Asymmetry between the two acidic patches dictates the direction of nucleosome sliding by the ISWI chromatin remodeler
Figures
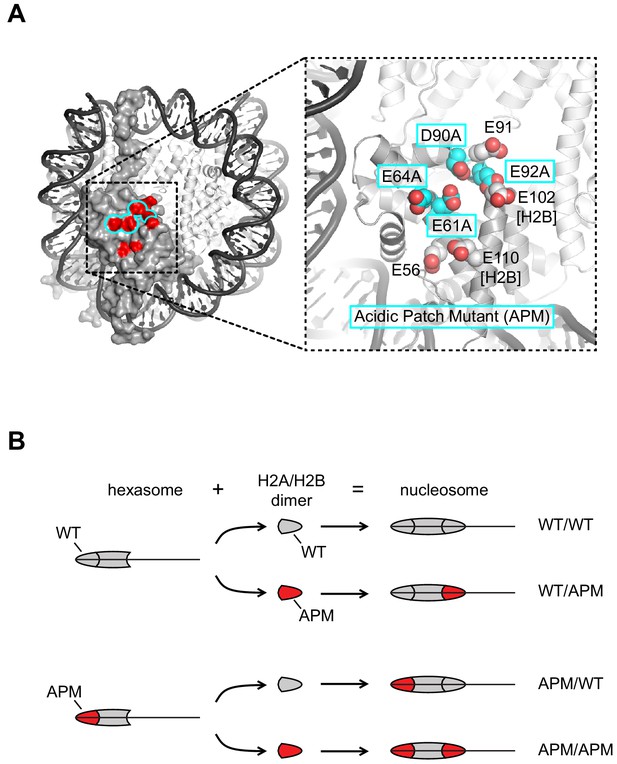
Design of nucleosomes with acidic patch mutations (APMs).
(A) The acidic patch of the nucleosome. In the overview, the surface is shown for one H2A/H2B dimer, with eight residues comprising the acidic patch highlighted in red. The acidic patch mutation (APM) consists of four alanine substitutions on H2A (E61A/E64A/D90A/E92A), highlighted in cyan. The structure shown is PDB code 1KX5 (Davey et al., 2002). (B) Strategy for making asymmetric nucleosomes using oriented hexasomes. As previously shown (Levendosky et al., 2016), hexasomes made with the Widom 601 (Lowary and Widom, 1998) preferentially lack an H2A/H2B dimer on the TA-poor side (here, the right side). Addition of either a wild type (gray) or APM H2A/H2B dimer (red) converts hexasomes to nucleosomes, allowing for production of all four combinations of H2A/H2B dimers with and without the APM. The locations of the H2A/H2B dimers and the H3/H4 tetramer on the Widom 601 is depicted in Figure 1—figure supplement 1.
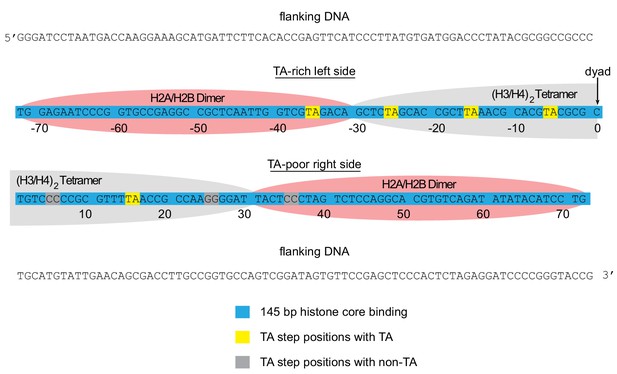
Arrangement of histone contacts with the Widom 601.
The approximate footprint from the histone core contacts of the H2A/H2B dimers (pink) and (H3/H4)2 tetramer (gray) are shown on the Widom 601 sequence (Lowary and Widom, 1998). In hexasomes, the sole H2A/H2B dimer preferentially resides on the TA-rich side of the Widom 601, shown here on the left.
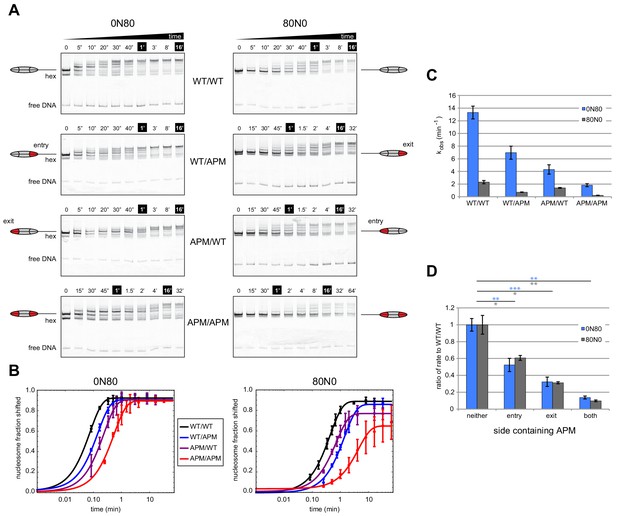
Mutations in the acidic patch slow down but do not disrupt centering ability of Chd1.
(A) Native gel nucleosome sliding assays using Chd1, with all four arrangements of APM and WT H2A/H2B dimers. Depending on which side the flanking DNA was on, the unique APM dimers were either on entry or exit sides, as indicated. The positions of the nucleosome cartoons indicate end-positioned nucleosomes in the gel, with the lower band representing residual hexasomes. Nucleosome centering is evidenced by slower migration in the gel. Reactions contained 40 nM hexasome, 60 nM H2A/H2B dimer, 200 nM Chd1, and 100 µM ATP. Note the different time series used (indicated above each gel), which helped capture sliding intermediates given the different reaction rates. Results are representative of three independent replicates. (B) Quantified data from (A) plotting the disappearance of end-positioned nucleosomes, overlaid with single exponential fits. The end-positioned band intensity was normalized to the total band intensity within each lane. Error bars show standard deviations from three replicates. (C) Mean observed rates ± standard deviations from fits obtained in (B). (D) Relative impact of APMs on Chd1 remodeling rates. Remodeling rates for WT/WT 0N80 and 80N0 were each scaled to 1. Rates for nucleosomes containing APM substitutions were scaled relative to WT/WT made with the same DNA construct. P-values *≤0.02; **<0.005; ***<0.0005.
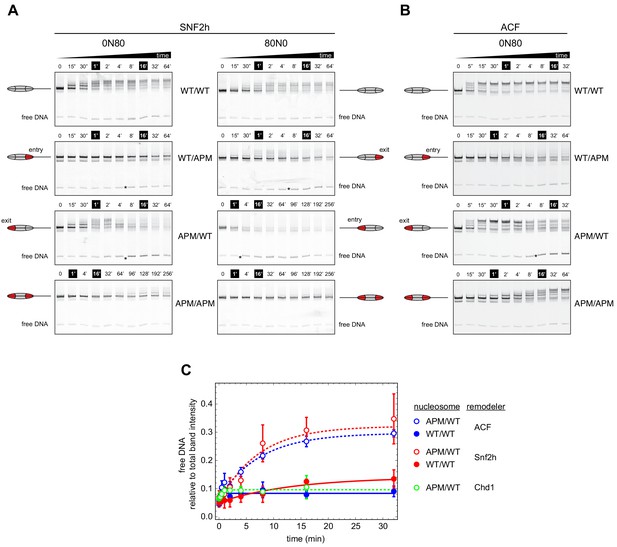
Asymmetry between the two nucleosome acidic patches disrupts nucleosome centering by ISWI remodelers.
(A) Native gel nucleosome sliding assays using SNF2h, with all four arrangements of WT and APM H2A/H2B dimers for 0N80 and 80N0 nucleosomes. Nucleosome centering corresponds to slower migration of nucleosome species. Asterisks highlight noticeable increases in free DNA. Remodeling reactions contained 40 nM hexasome, 80 nM H2A/H2B dimer, 1 µM SNF2h, and 2 mM ATP. Results are representative of two independent replicates. (B) Native gel nucleosome sliding experiments of 0N80 nucleosomes as in (A), but using Drosophila ACF. Remodeling reactions contained 40 nM hexasome, 70 nM H2A/H2B dimer, 100 nM ACF, and 2 mM ATP. Results are representative of two or more replicates. (C) ISWI remodelers but not Chd1 stimulate release of DNA from 0N80 nucleosomes with APM/WT histone cores. Shown is the accumulation of free DNA normalized to total band intensity from native gel experiments. All reactions tracking free DNA contained 40 nM hexasome, 70 nM H2A/H2B dimer, and 2 mM ATP with either 1 µM Snf2h, 100 nM ACF, or 200 nM Chd1. The mean and standard deviation of three or four replicates is plotted with single exponentials fits.
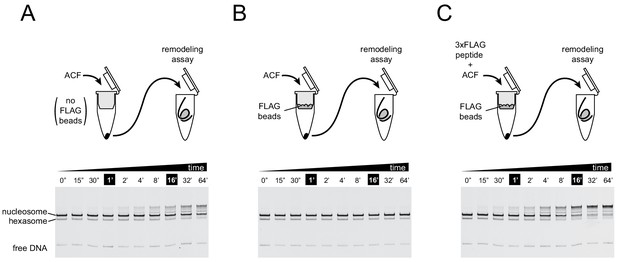
For the ACF complex, association of the Acf1 subunit tracks with nucleosome sliding activity.
To ensure that the remodeling activity observed in experiments using ACF was not caused by the Iswi ATPase subunit independently, nucleosome sliding activity was monitored after retention of the FLAG-tagged Acf1 regulatory subunit with anti-FLAG beads. Native gel nucleosome sliding experiments (see Materials and methods section) were conducted using 10 nM ACF complex that was incubated for 30 min on spin columns either without anti-FLAG beads (A), with anti-FLAG beads (B), or with anti-FLAG beads that were blocked with 3xFLAG peptide (C). The loss of activity observed in (B) indicated that remodeling activity from ACF was due to the complete ACF complex. Experiments were carried out in duplicate.
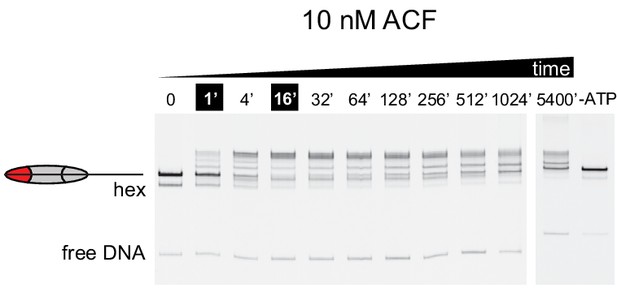
At sub-saturating concentrations, ACF shifts nucleosomes with asymmetric acidic patch mutations toward DNA ends.
Nucleosome sliding experiments monitored by native acrylamide gels were carried out with 10 nM ACF, 40 nM 0N80 APM hexasomes, 70 nM WT H2A/H2B dimer and 2 mM ATP. To see if the remodeling reaction had reached equilibrium, additional time points were taken after ~17 and 90 hr. These extended time points show a different distribution, suggesting that nucleosome bands begin moving back down the gel toward the end position. Although much slower than reactions performed with 100 nM ACF, these results are consistent with the finding that ACF initially centers and then shifts asymmetric APM/WT nucleosomes toward an end position. The rightmost lane contains nucleosomes from a reaction assembled without ATP that stayed at room temperature for 5400 min alongside the remodeling reaction. Shown is a representative of three independent replicates.
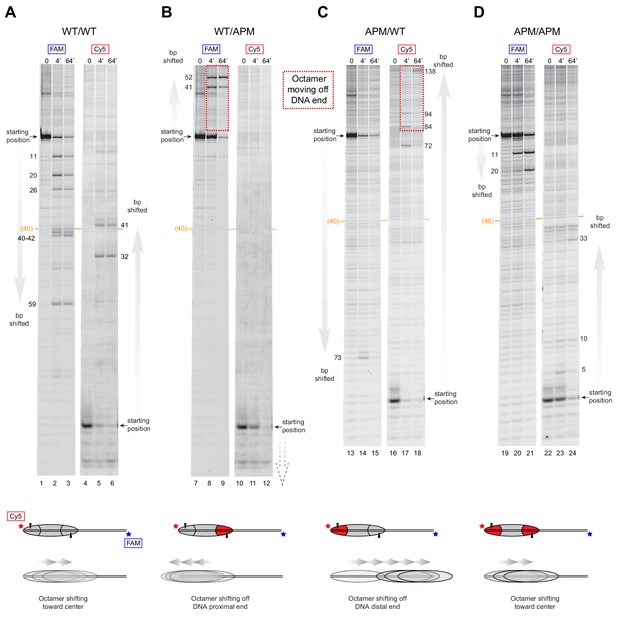
SNF2h slides histone cores with asymmetric acidic patch mutations off DNA ends.
Histone mapping of SNF2h sliding reactions, performed with 0N80 nucleosomes having the four combinations of wild type and APM H2A/H2B dimers: (A) WT/WT, (B) WT/APM, (C) APM/WT, and (D) APM/APM. Shown are scans of urea denaturing gels, which report on the locations of the histone octamer based on sites of H2B(S53C) cross-linking. Two scans are shown for each nucleosome, based on Cy5 and FAM labels on the top and bottom DNA strands, respectively. Numbering beside the gels indicates the distances (bp) the histone octamer shifted relative to the starting position, with the midpoint of the 80 bp flanking DNA highlighted with a central orange bar. Each nucleosome was generated by addition of 200 nM wild type (gray) or APM (red) H2A/H2B dimers to 100 nM wild type or APM hexasome. Cartoon schematics below each gel indicate the composition and orientation of APM and wild type H2A/H2B dimers, the sites of H2B(S53C) cross-linking (black triangles), and interpretations of sliding reactions. Sites where the histone octamer moves off DNA ends are highlighted by dotted red boxes. Sliding reactions contained 1 µM SNF2h and 2 mM ATP, and reactions were quenched at 0, 4, and 64 min. These results are representative of two independent experiments. Analogous experiments using 80N0 nucleosomes are shown in Figure 4—figure supplement 1. Extended gels are shown in Figure 4—figure supplement 2 and Figure 4—figure supplement 3.
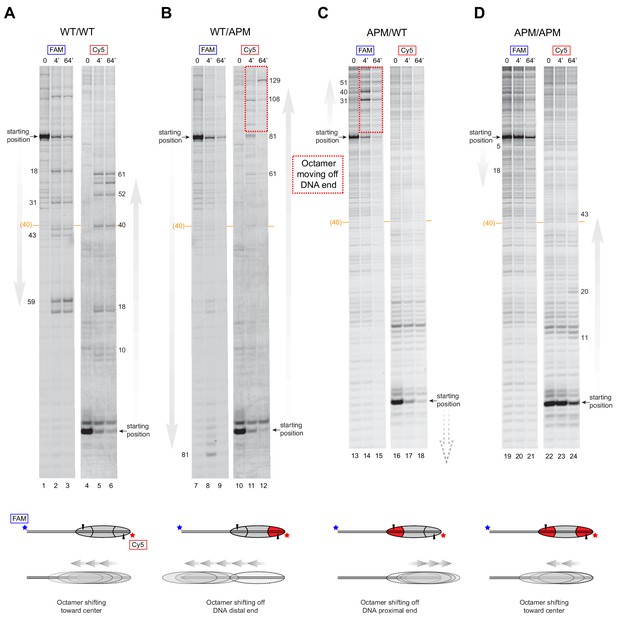
SNF2h slides histone cores with single acidic patch mutations off DNA ends regardless of Widom 601 orientation.
Histone mapping of SNF2h sliding reactions, performed with 80N0 nucleosomes having the four combinations of wild type and APM H2A/H2B dimers: (A) WT/WT, (B) WT/APM, (C) APM/WT, and (D) APM/APM. These experiments are analogous to those of Figure 4 but with the opposite sequence orientation of the Widom 601 relative to flanking DNA. These experiments were carried out in duplicate alongside those in Figure 4. Extended gels are shown in Figure 4—figure supplement 2 and Figure 4—figure supplement 3.
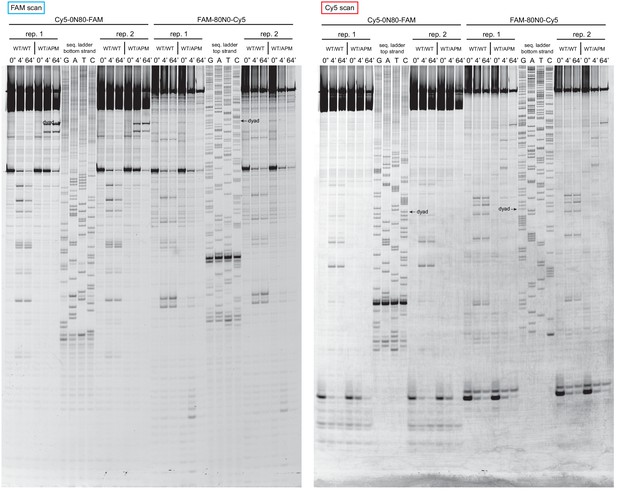
Extended gel images of histone mapping experiments using WT/WT and WT/APM nucleosomes.
These extended gels include the sequencing ladders used to determine the distances of shifted cross-links for experiments shown in Figure 4 and Figure 4—figure supplement 1. The dyad locations of unshifted nucleosomes are indicated.
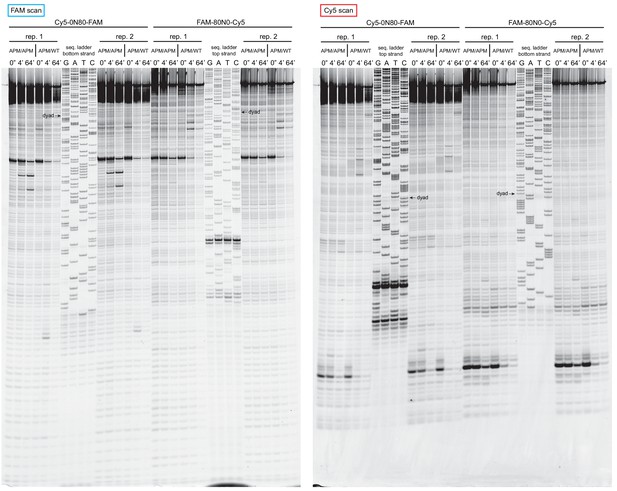
Extended gel images of histone mapping experiments using APM/WT and APM/APM nucleosomes.
These extended gels include the sequencing ladders used to determine the distances of shifted cross-links for experiments shown in Figure 4 and Figure 4—figure supplement 1. The dyad locations of unshifted nucleosomes are indicated.
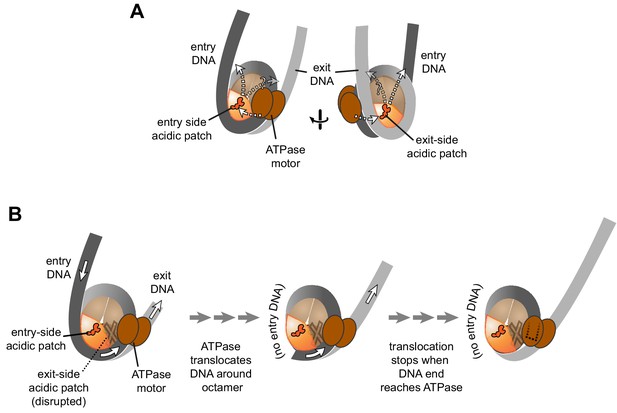
Schematics showing the spatial relationship between the remodeler ATPase motor, the two acidic patches, and flanking DNA of the nucleosome.
(A) A cartoon representation showing the two sides of a nucleosome with a remodeler ATPase motor bound at an SHL2 site. Broken arrows show paths connecting the ATPase motor with entry or exit DNA. (B) An illustration of how sliding the histone core off DNA ends impacts flanking DNA relative to the ATPase motor. As for ISWI, if action of the ATPase motor on one side of the nucleosome is severely limited due to an acidic patch mutation (brown "X"), DNA will be preferentially shifted in one direction due to ATPase action on the side with the wild type acidic patch (orange). Continued action on one side, with insufficient counteraction on the other side, will eventually pull all entry side DNA completely onto the histone core. If translocation continues without the need for flanking DNA on the entry side, the end of the DNA will eventually reach the ATPase motor at SHL2 (dotted outline).
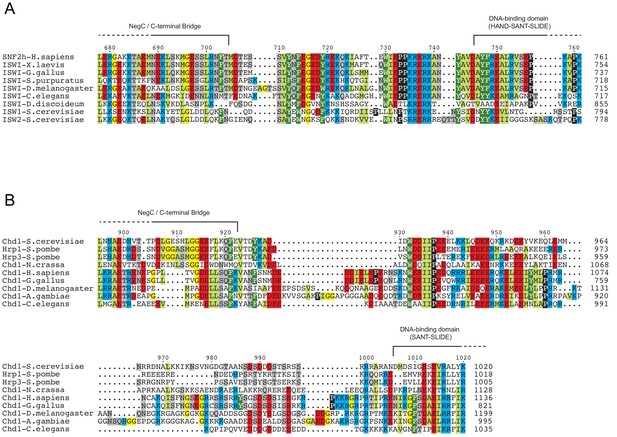
Chd1 remodelers have a longer linker connecting the ATPase motor and DNA-binding domain than ISWI.
Sequence alignments for a range of orthologs of ISWI (A) and Chd1 (B) chromatin remodelers, focused on the region between that ATPase motor and the DNA binding domain. Domain boundaries were taken from representative crystal structures: 3MWY (Hauk et al., 2010), 3TED (Sharma et al., 2011) and 1OFC (Grüne et al., 2003).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | mouse monoclonal ANTI-FLAG M2 Affinity Gel | Millipore Sigma | Cat #:2220 | (coupled to beads) |
| Recombinant DNA reagent | core Widom 601 (uppercase) and flanking DNA sequences (lowercase) | Lowary and Widom, 1998 | 5’_gggatcctaatgaccaaggaaa gcatgattcttcacaccgagttcatcc cttatgtgatggaccctatacgcggc cgcccTGGAGAATCCCGGTGCC GAGGCCGCTCAATTGGTCGTA GacAGCTCTAGCACCGCTTAAA CGCACGTACGCGCTGTCCCCCG CGTTTTAACCGCCAAGGGGAT TACTCCCTAGTCTCCAGGCACG TGTCAGATATATACATCCTGtgcat gtattgaacagcgaccttgccggtgccag tcggatagtgttccgagctcccactctaga ggatccccgggtaccg_3’ | |
| Sequence-based reagent | primer: Cy5-0-601 | IDT | 5’/5Cy5/TGGAGAATCCCGGTGCC GAGGCCGCTCAAT | |
| Sequence-based reagent | primer: 601–80-FAM | IDT | 5’/56-FAM/cggtacccggggatcctcta gagtgggagc | |
| Sequence-based reagent | primer: 601–0-Cy5 | IDT | 5’/5Cy5/CAGGATGTATATATCTGAC ACGTGCCTGGA | |
| Sequence-based reagent | primer: FAM-80–601 | IDT | 5’/56-FAM/gggatcctaatgaccaagg aaagcatgatt | |
| Peptide, recombinant protein (Saccharomyces cerevisiae) | ScChd1118-1274 | McKnight et al., 2011 | from Saccharomyces cerevisiae | |
| Peptide, recombinant protein (Homo sapiens) | HsSNF2h | Yang et al., 2006 | from Homo sapiens | |
| Peptide, recombinant protein (Drosophila melanogaster) | DmACF | Actif Motif | Cat #:31509 | from Drosophila melanogaster |
| Peptide, recombinant protein (Xenopus laevis) | XlHistone H2A | Luger et al., 1997 | from Xenopus laevis | |
| Peptide, recombinant protein (X. laevis) | XlHistone H2A- E61A/E64A/D90A/ E92A ‘APM’ | Girish et al., 2016 | from Xenopus laevis | |
| Peptide, recombinant protein (X. laevis) | XlHistone H2B | Luger et al., 1997 | from Xenopus laevis | |
| Peptide, recombinant protein (X. laevis) | XlHistone H2B-S53C | Kassabov et al., 2002 | from Xenopus laevis | |
| Peptide, recombinant protein (X. laevis) | XlHistone H3-C110A | Dechassa et al., 2010 | from Xenopus laevis | |
| Peptide, recombinant protein (X. laevis) | XlHIistone H4 | Luger et al., 1997 | from Xenopus laevis | |
| Commercial assay or kit | Thermo sequenase dye primer manual cycle sequencing kit | Affymetrix | Cat #:79260 | |
| Chemical compound, drug | NaCl | Fisher Scientific | Cat #:S641-500 | |
| Chemical compound, drug | Trizma base | Sigma | Cat #:T1503-1KG | |
| Chemical compound, drug | Boric Acid | Fisher Scientific | Cat #:A73-500 | |
| Chemical compound, drug | MgCl2 | Fisher Scientific | Cat #:BP214-500 | |
| Chemical compound, drug | EDTA | Thermo Fisher | Cat #:AM9260G | |
| Chemical compound, drug | KCl | Fisher Scientific | Cat #:P217-500 | |
| Chemical compound, drug | DTT | Sigma Aldrich | Cat #:D9779 | |
| Chemical compound, drug | Sucrose | Fisher Scientific | Cat #:S5-500 | |
| Chemical compound, drug | Nonidet P-40 | Fisher Scientific | Cat #:MP1RIST1315 | |
| Chemical compound, drug | BSA | New England Biolabs | Cat #:B9000S | |
| Chemical compound, drug | Salmon Sperm DNA | Invitrogen | Cat #:15632–011 | |
| Chemical compound, drug | 40% acrylamide/bis solution 19:1 | Bio-Rad | Cat #:1610144 | |
| Chemical compound, drug | Urea | Sigma Aldrich | Cat #:U1250 | |
| Chemical compound, drug | Acrylamide | Bio-Rad | Cat #:1610101 | |
| Chemical compound, drug | Bis N,N’-Methylene-Bis- Acrylamide | Bio-Rad | Cat #:1610201 | |
| Chemical compound, drug | 2-mercaptoethanol | Sigma Aldrich | Cat #:M6250 | |
| Chemical compound, drug | 4-Azidophenacyl bromide | Sigma Aldrich | Cat #:A6057 | |
| Chemical compound, drug | Phenol:Chloroform 5:1 | Sigma Aldrich | Cat #:P1944 | |
| Chemical compound, drug | Adenosine triphosphate disodium salt hydrate | Sigma Aldrich | Cat #:A1852 | |
| Chemical compound, drug | dNTPs | Invitrogen | Cat #:10297–018 | |
| Other | Multipurpose Mini Spin Columns | BioVision | Cat # 6572–50 | |
| Other | HisPrep FF 16/10 (Nickel affinity) | GE | Cat # 28-9365-51 | |
| Other | HisTrap HP, 5 ml (Nickel affinity) | GE | Cat # 17-5248-01 | |
| Other | HiTrap SP FF, 5 ml | GE | Cat # 17-5157-01 | |
| Other | HiTrap Q FF, 5 ml | GE | Cat # 17-5156-01 | |
| Other | HiLoad 16/600 Superdex 200, prep grade | GE | Cat # 28-9893-35 | |
| Other | HiLoad 16/600 Superdex 75, prep grade | GE | Cat # 28-9893-33 | |
| Other | HiPrep 26/10 Desalting | GE | Cat # 17-5087-01 | |
| Other | HiPrep 16/10 Q FF | GE | Cat # 17-5190-01 | |
| Other | HiPrep 16/10 SP FF | GE | Cat # 17-5192-01 | |
| Software, algorithm | ImageJ | imagej.nih.gov/ij/ | ||
| Software, algorithm | Mathematica | Wolfram |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.45472.014





