2018) and from before and after the transition, obtained when using the Kovalevsky protonation in the 90 Å box (bottom).
https://doi.org/10.7554/eLife.45318.007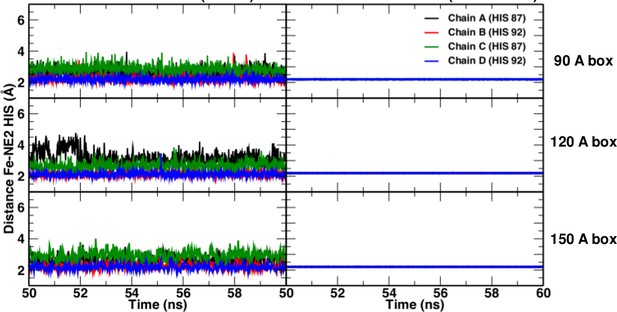
Fe-NHis bond lengths in simulations without (left) and with (right) explicit bond.
https://doi.org/10.7554/eLife.45318.008Tables
Histidine protonation states in hemoglobin as used in simulations by the present authors (Zheng et al., 2013; El Hage et al., 2018), and the histidine protonation states in the files that were supplied to us by the authors of the comment (originally from Kovalevsky et al., 2010).
https://doi.org/10.7554/eLife.45318.005| Zheng et al., 2013; El Hage et al., 2018 | Kovalevsky et al., 2010 | Zheng et al., 2013; El Hage et al., 2018 | Kovalevsky et al., 2010 | ||||
|---|---|---|---|---|---|---|---|
| Res. | Chain A/C | Chain A | Chain C | Res. | Chain B/D | Chain B | Chain D |
| 20 | HSE | HSP | HSE | 2 | HSE | HSE | HSE |
| 45 | HSE | HSE | HSE | 63 | HSE | HSP | HSE |
| 50 | HSD | HSD | HSP | 77 | HSE | HSE | HSE |
| 58 | HSE | HSP | HSE | 92 | HSD-Fe | HSD-Fe | HSD-Fe |
| 72 | HSD | HSP | HSP | 97 | HSE | HSP | HSP |
| 87 | HSD-Fe | HSD -Fe | HSD-Fe | 116 | HSE | HSP | HSP |
| 89 | HSE | HSE | HSP | 117 | HSE | HSE | HSE |
| 103 | HSE | HSP | HSP | 143 | HSE | HSE | HSP |
| 112 | HSE | HSP | HSP | 146 | HSP | HSP | HSP |
| 122 | HSE | HSE | HSE | ||||





