Enteropathogen antibody dynamics and force of infection among children in low-resource settings
Figures
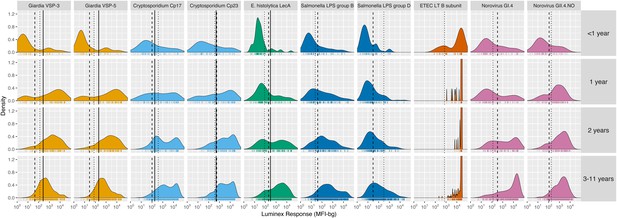
Age-stratified, IgG distributions among a longitudinal cohort of 142 children ages birth to 11 years in Leogane, Haiti, 1990 – 1999.
IgG response measured in multiplex using median fluorescence units minus background (MFI-bg) on the Luminex platform in 771 specimens, marked with rug plots below each distribution. Vertical lines mark seropositivity cutoffs are based on ROC analyses (solid), finite Gaussian mixture models (heavy dash), or distribution among presumed unexposed (light dash). Mixture models failed to converge for ETEC LT B subunit. Created with notebook (https://osf.io/dk54y) and data (https://osf.io/3nv98). Figure 1—figure supplement 1 shows similar distributions from the Tanzania study. Figure 1—figure supplement 2 contrasts Giardia VSP-3 distributions with trachoma pgp3 distributions. Figure 1—figure supplement 3 shows distributions from the Kenyan cohort.
Age-stratified, IgG distributions among 4989 children ages 1 to 9 years old in Kongwa, Tanzania, 2012−2015.
IgG response measured in multiplex using median fluorescence units minus background (MFI-bg) on the Luminex platform, marked with rug plots below each distribution. Vertical lines mark seropositivity cutoffs based on ROC analyses (solid) and finite Gaussian mixture models (heavy dash). Mixture models failed to converge for all antibody distributions except for Giardia and E. histolytica. Created with notebook (https://osf.io/dt9zu) and data (https://osf.io/kv4d3).

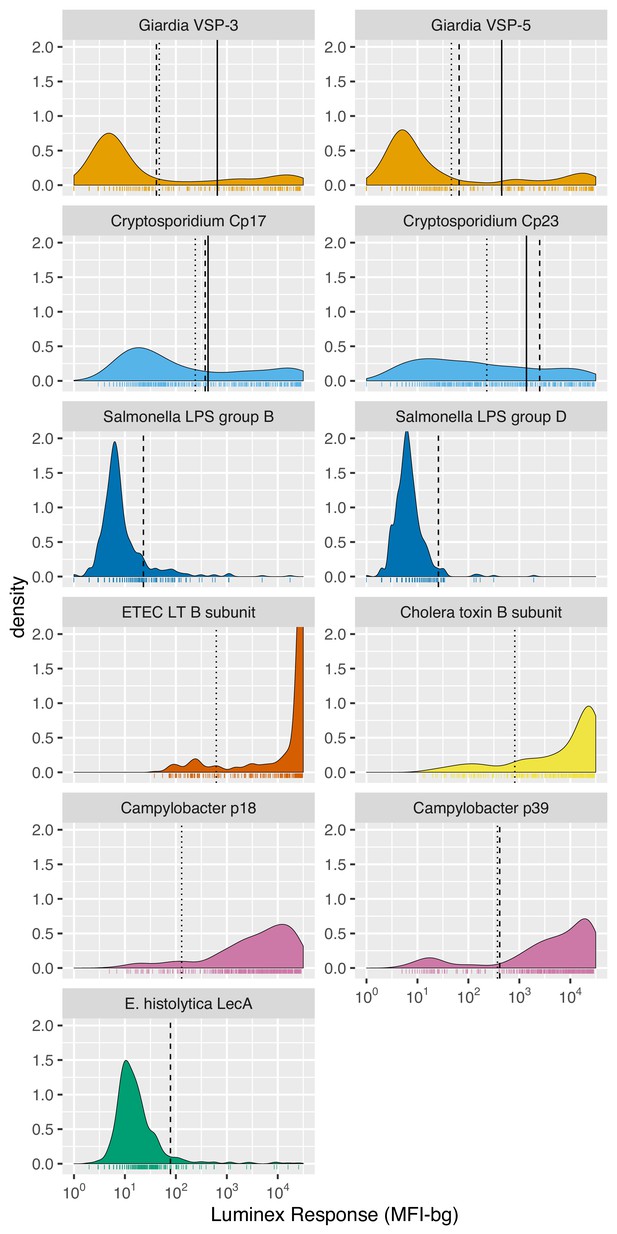
Contrasting age-dependent changes in distributions of IgG levels to Giardia VSP-3 and Chlamydia trachomatis pgp3 antigens among 4989 children ages 1 to 9 years old in Kongwa, Tanzania, 2012 – 2015.
IgG response measured in multiplex using median fluorescence units minus background (MFI-bg) on the Luminex platform, marked with rug plots below each distribution. Vertical lines mark seropositivity cutoffs based on ROC analyses (solid) and finite Gaussian mixture models (dash). Created with notebook (https://osf.io/dt9zu) and data (https://osf.io/kv4d3).
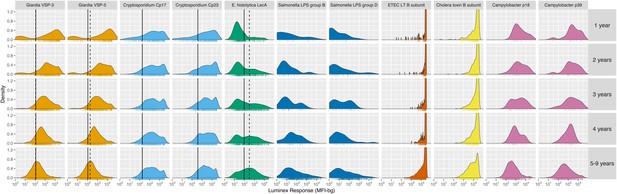
IgG distributions among children ages 4 to 17 months old in Asembo, Kenya, 2013.
IgG response measured in multiplex using median fluorescence units minus background (MFI-bg) on the Luminex platform. N = 439 samples from 240 children, marked with rug plots below each distribution. Vertical lines mark seropositivity cutoffs based on ROC analyses (solid), finite Gaussian mixture models (heavy dash), or distribution among presumed unexposed (light dash). Mixture models failed to converge for ETEC LT B subunit and Campylobacter p18. Created with notebook (https://osf.io/456jp) and data (https://osf.io/2q7zg).
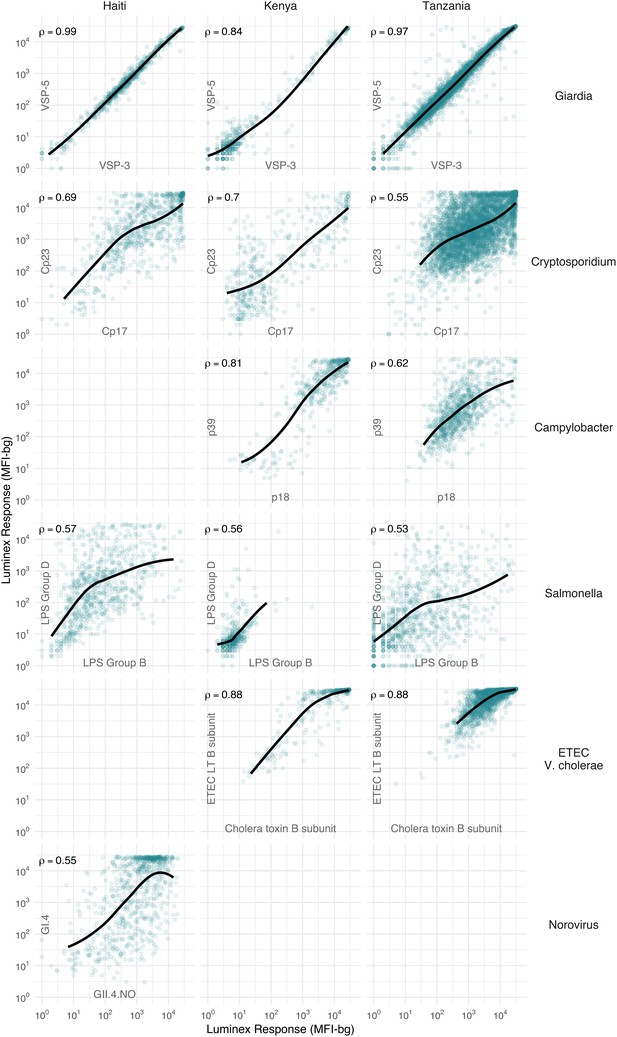
Joint distributions of select enteric pathogen antibody responses among children in three cohorts from Haiti, Kenya, and Tanzania.
Each panel includes Spearman rank correlations (ρ) and locally weighted regression smoothers with default parameters, trimmed to 95% of the data to avoid edge effects. Antibody response measured in multiplex using median fluorescence units minus background (MFI-bg) on the Luminex platform. Empty panels indicate that the antibodies were not measured in that cohort. Supplementary file 3 includes all pairwise comparisons. Created with notebook (https://osf.io/hv9ce) and data (https://osf.io/3nv98, https://osf.io/2q7zg, https://osf.io/kv4d3).
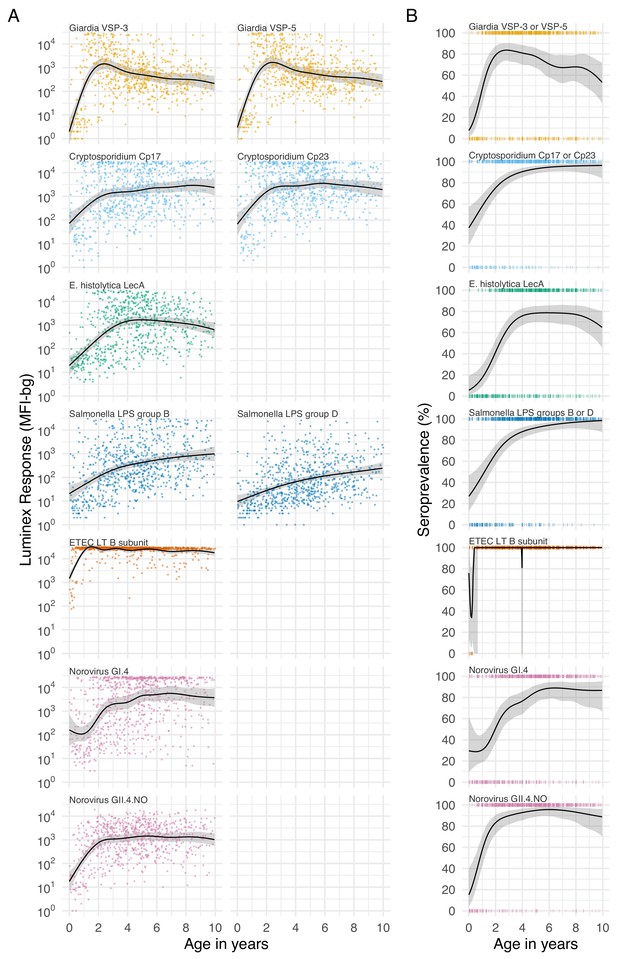
Age dependent mean IgG and seroprevalence in Haiti.
Geometric means (A) and seroprevalence (B), estimated with cubic splines among children ages birth to 10 years in Leogane, Haiti 1990–1999. Shaded bands are approximate, simultaneous 95% confidence intervals. IgG response measured in multiplex using median fluorescence units minus background (MFI-bg) on the Luminex platform (N = 771 measurements from 142 children). Created with notebook (https://osf.io/jeby3) and data (https://osf.io/3nv98). Data for some antigens measured among children < 5 years previously published (Arnold et al., 2017). Figure 3—figure supplement 1 includes similar curves from Kenya; Figure 3—figure supplement 2 includes similar curves from Tanzania.
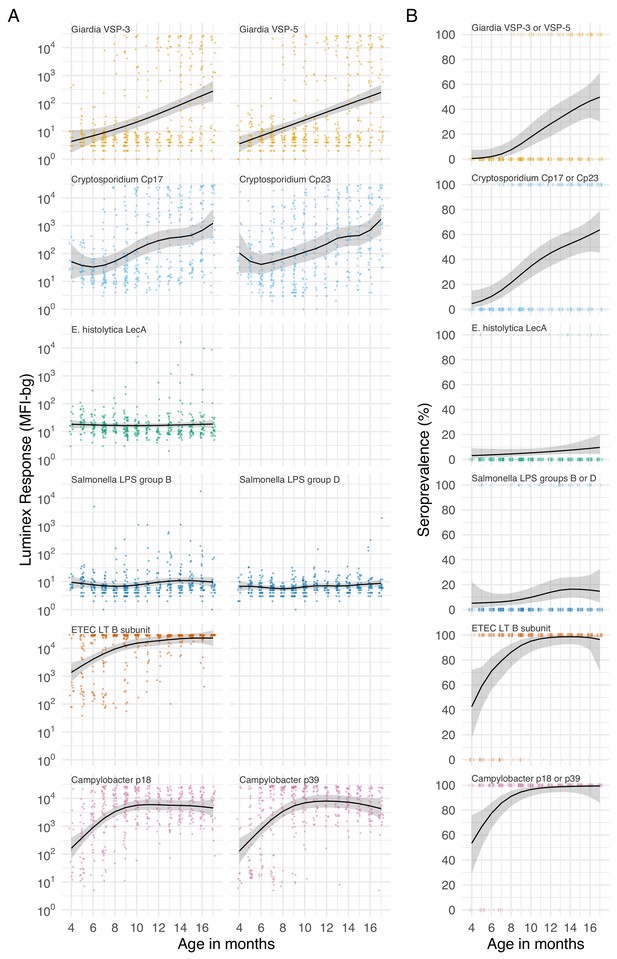
Age dependent mean IgG and seroprevalence in Kenya.
Geometric means (A) and seroprevalence (B), estimated with cubic splines among children ages 4 to 17 months in Asembo, Kenya, 2013. Shaded bands are approximate, simultaneous 95% confidence intervals. Ages were measured in months completed (rounded) so points in the figure are jittered. IgG response measured in multiplex using median fluorescence units minus background (MFI-bg) on the Luminex platform (N = 439 measurements from 240 children). Created with notebook (https://osf.io/jeby3) and data (https://osf.io/2q7zg).
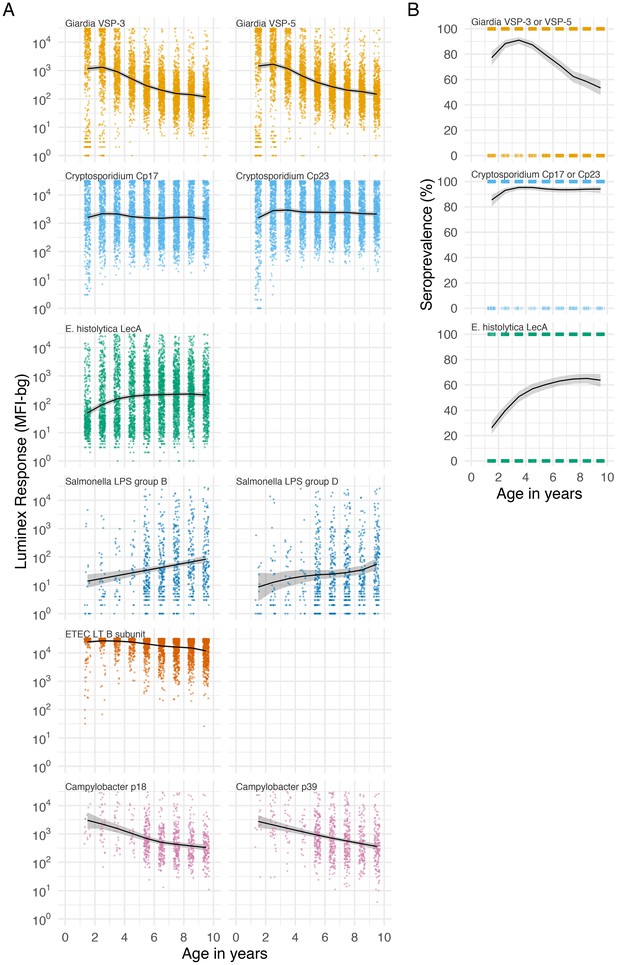
Age dependent mean IgG and seroprevalence in Tanzania.
Geometric means (A) and seroprevalence (B), estimated with cubic splines among children ages 1 to 9 years in Kongwa, Tanzania, 2012–2015. Shaded bands are approximate, simultaneous 95% confidence intervals. Ages were measured in years completed (rounded) so points in the figure are jittered. IgG response measured in multiplex using median fluorescence units minus background (MFI-bg) on the Luminex platform in 4989 specimens. Salmonella and Campylobacter antigens were only included in the multiplex in 2012 (N = 902 specimens). Seropositivity cutoffs could not be estimated for bacterial pathogens in this study, so seroprevalence curves are not shown. Created with notebook (https://osf.io/jeby3) and data (https://osf.io/kv4d3).
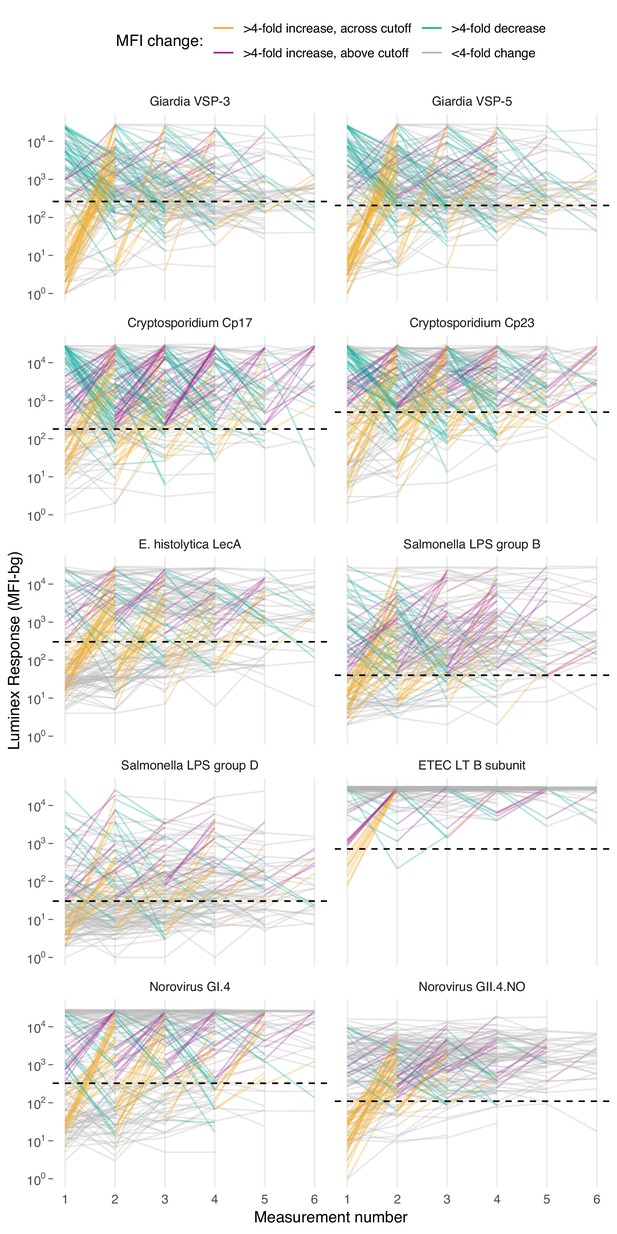
Longitudinal changes in IgG response over six repeated measurements among 142 children ages 0–11 years in Leogane, Haiti, 1990 – 1999.
Measurements were spaced by approximately 1 year (median spacing = 1, IQR = 0.7, 1.3). Horizontal dashed lines mark seropositivity cutoffs for each antibody. The number of children measured at each visit was: n1 = 142, n2 = 142, n3 = 140, n4 = 131, n5 = 111, n6 = 66); 29 children had >6 measurements that are not shown. IgG response measured in multiplex using median fluorescence units minus background (MFI-bg) on the Luminex platform. Created with notebook (https://osf.io/vyhra), which includes additional visualizations, and data (https://osf.io/3nv98). Figure 4—figure supplement 1 includes a similar figure from the Kenya cohort. Figure 4—figure supplements 2 and 3 summarize the proportion of children in each category across measurement rounds in Haiti and Kenya.

Longitudinal changes in IgG response between enrollment and follow-up among 205 children ages 4–17 months in Asembo, Kenya, 2013.
Horizontal dashed lines mark seropositivity cutoffs for each antibody. IgG response measured in multiplex using median fluorescence units minus background (MFI-bg) on the Luminex platform. Created with notebook (https://osf.io/6gk2q), which includes additional visualizations, and data (https://osf.io/2q7zg).
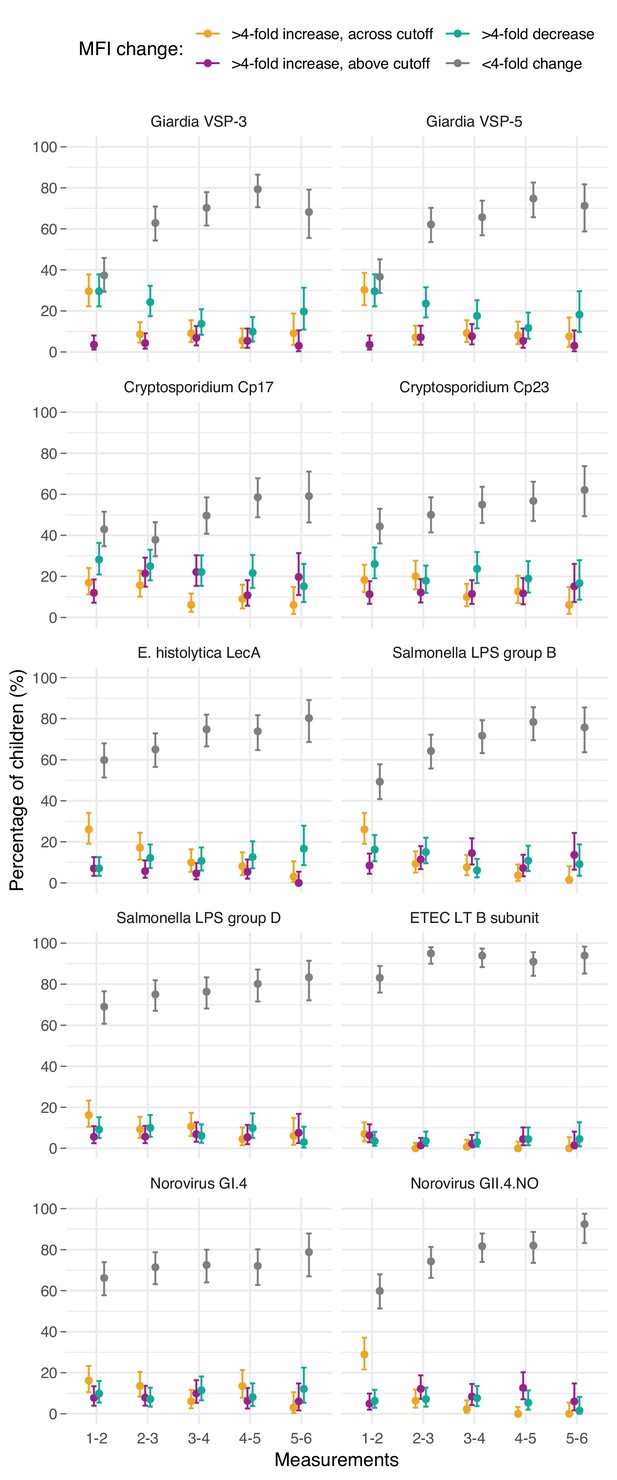
Proportion of children ages 0–11 years with different longitudinal changes in IgG response over six repeated measurements in Leogane, Haiti, 1990 – 1999.
Measurements were spaced by approximately 1 year (median spacing = 1, IQR = 0.7, 1.3). The number of children measured at each visit was: n1 = 142, n2 = 142, n3 = 140, n4 = 131, n5 = 111, n6 = 66); 29 children had >6 measurements that are not shown. IgG response measured in multiplex using median fluorescence units minus background (MFI-bg) on the Luminex platform. Created with notebook (https://osf.io/vyhra) and data (https://osf.io/3nv98).
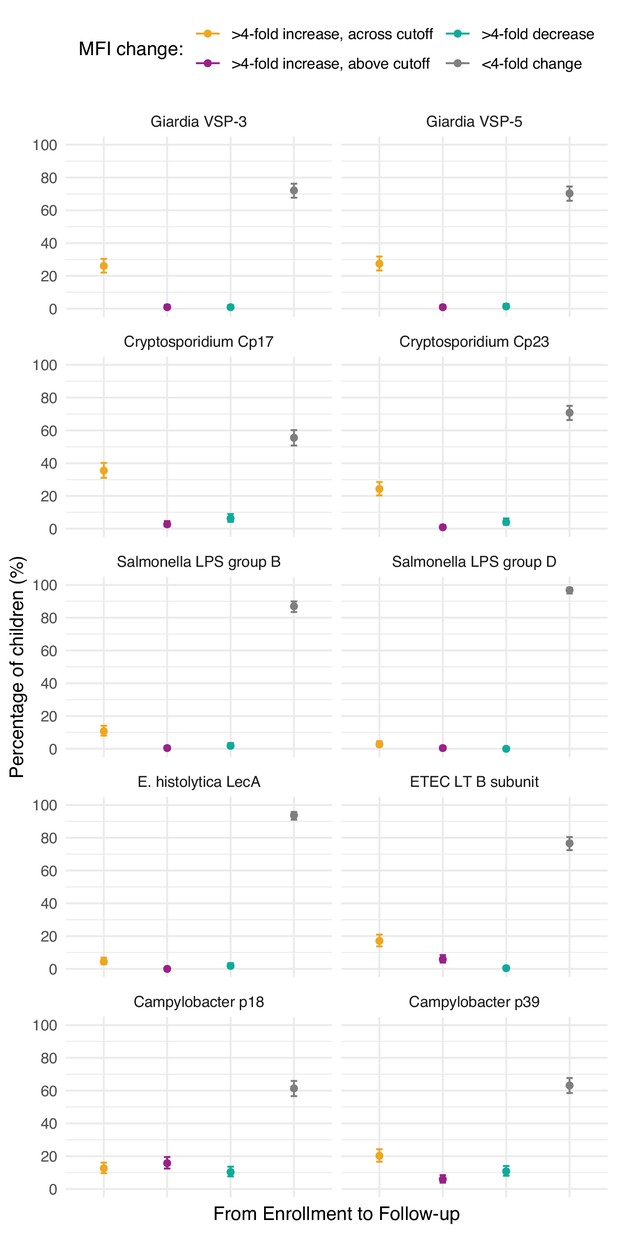
Proportion of children ages 4–17 months with different longitudinal changes in IgG response between enrollment and follow-up 6 months later in Asembo, Kenya, 2013.
N = 205 children measured longitudinally. IgG response measured in multiplex using median fluorescence units minus background (MFI-bg) on the Luminex platform. Created with notebook (https://osf.io/6gk2q) and data (https://osf.io/2q7zg).
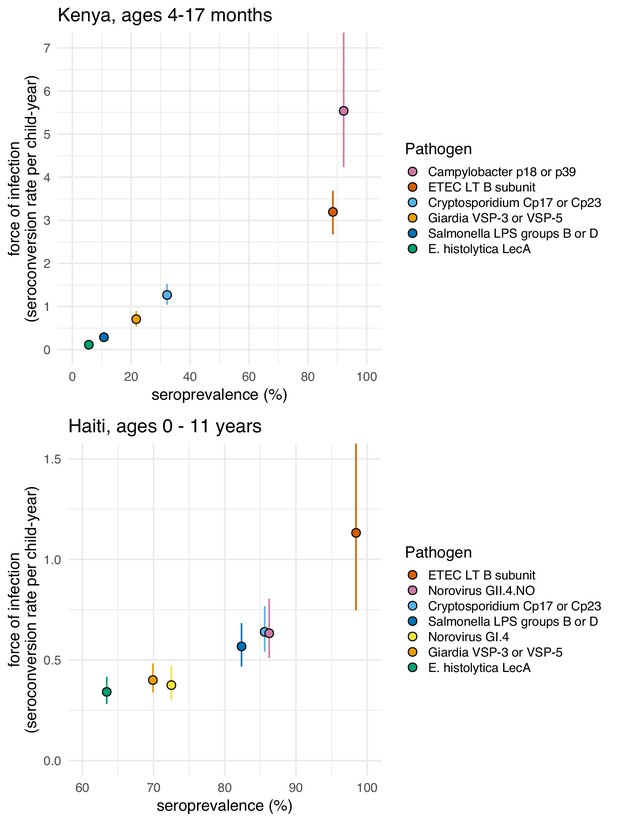
Average force of infection versus seroprevalence for enteropathogens measured in the Kenya and Haiti cohorts.
Force of infection estimated from prospective seroconversion rates. Vertical lines indicate 95% confidence intervals. Created with notebook (https://osf.io/jp9kf) and data (https://osf.io/2q7zg, https://osf.io/3nv98). Figure 5—figure supplement 1 includes estimates from Haiti stratified by age bands.
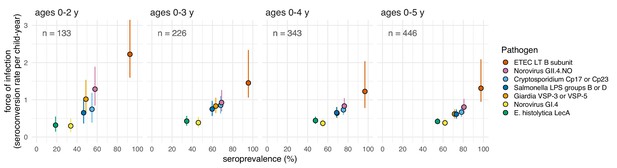
Average force of infection versus seroprevalence for enteropathogens measured in the Haiti cohort, stratified by different age bands.
Force of infection estimated from prospective seroconversion rates. Vertical lines indicate 95% confidence intervals. Created with notebook (https://osf.io/jp9kf) and data (https://osf.io/3nv98).
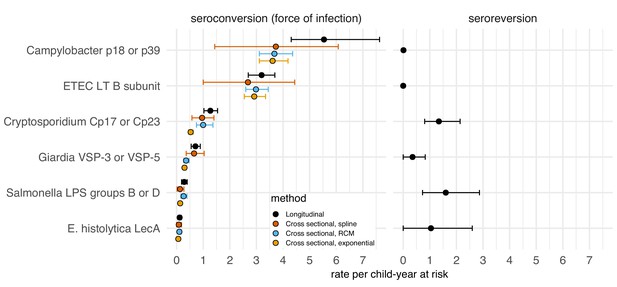
Enteropathogen seroconversion and seroreversion rates among 205 children ages 4 to 17 months measured longitudinally in Asembo, Kenya, 2013.
The seroconversion rate is a measure of a pathogen's force of infection. Longitudinal estimates are non-parametric rates of incident seroconversions and seroreversions among children at risk, assumed to occur at the midpoint of the measurement interval. Cross-sectional estimators were derived from age-specific seroprevalence curves using semiparametric cubic splines (spline), a reversible catalytic model (RCM) that assumed constant seroconversion and seroreversion rates with the seroreversion rate estimated from prospective data, and a parametric constant rate survival model (exponential). Error bars mark 95% confidence intervals. IgG response measured in multiplex using median fluorescence units minus background (MFI-bg) on the Luminex platform (N = 410 measurements from 205 children). Created with notebooks (https://osf.io/sqvj7, https://osf.io/j9nh3) and data (https://osf.io/2q7zg).
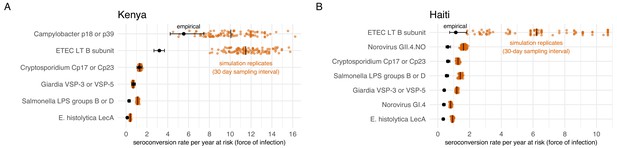
Empirical seroconversion rates compared with estimates from 100 simulated datasets with daily resolution IgG trajectories that were sampled at a 30 day interval before estimating seroconversion rates.
Vertical lines in the simulation results indicate medians. (A) In the Kenya cohort, children were ages 4–18 months and empirical IgG measurements were measured every 6 months (approximately 180 days). (B) In the Haiti cohort, children were ages 0–11 years, and empirical IgG measurements were measured approximately each year (median spacing = 1 year, IQR = 0.7, 1.3). Created with notebooks (https://osf.io/qmdf2, https://osf.io/9fxhb) and data (https://osf.io/2q7zg, https://osf.io/3nv98).
Tables
Number of children and samples tested, and estimated seropositivity cutoffs by country and antigen included in the seroepidemiologic analyses.
https://doi.org/10.7554/eLife.45594.003| Seropositivity cutoff, log10 IgG (MFI-bg) * | |||||
|---|---|---|---|---|---|
| N | N | External | Mixture | Presumed | |
| children | samples | Reference | Model | Unexposed | |
| Leogane, Haiti | |||||
| Giardia VSP-3 | 142 | 771 | 2.42 | 1.64 | 2.11 |
| Giardia VSP-5 | 142 | 771 | 2.31 | 1.46 | 1.88 |
| Cryptosporidium Cp17 | 142 | 771 | 2.26 | 2.00 | 2.58 |
| Cryptosporidium Cp23 | 142 | 771 | 2.70 | 2.75 | 2.57 |
| E. histolytica LecA | 142 | 771 | 2.48 | 2.30 | 1.93 |
| Salmonella LPS group B | 142 | 771 | 1.60 | 1.37 | |
| Salmonella LPS group D | 142 | 771 | 1.48 | 2.48 | |
| ETEC LT B subunit | 142 | 771 | 2.86 | ||
| Norovirus GI.4 | 142 | 771 | 2.51 | 2.09 | |
| Norovirus GII.4.NO | 142 | 771 | 2.04 | 2.24 | |
| Asembo, Kenya | |||||
| Giardia VSP-3 | 240 | 445 | 2.81 | 1.62 | 1.67 |
| Giardia VSP-5 | 240 | 445 | 2.65 | 1.82 | 1.67 |
| Cryptosporidium Cp17 | 240 | 445 | 2.63 | 2.58 | 2.38 |
| Cryptosporidium Cp23 | 240 | 445 | 3.14 | 3.40 | 2.36 |
| E. histolytica LecA | 240 | 445 | 1.89 | ||
| Salmonella LPS group B | 240 | 445 | 1.36 | ||
| Salmonella LPS group D | 240 | 445 | 1.41 | ||
| ETEC LT B subunit | 240 | 445 | 2.79 | ||
| Cholera toxin B subunit | 240 | 445 | 2.91 | ||
| Campylobacter p18 | 240 | 445 | 2.11 | ||
| Campylobacter p39 | 240 | 445 | 2.61 | 2.57 | |
| Kongwa, Tanzania | |||||
| Giardia VSP-3 | 4989 | 4989 | 2.23 | 2.04 | |
| Giardia VSP-5 | 4989 | 4989 | 2.15 | 2.27 | |
| Cryptosporidium Cp17 | 4989 | 4989 | 2.26 | ||
| Cryptosporidium Cp23 | 4989 | 4989 | 2.58 | ||
| E. histolytica LecA | 4989 | 4989 | 1.97 | 2.50 | |
| Salmonella LPS group B† | 902 | 902 | |||
| Salmonella LPS group D† | 902 | 902 | |||
| ETEC LT B subunit | 4989 | 4989 | |||
| Cholera toxin B subunit‡ | 4087 | 4087 | |||
| Campylobacter p18† | 902 | 902 | |||
| Campylobacter p39† | 902 | 902 | |||
-
*Seropositivity cutoffs determined using external reference samples (typically ROC curves except for Giardia and E. hystolitica in Haiti), finite Gaussian mixture models, or distribution among the presumed unexposed (see Materials and methods for details). External reference cutoffs vary across cohorts for the same antigen due to use of different bead sets in each cohort. External reference cutoffs reported from years (2013–2015) in Tanzania, estimated among 4087 samples. Cutoff values are missing if they could not be estimated in each method; cutoff values based on the presumed unexposed required longitudinal measurements within individual children and therefore could not be estimated for any antigen in the repeated cross-sectional design in Tanzania.
† Measured only in year 1 of the study (2012).
-
‡ Measured only in years 2–4 of the study (2013–2015).
Incidence rates of seroconversion and seroreversion per child year among children ages 0–11 years in Haiti, 1990–1999.
https://doi.org/10.7554/eLife.45594.018| Seropositivity cutoff * | 4-Fold change in IgG levels † | |||||||
|---|---|---|---|---|---|---|---|---|
| Pathogen | Child- years | Incident cases | Rate (95% CI) | Child- years | Incident cases | Rate (95% CI) | Ratio of cases | Ratio of rates |
| Seroconversion/boosting | ||||||||
| Giardia VSP-3 or VSP-5 | 269.6 | 108 | 0.40 (0.34, 0.48) | 277.2 | 120 | 0.43 (0.35, 0.54) | 1.1 | 1.1 |
| Cryptosporidium Cp17 or Cp23 | 109.3 | 70 | 0.64 (0.54, 0.77) | 241.0 | 204 | 0.85 (0.73, 0.97) | 2.9 | 1.3 |
| E. histolytica LecA | 283.7 | 97 | 0.34 (0.28, 0.42) | 297.1 | 107 | 0.36 (0.29, 0.45) | 1.1 | 1.1 |
| Salmonella LPS groups B or D | 132.1 | 75 | 0.57 (0.47, 0.68) | 226.8 | 149 | 0.66 (0.54, 0.80) | 2.0 | 1.2 |
| ETEC LT B subunit | 9.7 | 11 | 1.13 (0.75, 1.82) | 32.1 | 32 | 1.00 (0.70, 1.45) | 2.9 | 0.9 |
| Norovirus GI.4 | 213.0 | 80 | 0.38 (0.30, 0.47) | 254.3 | 107 | 0.42 (0.34, 0.53) | 1.3 | 1.1 |
| Norovirus GII.4.NO | 105.8 | 67 | 0.63 (0.51, 0.80) | 147.2 | 100 | 0.68 (0.54, 0.86) | 1.5 | 1.1 |
| Seroreversion/waning | ||||||||
| Giardia VSP-3 or VSP-5 | 441.6 | 91 | 0.21 (0.17, 0.25) | 290.9 | 127 | 0.44 (0.35, 0.54) | 1.4 | 2.1 |
| Cryptosporidium Cp17 or Cp23 | 586.1 | 29 | 0.05 (0.03, 0.07) | 273.5 | 171 | 0.63 (0.53, 0.74) | 5.9 | 12.6 |
| E. histolytica LecA | 395.2 | 43 | 0.11 (0.08, 0.15) | 310.4 | 67 | 0.22 (0.16, 0.27) | 1.6 | 2.0 |
| Salmonella LPS groups B or D | 544.3 | 25 | 0.05 (0.03, 0.07) | 344.5 | 89 | 0.26 (0.20, 0.32) | 3.6 | 5.6 |
| ETEC LT B subunit | 702.1 | 2 | 0.00 (0.00, 0.01) | 649.7 | 22 | 0.03 (0.02, 0.05) | 11.0 | 11.9 |
| Norovirus GI.4 | 464.9 | 28 | 0.06 (0.03, 0.09) | 362.2 | 56 | 0.15 (0.11, 0.21) | 2.0 | 2.6 |
| Norovirus GII.4.NO | 574.3 | 19 | 0.03 (0.02, 0.05) | 477.9 | 39 | 0.08 (0.06, 0.11) | 2.1 | 2.5 |
-
*Incident changes in serostatus defined by crossing seropositivity cutoffs.
†Incident changes in serostatus defined by a 4-fold increase or decrease in IgG levels (MFI-bg), with incident boosting episodes restricted to changes that ended above the seropositivity cutoff and incident waning episodes restricted to changes that started from above the seropositivity cutoff.
| Reagent type or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Peptide, recombinant protein | Giardia intestinalis VSP-3 | PMID: 17901334 PMID: 20876825 | GenBank: XM_001707314 | Dr. Jeffrey Priest (CDC) |
| Peptide, recombinant protein | Giardia intestinalis VSP-5 | PMID: 11500396 PMID: 20876825 | GenBank: AF354538.1 | Dr. Jeffrey Priest (CDC) |
| Peptide, recombinant protein | Cryptosporidium Cp17 | PMID: 10699255 PMID: 15165066 | GenBank: AF114166 | Dr. Jeffrey Priest (CDC) |
| Peptide, recombinant protein | Cryptosporidium Cp23 | PMID: 8892291 PMID: 10203492 | GenBank: U34390 | Dr. Jeffrey Priest (CDC) |
| Peptide, recombinant protein | Entamoeba histolytica LecA | PMID: 2000392 PMID: 14741152 PMID: 24591430 | GenBank: M60498 | Dr. William Petri (University of Virginia) and Dr. Joel Herbein (TechLab) |
| Peptide, recombinant protein | Campylobacter jejuni p18 | PMID: 8576327 PMID: 16014430 | GenBank: X83374 | Dr. Jeffrey Priest (CDC) |
| Peptide, recombinant protein | Campylobacter jejuni p39 | PMID: 10688204 PMID: 16014430 | GenBank: CAL34198.1 | Dr. Jeffrey Priest (CDC) |
| Peptide, recombinant protein | ETEC heat labile toxin B subunit | PMID: 3882744 PMID: 18494692 | Sigma-Aldrich | |
| Peptide, recombinant protein | Cholera toxin B subunit | PMID: 3882744 | Sigma-Aldrich | |
| Peptide, recombinant protein | Salmonella enterica LPS group B | PMID: 2567429 PMID: 17329442 | Sigma-Aldrich | |
| Peptide, recombinant protein | Salmonella enterica LPS group D | PMID: 2567429 PMID: 17329442 | Sigma-Aldrich | |
| Peptide, recombinant protein | Norovirus GI.4 Virus Like Particles | This paper | Dr. Jan Vinje (CDC) | |
| Peptide, recombinant protein | Norovirus GII.4.NO Virus Like Particles | This paper | Dr. Jan Vinje (CDC) |
Additional files
-
Supplementary file 1
Effect of intervention, bead lot, and season on enteropathogen antibody response (osf.io/6br2f).
- https://doi.org/10.7554/eLife.45594.021
-
Supplementary file 2
Classification agreement between different seropositivity cutoff approaches (osf.io/7x6sw).
- https://doi.org/10.7554/eLife.45594.022
-
Supplementary file 3
Joint distributions of antibody response (osf.io/wchzq).
- https://doi.org/10.7554/eLife.45594.023
-
Supplementary file 4
IgG measurements in the Kenya cohort among children with- and without confirmed Cryptosporidium and Giardia infections in diarrheal stools (osf.io/e4tbg).
- https://doi.org/10.7554/eLife.45594.024
-
Supplementary file 5
Sensitivity analyses: fold-changes in IgG used to identify presumed unexposed measurements and force of infection in Haiti and Kenya (osf.io/u79bm).
- https://doi.org/10.7554/eLife.45594.025
-
Supplementary file 6
Estimation of age-dependent means and seroprevalence using multiple approaches (osf.io/r25hp).
- https://doi.org/10.7554/eLife.45594.026
-
Supplementary file 7
Estimation of force of infection from age-structured seroprevalence in Kenya (osf.io/9wbh5).
- https://doi.org/10.7554/eLife.45594.027
-
Supplementary file 8
Simulation study to assess the influence of sampling intervals on serological estimates of force of infection (osf.io/9zt4d).
- https://doi.org/10.7554/eLife.45594.028
-
Transparent reporting form
- https://doi.org/10.7554/eLife.45594.029





