Dual separable feedback systems govern firing rate homeostasis
Figures
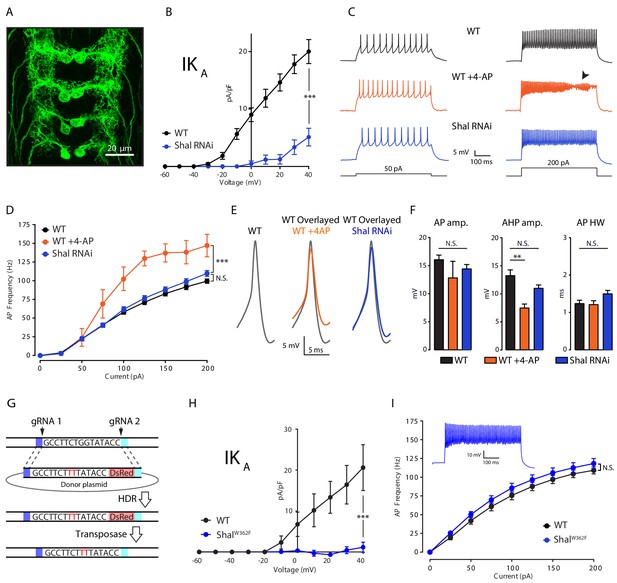
Firing rate homeostasis in Drosophila motoneurons.
(A) Confocal max projection of Drosophila 3rd instar larval VNC shows selective Gal4 expression in MN1 (MN1-GAL4 > UAS-CD8:GFP). (B) I-V plots of MN1 IKA in WT (black, n = 20) and Shal-RNAi (blue, n = 10). (C) Representative voltage traces from WT (black), WT + 4 AP (orange) and Shal-RNAi (blue) at 50 pA (left) and 200 pA (right) current injections. Arrow indicates impaired action potentials due to depolarization block. (D) F-I curves of WT (n = 10), WT + 4 AP (n = 4) and Shal-RNAi (n = 15). (E) Example action potential waveforms (left) and overlay (right). (F) Quantification of action potential, after-hyperpolarization amplitudes (AP amp. AP AHP amp., respectively) and action potential half-width (AP HW). (G) CRISPR strategy for generating a targeted pore-blocking point mutation in Shal. Dark and light blue regions represent 5’ and 3’ 1 kb homology arms designed for recombination of mutated segment from pHD-ScarlessDsRed donor vector into the endogenous Shal gene locus. Selection marker represented in pink. (H) Elimination of IKA across all motoneurons in the ShalW362F mutant. WT (black, n = 7) and ShalW362F (blue, n = 13). (I) F-I curves of WT (n = 19) and ShalW362F (n = 15). Inset: Representative voltage trace from ShalW362F motoneuron at 200 pA current injection. Mean ± S.E.M.; *p<0.05; **p<0.005; ***p<0.0005. N.S., not significant; two-way RM-ANOVA with post-hoc tests (I-V plots and F-I curves) or one-way ANOVA with Bonferoni post-hoc tests (AP waveform measurements).
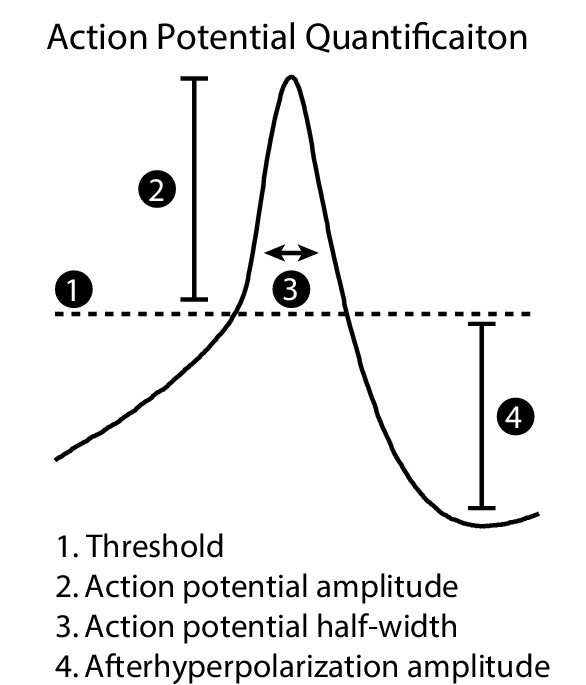
Action potential waveform measurements.
Action potential threshold was the membrane potential at the first inflection point, calculated as 1.5 standard deviations away from the mean second derivative of the AP wave at baseline. Action potential amplitude (AP amp) was the difference in voltage between the threshold and the maximum potential. Action potential half-width (AP HW) was the AP width at half-max amplitude. After-hyperpolarization amplitude (AHP) was the difference in voltage between threshold and the minimum potential after the peak. AP amp, AP HW, and AHP measurements were averaged from all action potentials elicited from a standard current step protocol for each cell.
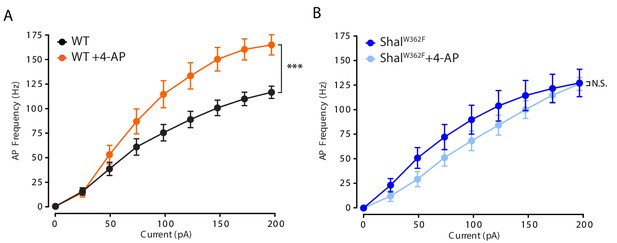
4-AP Does Not Increase Firing Rates in ShalW362F Motoneurons.
(A) F-I curves of WT (black, n = 9) and WT +4 AP (orange, n = 9). (B) F-I curves of ShalW362F (dark blue, n = 5) and ShalW362F +4-AP (light blue, n = 9). Mean ± S.E.M.; two-way RM-ANOVA and Bonferroni's multiple comparisons test.
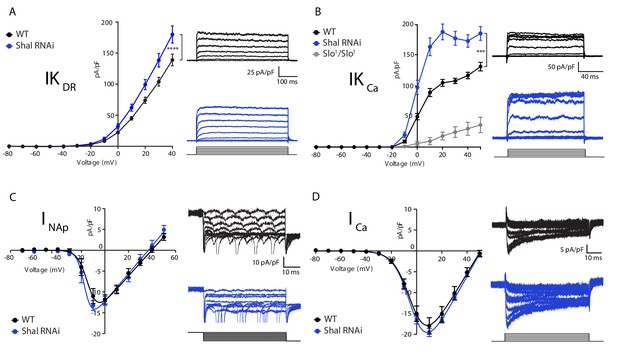
Homeostatic potassium current rebalancing stabilizes MN1 excitability in the absence of Shal-dependent IKA.
(A, B) I-V plots and representative traces of voltage-dependent delayed rectifier (IKDR, (A) and Ca2+-dependent (IKCa, (B) potassium currents in WT (black; n = 7 and n = 9 for IKDR and IKCa, respectively) and Shal-RNAi (blue; n = 12 and n = 8 for KDR and KCa, respectively) MN1. The specificity of the IKCa current protocol was confirmed in slo1 mutants (B, grey; n = 4), which exhibited minimal Ca2+-dependent potassium currents. (C, D) I-V plots and representative traces of persistent sodium currents (INaP, C, WT: n = 9; Shal-RNAi: n = 9) and voltage dependent calcium currents (ICa, D; WT, n = 8; Shal-RNAi, n = 6) in WT (black) and Shal-RNAi (blue) MN1. Mean ± S.E.M.; *p<0.05; **p<0.005; ***p<0.0005. N.S., not significant; two-way RM-ANOVA with Bonferoni post-hoc tests (I-V plots).
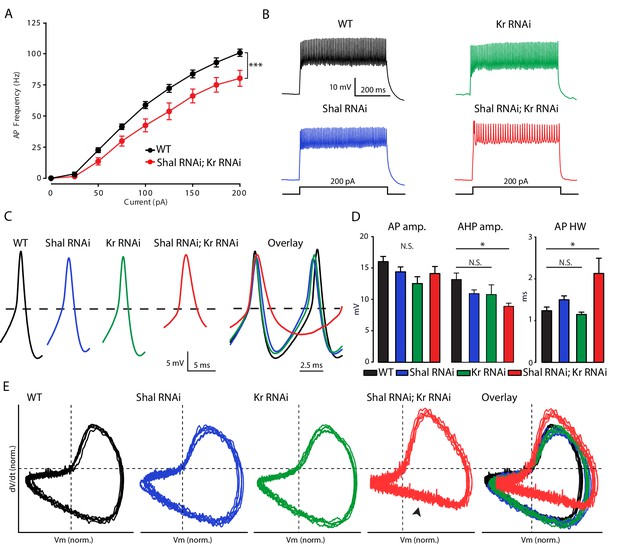
Krüppel is necessary for firing rate homeostasis and preservation of AP waveform.
(A) F-I curves of WT (black; n = 10) and double Shal-RNAi; Kr-RNAi (red; n = 16). (B) Representative voltage traces from WT (black), Shal-RNAi (blue), Kr-RNAi (green) and double Shal-RNAi; Kr-RNAi (red) at 200 pA current injections. (C) Left: Example action potential waveforms for WT (black), Shal-RNAi (blue), Kr-RNAi (green) and double Shal-RNAi; Kr-RNAi (red). Right: Action potential overlays. (D) Action potential waveform quantification. (E) Phase plane plots of normalized example action potential waveforms for each genotype (left four panels) and overlays (far right panel). Each plot contains five sequential action potentials from a representative cell to illustrate AP to AP consistency. Mean ± S.E.M.; *p<0.05; **p<0.005; ***p<0.0005. N.S., not significant; two-way RM-ANOVA (F-I curves), one-way ANOVA with Bonferoni post-hoc tests (AP waveform comparisons).
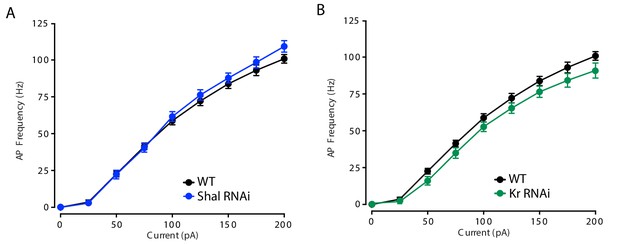
Kr does not contribute to setting MN1 baseline firing rate.
(A) F-I curves of WT (black, n = 10) and Shal-RNAi (blue, n = 15). (B) F-I curves of WT (black, n = 10) and Kr-RNAi (green, n = 8). Mean ± S.E.M.; two-way RM-ANOVA and Bonferroni's multiple comparisons test.
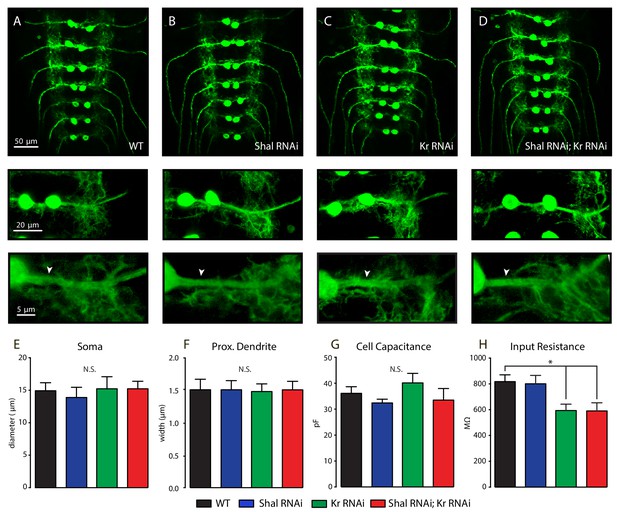
Normal motoneuron morphology.
(A–D) Confocal max projections of MN1-Ib motorneurons (MN1-GAL4 > UAS-CD8:GFP) in WT (A), Shal-RNAi (B), Krüppel (Kr)-RNAi, (C) and combined Shal and Krüppel-RNAi (Shal-RNAi; Kr-RNAi, (D). Representative MN1-Ibs shown at higher magnification in inserts to reveal single neuron morphology. Arrows indicate MN1-Ib primary dendrite. (E, F) Quantification of somatic diameter (E) and proximal dendrite width (F) in WT (back), Shal-RNAi (blue), Krüppel-RNAi (Kr, green) and combined Shal and Krüppel-RNAi (Shal-RNAi; Kr-RNAi, red). (G, H) Whole-cell capacitance (G) and input resistance (H) in WT (back), Shal-RNAi (blue), Krüppel-RNAi (Kr, green) and combined Shal-RNAi and Krüppel-RNAi (Shal-RNAi; Kr-RNAi, red). Mean ± S.E.M.; *p<0.05; **p<0.005; ***p<0.0005. N.S., not significant; one-way ANOVA with Bonferoni post-hoc tests.
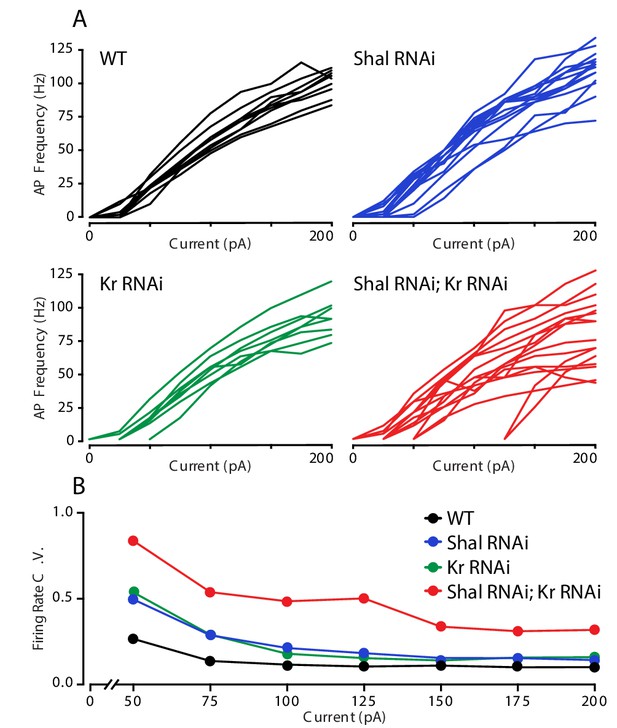
Krüppel constrains cell-to-cell firing rate variance.
(A) Individual motorneuron F-I curves for WT (black), Shal-RNAi (blue), Kr-RNAi (green) and double Shal-RNAi; Kr-RNAi (red) groups. (B) The coefficient of variation across cells is calculated for each genotype at each current injection step and plotted. Data at 25 pA are not shown since a large fraction of cells in each genotype failed to fire an action potential.
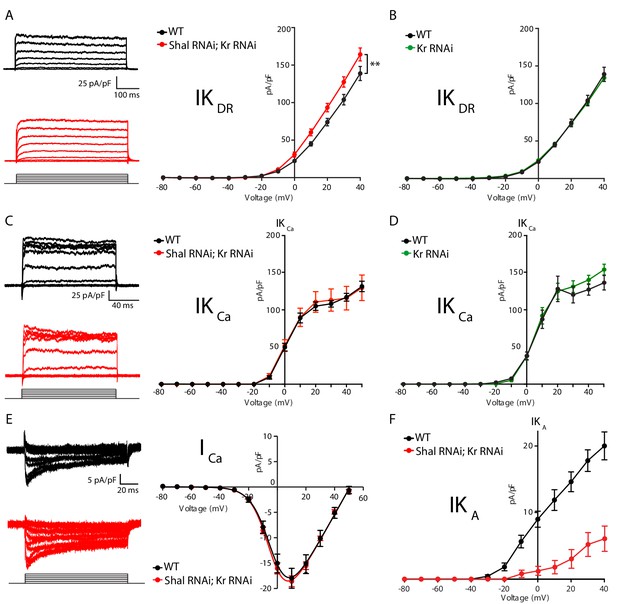
Krüppel controls IKCa but not IKDRduring firing rate homeostasis.
(A) IKDR I-V plots (right) and representative traces (left) in WT (black, n = 7) and Shal-RNAi; Kr-RNAi (red, n = 19) MN1. (B) I-V plots of WT (black, n = 7) and Kr-RNAi (green, n = 14) MN1. (C) IKCa I-V plots (right) and representative traces (left) in WT (black) and Shal-RNAi; Kr-RNAi (red) MN1. (D) I-V plots of WT (black, n = 7) and Kr-RNAi (green, n = 9) MN1. (E) I-V plots (right) and representative traces (left) of voltage dependent calcium currents (ICa) in WT (black; n = 8) and double Shal-RNAi; Kr-RNAi (red; n = 6) MN1. (F) I-V plots of MN1 IKA currents in WT (black, n = 20) and double Shal-RNAi, Kr-RNAi (red, n = 10). Mean ± S.E.M.; *p<0.05; **p<0.005; ***p<0.0005; two-way RM-ANOVA with Sidak post-hoc tests. Mean ± S.E.M.; *p<0.05; **p<0.005; ***p<0.0005. N.S., not significant; two-way RMANOVA (I-V plots), one-way ANOVA with Bonferoni post-hoc tests.
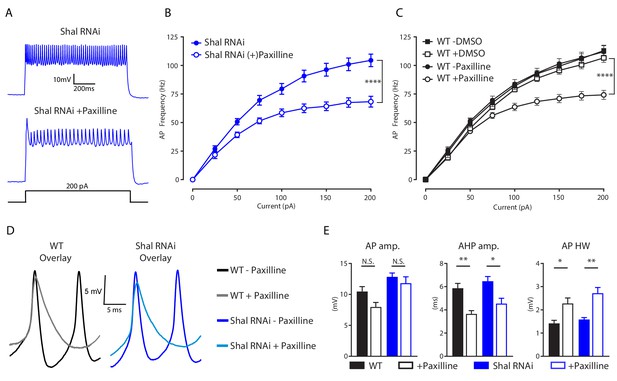
IKCa is necessary to maintaining WT firing rates.
(A) Representative voltage traces from Shal-RNAi and Shal-RNAi +Paxilline at 200 pA step current injection. (B) F-I curves of Shal-RNAi at baseline (filled circles) and Shal-RNAi +Paxilline (open circles), paired recordings, n = 8. (C) F-I curves of WT at baseline (filled circles) and WT + Paxilline (open circles), paired recordings, n = 10; WT at baseline (filled squares) and WT + DMSO (open squares), paired recordings, n = 6. Paxilline was dissolved in DMSO. (D) Example action potential waveforms for WT ± Paxilline overlayed (black, grey) and Shal-RNAi ± Paxilline overlayed (dark blue, light blue). (E) Action potential waveform quantification. Mean ± S.E.M.; *p≤0.05; **p<0.01; ***p<0.0001 ****p≤0.0001. N.S., not significant; two-way RM-ANOVA (F-I curves), one-way ANOVA (AP waveform comparisons) with Tukey post-hoc tests.
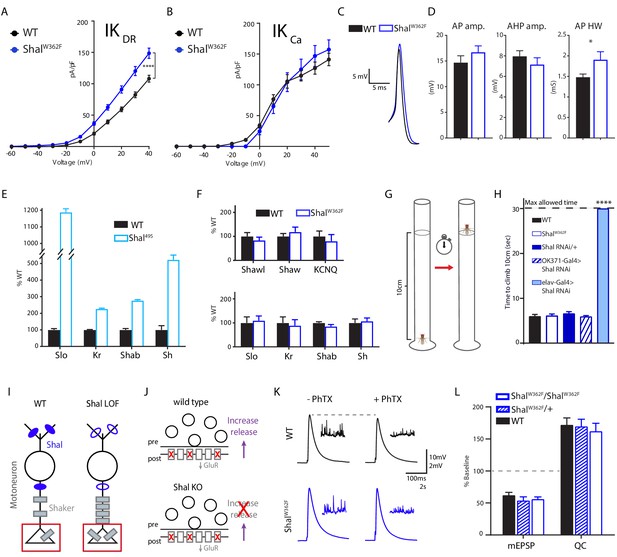
Shal activity block engages distinct homeostatic rebalancing mechanism.
(A) IKDR I-V plots (left) and representative traces (right) in WT (black, n = 7) and ShalW362F (blue, n = 13) motoneurons. (B) IKCa I-V plots (left) and representative traces (right) in WT (black; n = 12) and ShalW362F (blue; n = 10) motoneurons. (C) Example action potential waveforms for WT (black) and Shal495 (blue) overlaid. (D) Action potential waveform quantification. (E) Quantitative PCR from WT (black) and Shal495 (blue) whole third instar larval brains (≥3 biological replicates, each). (F) Quantitative PCR from WT (black) and ShalW362F (blue) whole third instar larval brains (≥3 biological replicates, each). (G) Cartoon representation of negative geotaxis assay. A single animal was placed in a 20 cm tall clean glass tube. The fly was startled by tapping and time to climb to 10 cm high mark was recorded. (H) Results of climbing assay. WT n = 10, ShalW362F n = 10, Shal-RNAi/+ n = 10, Shal-RNAi/OK371 n = 10, elav-Gal4;Shal-RNAi n = 10, elav-Gal4;Shal-RNAi;Kr-RNAi n = 10. (I) Cartoon diagramming how loss of Shal induces compensatory upregulation of Shaker in motoneuron axons and nerve terminals. (J) Diagrams illustrating the consequences of ion channel rebalancing on presynaptic homeostatic potentiation (PHP). Top: reduced postsynaptic receptor sensitivity triggers compensatory upregulation of presynaptic neurotransmitter release during PHP. Bottom: Shal knockout animals fail to express PHP. (K) Sample traces showing representative EPSP and mEPSP amplitudes in the absence and presence of PhTX for WT (black) and ShalW362F (blue). (L) Reduction in mEPSP amplitudes and increase in quantal content (QC) following incubation in PhTX expressed as percent change relative to baseline for each genotype in absence of PhTX. WT baseline n = 9,+PhTX n = 12; ShalW362F/+baseline n = 6,+PhTX n = 6; ShalW362F/ShalWW362F baseline n = 9,+PhTX n = 10. Mean ± S.E.M.; *p<0.05; **p<0.005; ***p<0.0005, ****p<0.0001. N.S., not significant; two-way RM-ANOVA with Bonferoni post-hoc tests (I-V plots); Student’s t test (AP waveform comparisons); one-way ANOVA (climbing assay) with Tukey post-hoc tests.
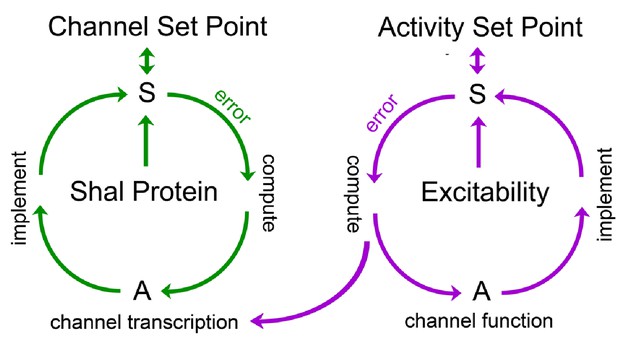
Model for FRH in Drosophila motoneurons.
A set point is operationally defined as the level of a variable that is retargeted by a homeostatic signaling system. We propose the existence of a set point for abundance of the Shal protein and a separate set point for neuronal activity. Each set point is connected to a dedicated sensor (S) that monitors either protein levels or neuronal activity. If the sensor detects deviation from the set point, an error signal is produced. The sign and magnitude of the error is computed, over time, and drives changes in actuators (A) that implement negative feedback and restoration of the set point. Actuators in the Shal proteostasis feedback system (green) include the transcription factor Kruppel and downstream transcriptional changes in ion channels that include, but are not be limited to, Shaker and slo. Actuators for the activity-dependent homeostat (purple) remain unknown in Drosophila, but could include changes in ion channel transcription in other systems. We propose that the activity-dependent homeostat (purple) could be connected to the mechanisms of ion channel homeostasis. In this manner, activity-dependent homeostatic signaling could act primarily on the ion channel proteostatic program. If actuators of the activity-dependent proteostat include changes in ion channel gene expression, then channel proteostasis mechanisms could be secondarlily engaged. This might suggest the existence of repressors that couple channel and activity-dependent homeostatic systems, though there remains no experimental evidence to date.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Chemical compound, drug | Protease (Type XIV, Streptomyces griseus) | Sigma | P5147; CAS 9036-06-0 | |
| Chemical compound, drug | 1-naphthylacetyl spermine trihydrochloride (NASP) | Sigma | N193; CAS 1049731-36-3 | |
| Chemical compound, drug | Tetrodotoxin citrate (TTX) | Tocris | 1069; CAS 18660-81-6 | |
| Chemical compound, drug | Tetraethylamonium chloride (TEA-Cl) | Sigma | T2265; CAS 56-34-8 | |
| Chemical compound, drug/drug | 4-Aminopyridine (4-AP) | Sigma | A78403; CAS 504-24-5 | |
| Chemical compound, drug | Paxilline | Tocris | 2006; CAS 57186-25-1 | |
| Chemical compound, drug | XE-991 dihydrochloride | Tocris | 2000; CAS 122955-13-9 | |
| Chemical compound, drug | Philanthotoxin-433 (PhTX) | Santa Cruz Biotechnology | sc-255421; CAS 276684-27-6 | |
| Gene (Drosophila melanogaster) | w1118 | N/A | FLYB: FBal0018186 | |
| Genetic reagent (D. melanogaster) | MN1-Ib-GAL4 | Kim et al., 2009 | Yuh-Nung Jan (UCSF, San Francisco, CA) | |
| Genetic reagent (D. melanogaster) | UAS-Shal-RNAi | Vienna Drosophila RNAi Center (VDRC) | VDRC:103363 | P{KK100264}VIE-260B |
| Gene (D. melanogaster) | ShalW362F | This paper | N/A | CRISPR-Cas9 engineered point mutation |
| Genetic reagent (D. melanogaster) | elavC155-GAL4 | Bloomington Drosophila Stock Center | BDSC:458 | P{w[+mW.hs]=GawB}elav[C155] |
| Genetic reagent (D. melanogaster) | OK371-GAL4 | Bloomington Drosophila Stock Center | BDSC:26160 | P{GawB}VGlut[OK371] |
| Gene (D. melanogaster) | Slo1 | Bloomington Drosophila Stock Center | BDSC:4587 | |
| Genetic reagent (D. melanogaster) | UAS-Kr-RNAi | Bloomington Drosophila Stock Center | BDSC:27666 | P{TRiP.JF02745}attP2 |
| Genetic reagent (D. melanogaster) | UAS-CD8:GFP/UASmCD8:GFP | N/A | FLYB: FBti0012686 | |
| Gene (D. melanogaster) | Shal495 | Bloomington Drosophila Stock Center | BDSC:18338 | PBac{WH}Shal[f00495] |
| Sequence-based reagent | Forward primer to clone Shal upstream gRNA into pCFD4 | This paper | N/A | TATATAGGAAAGATATCCGGGTGAACTTCGCAACTTCACATCGATTCCGGGTTTTAGAGCTAGAAATAGCAAG |
| Sequence-based reagent | Reverse primer to clone Shal downstream gRNA into pCFD4 | This paper | N/A | ATTTTAACTTGCTATTTCTAGCTCTAAAACTCTGGCATTAGAGAACGATTCGACGTTAAATTGAAAATAGGTC |
| Sequence-based reagent | Forward primer for Shal 5’ homology arm amplification, for insertion into pHD-ScarlessDsRed | This paper | N/A | GGAGACCTATAGTGTCTTCGGGGCCGAgcataattgctcccaagaac |
| Sequence-based reagent | Reverse primer for Shal 5’ homology arm amplification, for insertion into pHD-ScarlessDsRed | This paper | N/A | CGTCACAATATGATTATCTTTCTAGGGTTAACAAAATGCACATACAAAAGATGC |
| Sequence-based reagent | Forward primer for Shal 3’ homology arm amplification, for insertion into pHD-ScarlessDsRed | This paper | N/A | CGCAGACTATCTTTCTAGGGTTAAGCGTTTTAGTTTTATCGATTTATTTG |
| Sequence-based reagent | Reverse primer for Shal 3’ homology arm amplification, for insertion into pHD-ScarlessDsRed | This paper | N/A | GGAGACGTATATGGTCTTCTTTTCCcgggaaacagccagggggcgaggc |
| Sequence-based reagent | Primer for mutagenesis: Shal W362F and upstream PAM | This paper | N/A | CTTCACATCGATTCCGGCCGCCTTCTTTTATACCATCGTCACAATG |
| Sequence-based reagent | Primer for mutagenesis: downstream PAM | This paper | N/A | gttttttgttgatttcaaatacactctggcattagagaacg |
| Recombinant DNA reagent | pHD-ScarlessDsRed | Drosophila Genomics Resource Center | DGRC:1364 | |
| Recombinant DNA reagent | pCFD4: U6:1-gRNA U6:3-gRNA | Addgene | 49411 | |
| Commercial assay or kit | RNeasy Plus Micro Kit | Qiagen | 74034 | |
| Commercial assay or kit | Turbo DNA-free Kit | Ambion | AM1907 | |
| Commercial assay or kit | SuperScript III First-Strand | Invitrogen | 18080–051 | |
| Commercial assay or kit | TaqMan Fast Universal PCR Master Mix (2X), no AmpErase UNG | Applied Biosystems | 4352042 | |
| Commercial assay or kit | KCNQ FAM Taqman gene expression assay | Applied Biosystems | Dm01846741_g1 | |
| Commercial assay or kit | Kr FAM Taqman gene expression assay | Applied Biosystems | Dm01821853_g1 | |
| Commercial assay or kit | RpL32 FAM Taqman gene expression assay | Applied Biosystems | Dm02151827_g1 | |
| Commercial assay or kit | Sh FAM Taqman gene expression assay | Applied Biosystems | Dm01828717_m1 | |
| Commercial assay or kit | Shab FAM Taqman gene expression assay | Applied Biosystems | Dm01821965_m1 | |
| Commercial assay or kit | Shaw FAM Taqman gene expression assay | Applied Biosystems | Dm01841512_g1 | |
| Commercial assay or kit | Shawl FAM Taqman gene expression assay | Applied Biosystems | Dm01809871_m1 | |
| Commercial assay or kit | Slo FAM Taqman gene expression assay | Applied Biosystems | Dm02150795_m1 | |
| Software, algorithm | Clampex 10.3 | Molecular Devices | https://www.moleculardevices.com | |
| Software, algorithm | Igor Pro 7.02 | WaveMetrics | https://www.wavemetrics.net/ | |
| Software, algorithm | MiniAnalysis 6.0.7 | Synapsoft | http://www.synaptosoft.com/MiniAnalysis/ | |
| Software, algorithm | SDS 2.4 | Applied Biosystems | https://www.thermofisher.com/order/catalog/product/4350490 | |
| Software, algorithm | Excel 2013 | Microsoft | https://www.microsoft.com/ | |
| Software, algorithm | GraphPad Prism 7 | GraphPad | https://www.graphpad.com/ | |
| Software, algorithm | Adobe Illustrator CC 2018 | ADOBE ILLUSTRATOR CC | https://www.adobe.com |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.45717.014





