Symmetry transitions during gating of the TRPV2 ion channel in lipid membranes
Figures
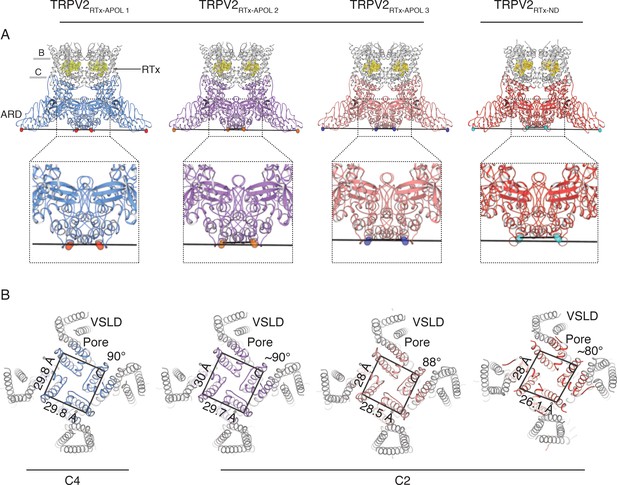
Overview of TRPV2RTx-APOL and TRPV2RTx-ND structures.
(A) Orthogonal view of TRPV2RTx-APOL 1-3 and TRPV2RTx-ND structures. TM domains are colored in grey and the cytoplasmic domains (ARD and C-terminal domain) are colored in blue, violet, salmon and red, respectively. RTx is shown in stick and sphere representation and colored in yellow. Lines drawn between diagonally opposite ARDs (residue E95, shown in red, orange, blue and cyan spheres, respectively) illustrate the relative position of ARDs in the tetramer. The close-up shows that the ankyrin repeats of diagonally opposing subunits in TRPV2RTx-APOL 2 and TRPV2RTx-ND are positioned in different planes. (B) Top view of the channel (S5, S6 and PH are colored in blue, violet, salmon and red, respectively). Lines drawn between residues V620 in the S6 helix illustrate the symmetry within the pore domain. Distances and angles indicate the presence of two-fold symmetry.
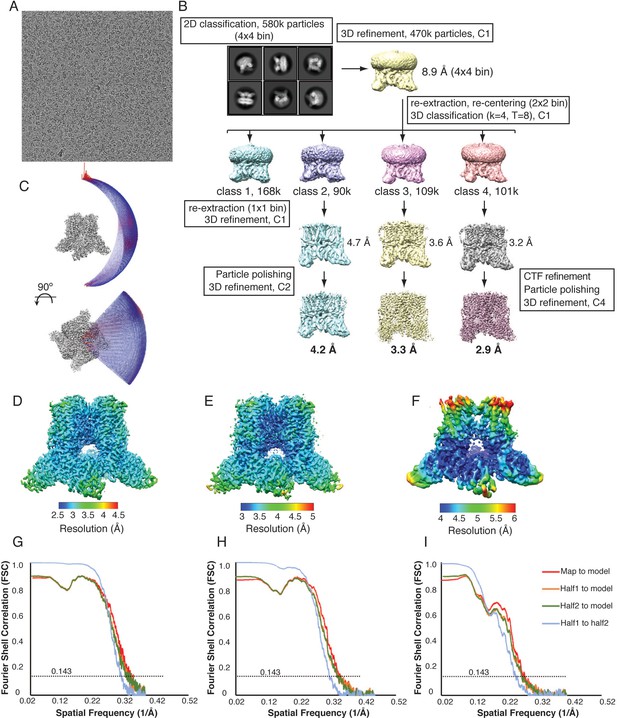
Cryo-EM data collection and processing, TRPV2RTx-APOL.
(A) Representative micrograph from the TRPV2RTx-APOL dataset. (B) 3D reconstruction workflow resulting in three distinct TRPV2RTx-APOL structures. (C) Euler plot distribution. Red regions signify the best represented views. (D–F) Local resolution estimates calculated in RELION for TRPV2RTx-APOL 1 (D), TRPV2RTx-APOL 2 (E), TRPV2RTx-APOL 3 (F). (G-I) FSC curves calculated between the half maps (blue), atomic model and the final map (red), and between the model and each half-map (orange and green) for TRPV2RTx-APOL 1(G), TRPV2RTx-APOL 2(H), TRPV2RTx-APOL 3(I).
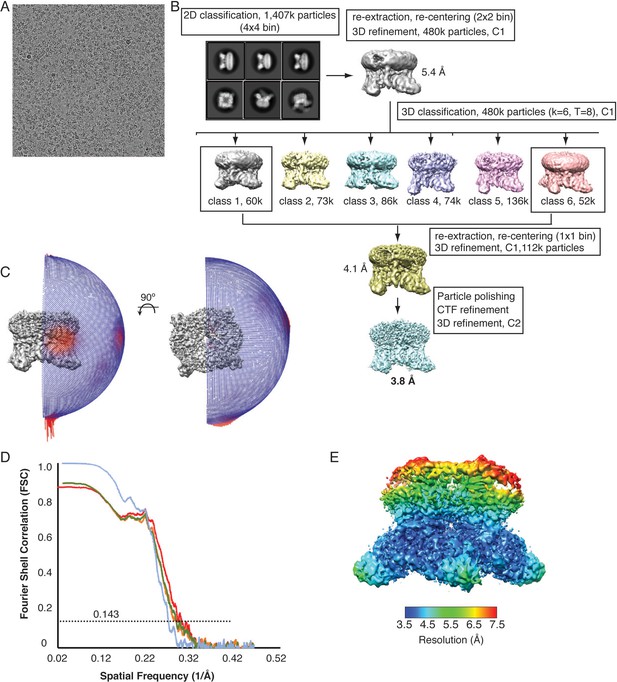
Cryo-EM data collection and processing, TRPV2RTx-ND.
(A) Representative micrograph from the collected TRPV2RTx-ND dataset. (B) 3D reconstruction workflow. (C) Euler distribution plot. Red regions indicate the best represented views. (D) FSC curves calculated between the half maps (blue), atomic model and the final map (red), and between the model and each half-map (orange and green). (E) Local resolution estimate, calculated in RELION.

C2 symmetry in the TRPV2RTx-ND.
(A) Classes 1 and 6 from the 3D classification of partcles from the TRPV2RTx-ND dataset (Figure 1—figure supplement 2) refined individually with no symmetry imposed. (B) To evaluate the C2 symmetry in TRPV2RTx-ND, FSC curves were calculated between the non-symmetrized (C1) combined class 1 and 6 map. This map contains the same particles as the final map in Figure 1—figure supplement 2. Half maps (blue), atomic model and the full map (red), and between the model and each half-map (orange and green). (C) Fit of TRPV2RTx-ND coordinates into the refined, non-symmetrized map of class 1. FSC curves between half maps (blue), atomic model and the full map (red), and between the model and each half-map (orange and green). (D) Fit of TRPV2RTx-ND coordinates into the refined, non-symmetrized map of class 6. FSC curves between half maps (blue), atomic model and the full map (red), and between the model and each half-map (orange and green). (E–F) Electron density around the S6 helices (E) and the pore helices (F) in the combined class 1 and class 6 map with no symmetry imposed. The density is contoured at level 0.01 and radius 2.
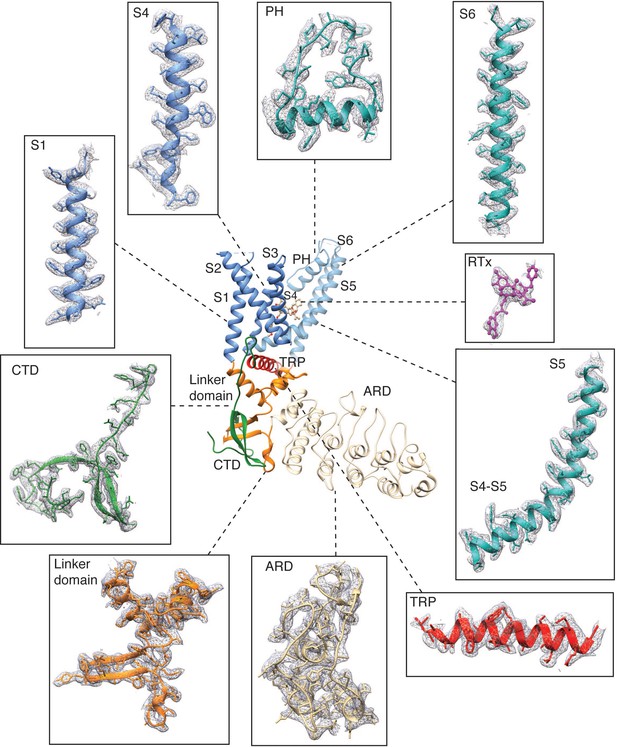
Representative electron densities in the TRPV2RTx-APOL 1 cryo-EM map.
Densities are contoured at level 0.06 and radius 2.

Representative electron densities in the TRPV2RTx-APOL 2 cryo-EM map.
Densities are contoured at level 0.06 and radius 2.

Representative electron densities in the TRPV2RTx-APOL 3 cryo-EM map.
Densities are contoured at level 0.02 and radius 2.
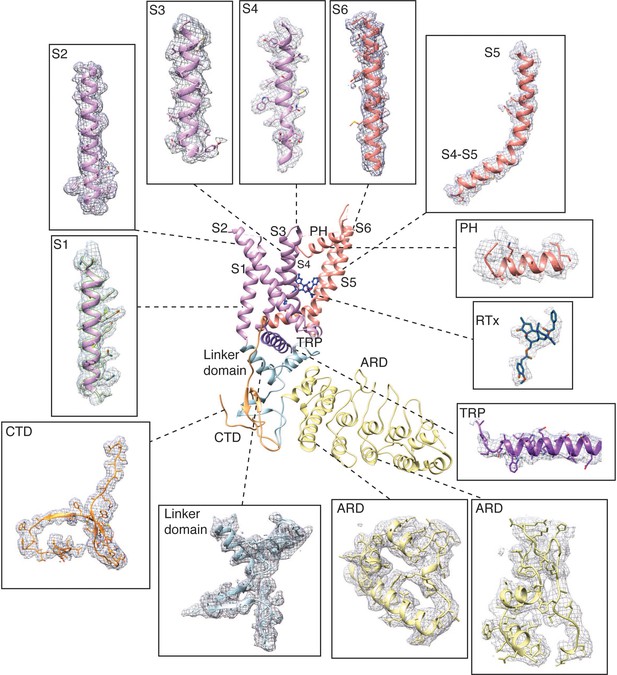
Representative electron densities in the TRPV2RTx-ND cryo-EM map.
Densities are contoured at level 0.01–0.03 and radius 2.
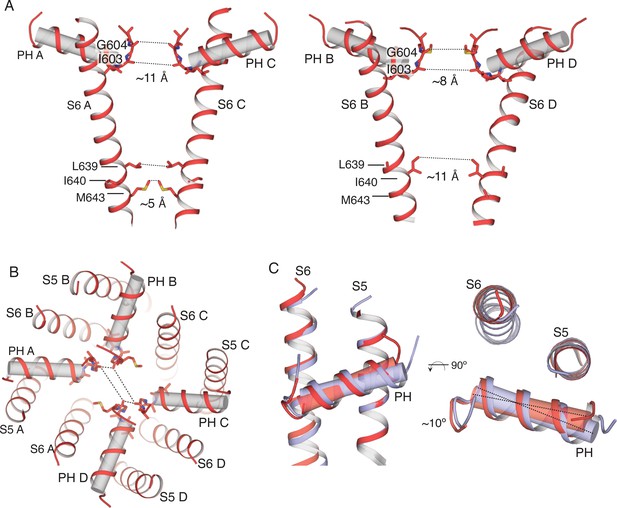
Overview of the pore in the TRPV2RTx-ND structure.
(A) S6 and pore helices of subunits A and C (left) and subunits B and D (right). Pore helices are shown in both cartoon and cylinder representation (grey). Dashed lines and values represent distances between the indicated residues. S6 helices in A and C are straight and α-helical, while the S6 in subunits B and D is bent. (B) Top view of the TRPV2RTx-ND pore, with pore helices shown in both cartoon and cylinder representation. Dashed lines illustrate the distances between residues G604 in the selectivity filter. (C) Overlay of the TRPV2RTx-ND pore domains (S5, S6 and pore helices). Subunit A is shown in red and subunit B in violet. The pore helix of subunit A swivels by ~10° relative to subunit B.

Pore comparison of TRPV2APO (orange) and TRPV2RTx-APOL 1-3 (blue, purple and salmon, respectively).
HOLE profiles (dots and graph) indicate that both the selectivity filter and the common gate are closed in TRPV2RTx-APOL 1-3.
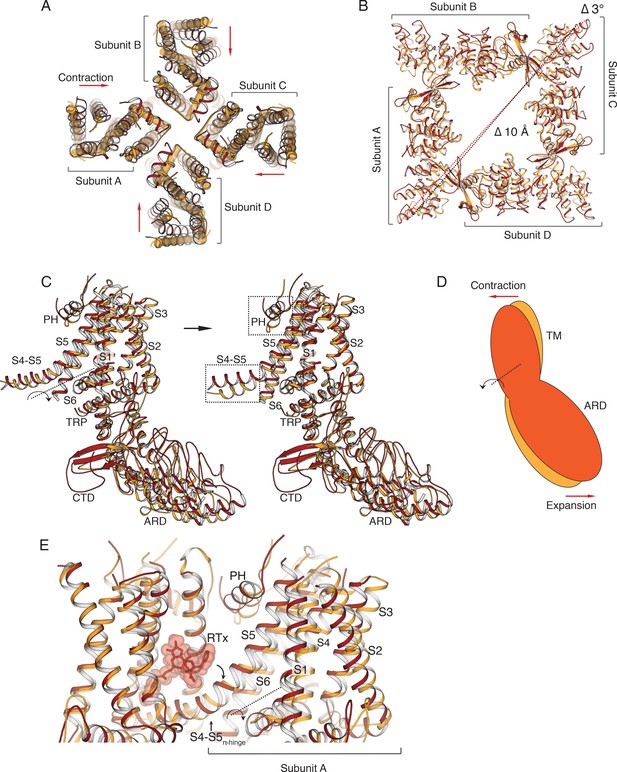
Comparison of TRPV2RTx-ND (red) and TRPV2APO (orange).
(A) Overlay of TRPV2RTx-ND and TRPV2APO, top view. TRPV2RTx-ND is shown in cartoon representation and TRPV2APO as cylinders. Relative to TRPV2APO, the TM subunits of TRPV2RTx-ND exhibit contraction (red arrows). (B) Top view of the ARDs in TRPV2RTx-ND and TRPV2APO. TM helices are removed for ease of viewing. Dashed lines represent distances between residues T100, showing a 10 Å expansion (Δ 10 Å) and 3° rotation (Δ 3o) of the TRPV2RTx-ND ARD assembly relative to TRPV2APO. (C) A rigid-body rotation of TRPV2RTx-ND subunit B around the S4-S5 linker achieves alignment with the subunit B from TRPV2APO. Following alignment, only the S4-S5 linkers and the pore helices (PH) diverge in the two subunits (dashed box). (D) Cartoon illustrating how the movements of the TM and the ARD in TRPV2RTx-ND are coupled. The red and orange shapes represent a single subunit of TRPV2RTx-ND and TRPV2APO, respectively. The rotation of the subunit is manifested as ‘contraction’ in the TM domains and ‘expansion’ of the ARD. (E) RTx binding in the vanilloid binding pocket exerts force on the S4-S5 linker, changing the conformation of the junction from α- to π-helix, and induces the rotation of the subunit around the S4-S5π-hinge.
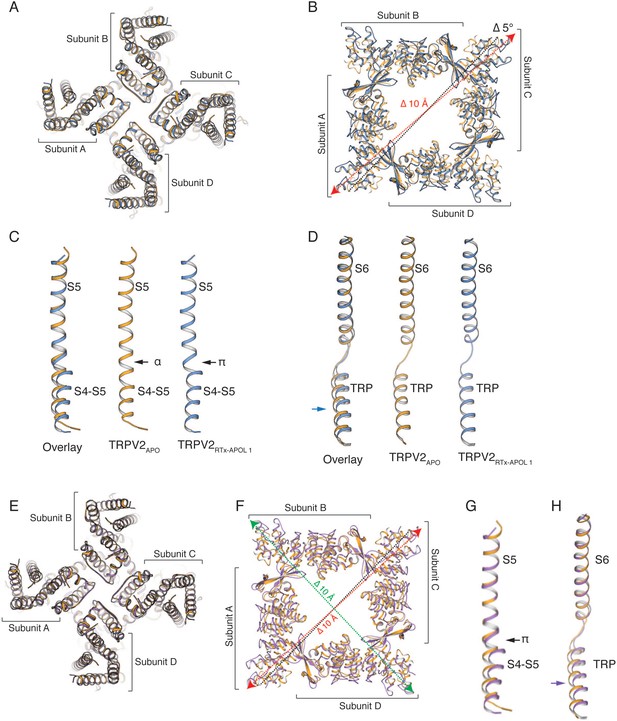
Comparison of TRPV2RTx-APOL 1-2 and TRPV2APO.
(A–D) Comparison of TRPV2RTx-APOL 1 (blue) and TRPV2APO (orange). (A), Overlay of the TM helices. Individual subunits are indicated. (B) Top view of the cytoplasmic domains. The TMs are removed for ease of viewing. Distance measured between residues T100 in TRPV2APO (black dotted line) and TRPV2RTx-APOL 1 (red dotted line). The cytoplasmic assembly rotates by 5° and widens by 10 Å in the presence of RTx. (C) Overlay of S5 helices. In the presence of RTx, a π-helix is formed at the junction of the S4-S5 linker and the S5 helix changing the position of the S4-S5 linker. (D) Overlay of S6 helices and the TRP domain. The TRP domain is displaced in the presence of RTx. (E–H) Comparison of TRPV2RTx-APOL 2 (violet) and TRPV2APO (orange). (E) Top-view of the overlay of the TM helices. Individual subunits are indicated. (F) Top-view of the cytoplasmic domains. The TMs are not shown for ease of viewing. Distances measured between residues T100 of diagonally opposing subunits (TRPV2APO (black dotted line) and TRPV2RTx-APOL 2 (red and green dotted lines). (G) Overlay of the S5 helices shows that the S4-S5 linker adopts different conformations. The divergence originates at the π-helix. (H) Overlay of the S6 helices and the TRP domain shows a displacement of the TRP helix in the TRPV2RTx-APOL 2.

Two-fold symmetry in TRPV2RTx-APOL 3 (salmon).
(A) Pore of the four-fold symmetric TRPV2APO (orange) compared to the pore of the two-fold symmetric TRPV2RTx-APOL 3 (salmon) subunits A and C (middle) and subunits B and D (right). (B) Position of the pore helix in TRPV2RTx-APOL 3 subunit A (salmon) compared to the subunit B (grey). (C) Conformation of the S4-S5 linker in TRPV2RTx-APOL 3 subunit A (salmon) compared to subunit B (grey). (D) Comparison of TRPV2RTx-APOL 3 (salmon) and TRPV2APO (orange) ARD. The TMs are removed for ease of viewing. The dashed lines represent the distance between diagonally opposite residues T100 in TRPV2APO (black line) and TRPV2RTx-APOL 3 (red line). The ARD are rotated and expanded in TRPV2RTx-APOL 3.

Symmetry breaking in the TRPV2RTx-APOL 2-3 ARD.
(A–B) Subunit A of the four-fold symmetric TRPV2RTx-APOL 1 overlaid with TRPV2APO shows good alignment of the TM domains, with divergence at the S4-S5 linker and the ARD. (C) Two-fold symmetry in the ARD and S4-S5 linker of the TRPV2RTx-APOL 2 structure. Subunit A (purple) overlaid with TRPV2APO (orange) (left). Subunit B (dark grey) overlaid with TRPV2APO (orange) (right). In both subunits, TM domains are aligned but ARD and the S4-S5 linker (dashed line box) diverge. (D) TRPV2RTx-APOL 2 subunits A and B (dark grey) assume distinct conformations in the ARD. (E), Two-fold symmetry in the ARD and S4-S5 linker of the TRPV2RTx-APOL 3 structure. Subunit A (salmon) overlaid with TRPV2APO (orange) (left). Subunit B (purple) overlaid with TRPV2APO (orange) (right). Similar to TRPV2RTx-APOL 2, TM domains are aligned but ARD and the S4-S5 linker (dashed line box) diverge. (D) TRPV2RTx-APOL 3 subunits A and B (purple) assume distinct conformations in the ARD, albeit to lesser extent than TRPV2RTx-APOL 2.
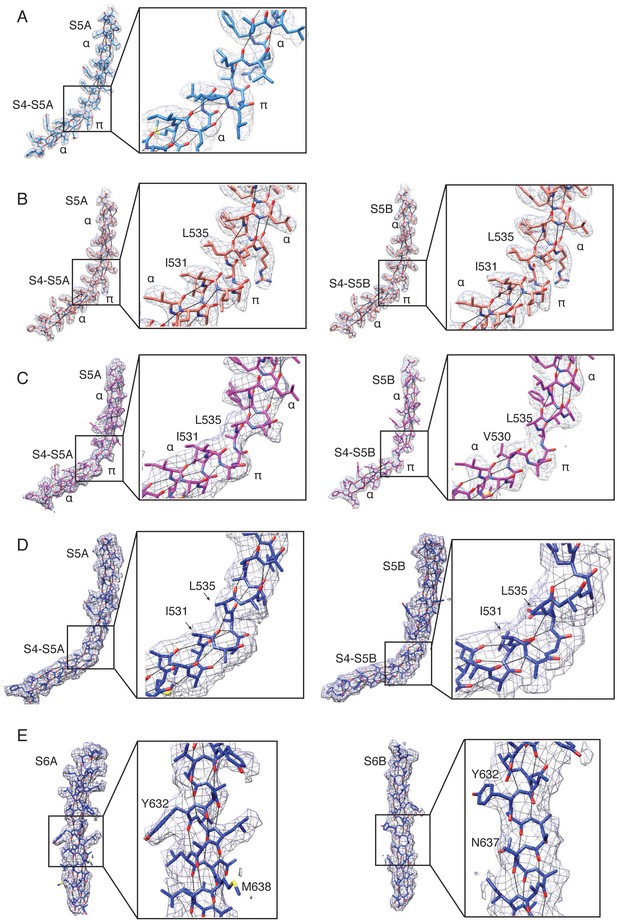
Electron density around the S4-S5 linker π-helices in TRPV2RTx-APOL 1 (A), TRPV2RTx-APOL 2 (B) and TRPV2RTx-APOL 3 (C).
(A) TRPV2RTx-APOL 1 density is contoured at level 0.06 and radius 2. Only subunit A is shown. The inset shows a close-up of the junction between the S4-S5 linker and the S5 helix. (B) TRPV2RTx-APOL 2 density is contoured at level 0.04 and radius 2. Subunits A and B are shown. The insets show a close-up of the junction between the S4-S5 linker and the S5 helix. (C) TRPV2RTx-APOL 1 density is contoured at level 0.015 and radius 2. Subunits A and B are shown. The insets show a close-up of the junction between the S4-S5 linker and the S5 helix. (D–E) Electron density in TRPV2RTx-ND contoured at level 0.01. (D) Density around the S5 helix and the S4-S5 linker in TRPV2RTx-ND subunits A and B. The insets show a close-up of the junction between the S4-S5 linker and the S5 helix. (E) Density around TRPV2RTx-ND helices S6A and S6B. The inset shows a close-up of the hydrogen bonds in the helix.
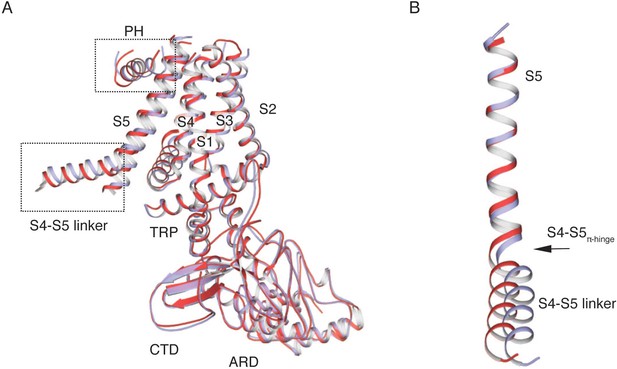
Comparison of TRPV2RTx-ND subunits A (red) and B (violet).
(A) Overlay of the subunits. The regions that diverge from the overlay, the S4-S5 linker and the pore helix (PH), are indicated by a dashed line box. (B) Overlay of S5 helices. The alignment diverges at the S4-S5 linker π-helix (S4-S5π-hinge) giving rise to different conformations of the S4-S5 linker in the two subunits.
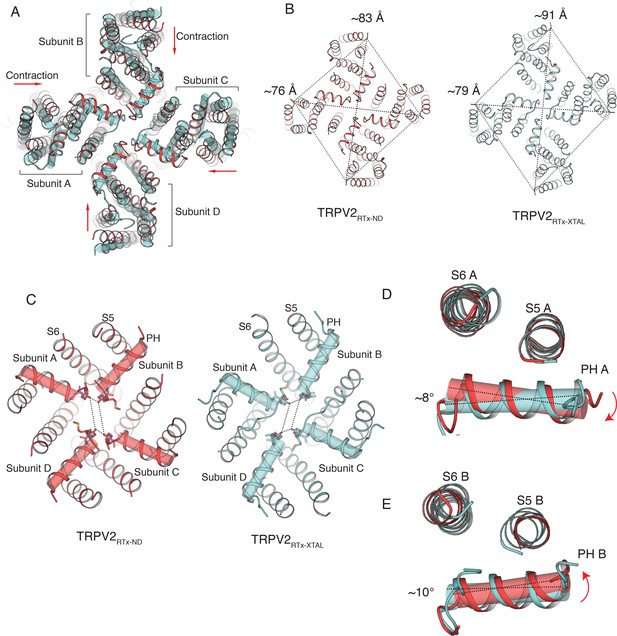
Comparison of TRPV2RTx-ND (red) and TRPV2RTx-XTAL (cyan).
(A) Overlay of TRPV2RTx-ND and TRPV2RTx-XTAL, top view. TRPV2RTx-ND is shown in cartoon representation and TRPV2RTx-XTAL as cylinders. Relative to TRPV2RTx-XTAL, the TM domains of TRPV2RTx-ND are contracted (red arrows). (B) Comparison of two-fold symmetry in TRPV2RTx-ND and TRPV2RTx-XTAL. Dashed lines represent distances between residues A427. The distances between diagonally opposing subunits are indicated. (C) Top view of the SF in TRPV2RTx-ND and TRPV2RTx-XTAL. Pore helices are shown in both cartoon and cylinder representation. Dashed lines represent distances between residues G604 in the selectivity filter. (D–E) Overlay of the pore domains of TRPV2RTx-ND and TRPV2RTx-XTAL subunit A (D) and subunit B (E) shows that the pore helices A and B in TRPV2RTx-ND swivel by ~8° and ~10°, respectively, compared to TRPV2RTx-XTAL.
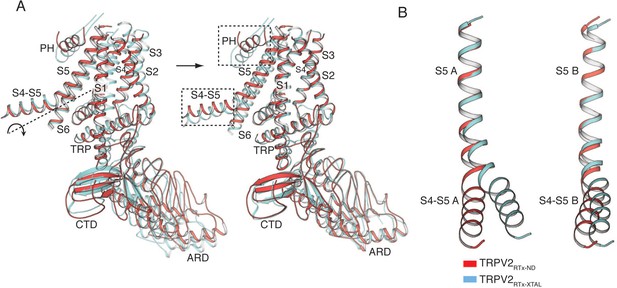
Comparison of subunits B in TRPV2RTx-ND (red) and TRPV2RTx-XTAL (cyan).
(A) Rotation of subunit B from TRPV2RTx-ND around the S4-S5π-hinge aligns it to subunit B from TRPV2RTx-XTAL. The S4-S5 linker and the PH (dashed box) diverge from the alignment. Rotation axis indicated with dashed line and arrow. (B) Overlay of TRPV2RTx-ND and TRPV2RTx-XTAL S5 helices from subunit A (left) and subunit B (right) show that the S4-S5 linkers assume different conformations.
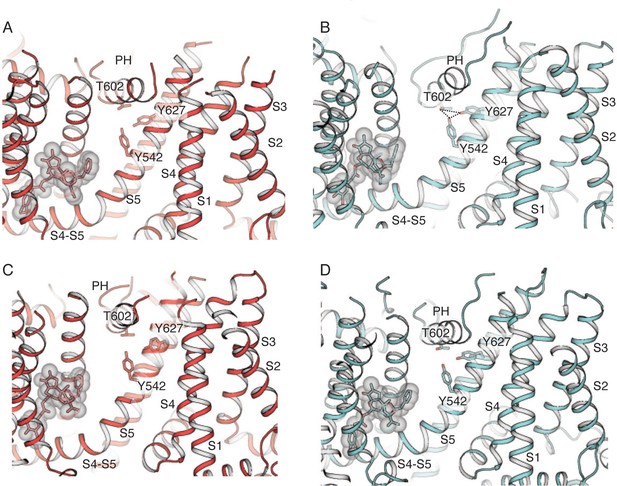
Interactions between the pore helix (PH) and S5 and S6.
RTx is shown in stick and transparent surface representations. (A–B) Side view of subunits A in TRPV2RTx-ND(A) and TRPV2RTx-XTAL(B). The putative hydrogen bond triad (Y542-T602-Y627) is present in subunit A of TRPV2RTx-XTAL. (C–D) The triad is broken in TRPV2RTx-ND. Side view of subunits B in TRPV2RTx-ND(C) and TRPV2RTx-XTAL(D). The interaction network is absent in both structures.
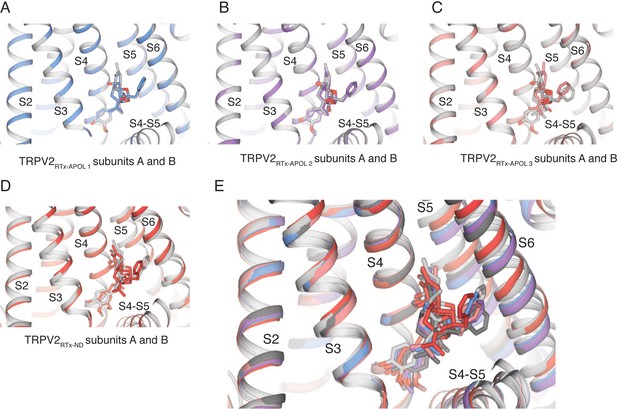
Binding of RTx in TRPV2RTx-APOL and TRPV2RTx-ND.
(A) Binding of RTx in subunit A (blue) and B (grey) of the C4 symmetric TRPV2RTx-APOL 1. (B) Binding of RTx in subunit A (violet) and B (grey) of the C2 symmetric TRPV2RTx-APOL 2. (C) Binding of RTx in subunit A (salmon) and B (grey) of the C2 symmetric TRPV2RTx-APOL 3. (D) Binding of RTx in subunit A (red) and B (grey) of the C2 symmetric TRPV2RTx-ND. (E) Overlay of TRPV2RTx-APOL 1 subunit A (blue), TRPV2RTx-APOL 2 subunit A (violet), TRPV2RTx-APOL 3 subunit A (salmon) and subunit B (dark grey), TRPV2RTx-ND subunit A (red) and subunit B (light grey) shows the different binding poses of RTx captured in this study.

The pore turret in TRPV2RTx-APOL 1 (blue) and rat TRPV2 (PDB 6BO4, purple).
(A) Top view of the map and model of TRPV2RTx-APOL 1 with the pore domain indicated by dashed red box. The density containing S5, S6, the PH, pore loop and turret is colored in red. (B) Side view of the map and model of the pore domain in TRPV2RTx-APOL 1. The density containing S5, S6, the PH, pore loop and turret is colored in red. S5, S6, PH and pore turret are indicated. (C) Top view of the map and model of rat TRPV2 with the pore domain indicated by dashed red box. (D) Side view of the map and model of the pore domain in rat TRPV2. S5, S6, PH and pore turret are indicated. (E) Position of the turret relative to the membrane (yellow lines) in TRPV2RTx-APOL 1 and rat TRPV2. The sequence of the turret shows conservation (grey boxes) and amino acids colored in red indicate charged or polar residues.
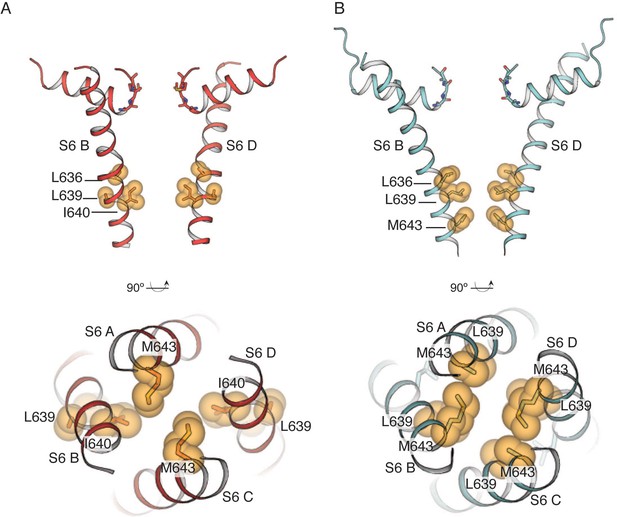
The common gate in TRPV2RTx-ND (red) and TRPV2RTx-XTAL (cyan).
(A) Side view of the TRPV2RTx-ND pore showing subunits B and D (top). Gate residue I640 is shown in yellow spheres, along with the hydrophobic residues L636 and L639 (side chains not built). Bottom-up view (bottom) shows the contribution of all four subunits to the common gate (M643 in subunits A and C, I640 in subunits B and D). (B) Side view of the TRPV2RTx-XTAL pore, showing subunits B and D (top). Gate residue M643 is shown in yellow spheres, along with hydrophobic pore lining residues L636 and L639. Bottom-up view of the common gate (bottom) shows gate residues M643 (side chain not built in subunits A and C).

The amphipol and nanodisc clouds.
(A) Top-view of the TRPV2RTx-APOL 1-3 and TRPV2RTx-ND maps. (B) Top-view of the TRPV2RTx-APOL 1-3 and TRPV2RTx-ND maps with the non-protein density colored in red. All maps were filtered to 5 Å and contoured at level 0.015.
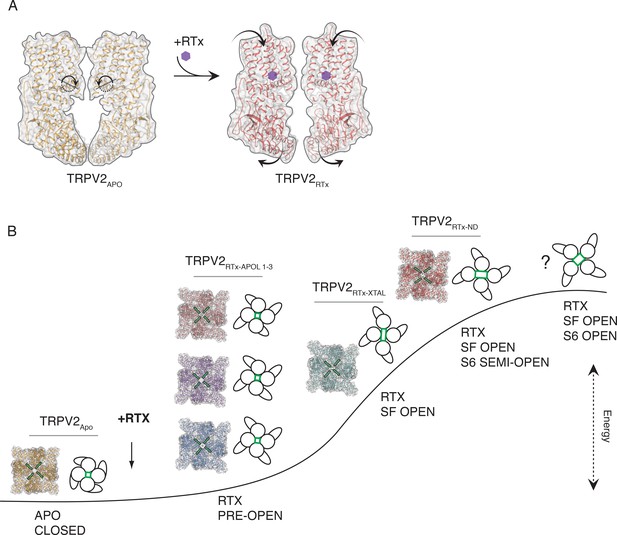
Conformational states associated with RTx-mediated gating of TRPV2.
(A) TRPV2 subunit rotation upon binding of RTx. Rotation axis and direction are indicated in dashed line and circular arrow in apo TRPV2 (left). The rotation results in contraction of the TM domains and widening of the cytoplasmic assembly (right). (B) Hypothetical trajectory of TRPV2 gating with associated conformational states. Upon addition of RTx, TRPV2 first enters low-energy pre-open states that are characterized by rotation, widening and symmetry breaking in the ARD (TRPV2RTx-APOL 1-3, models shown in cartoon and surface representation). In the next step, the channel assumes C2 symmetric state with an open SF, but closed common (S6) gate (TRPV2RTx-XTAL, model shown in cartoon and surface representation). This is followed by a less C2 symmetric state with an open SF and semi-open common (S6) gate (TRPV2RTx-ND, model shown in cartoon and surface representation). Finally, we propose that the channel assumes a high-energy fully open state that is C4 symmetric but might have C2 symmetry in the SF. The SF is indicated in green in models and cartoons.
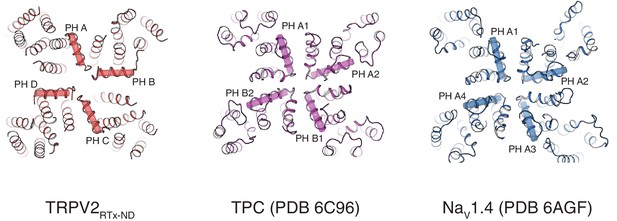
Comparison of TRPV2RTx-ND (red), TPC1 (PDB 6C96, purple) and NaV1.4 (PDB 6AGF, blue).
Top view, pore helices are indicated.
Tables
Data collection and refinement statistics
https://doi.org/10.7554/eLife.45779.010| Data collection and processing | TRPV2RTx-ND | TRPV2RTx-APOL 1 | TRPV2RTx-APOL 2 | TRPV2RTx-APOL 3 |
|---|---|---|---|---|
| Electron microscope | Titan Krios | Titan Krios | ||
| Electron detector | Falcon III | Falcon III | ||
| Magnification | 75,000x | 75,000x | ||
| Voltage (kV) | 300 | 300 | ||
| Electron exposure (e–/Å2) | 42 | 42 | ||
| Defocus range (μm) | −1.25 to −3.0 | −1.25 to −3.0 | ||
| Pixel size (Å) | 1.08 | 1.08 | ||
| Detector | Counting | Counting | ||
| Total extracted particles (no.) | 1,407,292 | 580,746 | ||
| Refined particles (no.) | 482,602 | 470,760 | ||
| Reconstruction | ||||
| Final particles (no.) | 112,622 | 101,570 | 109,623 | 90,862 |
| Symmetry imposed | C2 | C4 | C2 | C2 |
| Nominal Resolution (Å) | 3.8 | 2.9 | 3.3 | 4.19 |
| FSC 0.143 (masked/unmasked) | 3.7/3.9 | 2.9/3.05 | 3.2/3.5 | 4.0/4.3 |
| Map sharpening B factor (Å2) | −30 | −78 | −92 | −133 |
| Refinement | ||||
| Model composition Non-hydrogen atoms Protein residues Ligands | 16,878 2396 RTx: 4 | 18,236 2404 RTx: 4 | 18,452 2440 RTx: 4 | 17,548 2440 RTx: 4 |
| Validation MolProbity score Clashscore Poor rotamers (%) | 1.39 4 0 | 1.11 1.9 0 | 1.28 2.7 0 | 1.37 2.7 0 |
| Ramachandran plot | ||||
| Favored (%) Allowed (%) Disallowed (%) | 96.5 3.5 0 | 97.1 2.9 0 | 96.6 3.4 0 | 95.5 4.5 0 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line | DH10Bac E. coli | ThermoFisher Scientific | 10361012 | |
| Cell line | Sf9 | ATCC | CRL-1711 | RRID:CVCL_0549 |
| Recombinant DNA reagent | rabbit TRPV2 | Genscript | Pubmed Accession No. XM_017349044 | |
| Recombinant DNA reagent | Bac-to-Bac Baculovirus Expression System | ThermoFisher Scientific | 10359016 | |
| Recombinant DNA reagent | MSP2N2 scaffold protein | Stephen Sligar laboratory | Addgene:Cat#29520 | PMID:20817758 |
| Chemical compound, drug | n-dodecyl-β-d- maltopyranoside(DDM) | Anatrace | D310 | |
| Chemical compound, drug | Cholesteryl Hemisuccinate | Anatrace | CH210 | |
| Chemical compound, drug | Amphipol A8-35 | Anatrace | A835 | |
| Chemical compound, drug | TRIS | Fisher Scientific | BP152 | |
| Chemical compound, drug | NaCl | Fisher Scientific | S271 | |
| Chemical compound, drug | CaCl2 | Fisher Scientific | C70 | |
| Chemical compound, drug | leupeptin | GoldBio | L-010 | |
| Chemical compound, drug | pepstatin | GoldBio | P-020 | |
| Chemical compound, drug | aprotinin | GoldBio | A-655 | |
| Chemical compound, drug | DNase I | GoldBio | D-301 | |
| Chemical compound, drug | β-mercapto ethanol | Sigma Aldrich | M3148 | |
| Chemical compound, drug | PMSF | Sigma Aldrich | P7626 | |
| Chemical compound, drug | anti-FLAG resin | Sigma Aldrich | A4596 | |
| Chemical compound, drug | Resiniferatoxin | Sigma Aldrich | R8756 | |
| Chemical compound, drug | Bio-Beads SM-2 | BioRad | 152–8920 | |
| Chemical compound, drug | 1,2-dimyristoyl- sn-glycero-3- phosphocholine | Avanti Polar Lipids | 850345P | |
| Chemical compound, drug | 1-palmitoyl-2- oleoyl-sn-glycero-3- phosphocholine (POPC) | Avanti Polar Lipids | 850457C | |
| Chemical compound, drug | 1-palmitoyl-2-oleoyl -sn-glycero-3- phosphoethanolamine (POPE) | Avanti Polar Lipids | 850757C | |
| Chemical compound, drug | 1-palmitoyl-2-oleoyl- sn-glycero-3-phospho- (1'-rac-glycerol) (POPG) | Avanti Polar Lipids | 840457C | |
| Other | Whatman No. one filter paper | Sigma Aldrich | WHA1001325 | |
| Other | UltrAuFoil R1.2/1.3 300-mesh grid | Electron Microscopy Sciences | Q350AR13A | |
| Software, algorithm | MotionCor2 | Zheng et al. (2017) | http://msg.ucsf.edu/em/software/motioncor2.html | RRID:SCR_016499 |
| Software, algorithm | GCTF | Zhang (2016) | https://www.mrc-lmb.cam.ac.uk/kzhang/ | RRID:SCR_016500 |
| Software, algorithm | RELION 3.0 | Zivanov et al. (2018) | https://www2.mrc-lmb.cam.ac.uk/relion/ | RRID:SCR_016274 |
| Software, algorithm | Coot | Emsley and Cowtan (2004) | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ | RRID:SCR_014222 |
| Software, algorithm | Phenix | Adams et al. (2010) | http://phenix-online.org/ | RRID:SCR_014224 |
| Software, algorithm | Molprobity | Chen et al. (2010) | http://molprobity.biochem.duke.edu/index.php | RRID:SCR_014226 |
| Software, algorithm | UCSF Chimera | Pettersen et al. (2004) | https://www.cgl.ucsf.edu/chimera/ | RRID:SCR_004097 |
| Software, algorithm | Pymol | Shrödinger LLC | https://pymol.org/2/ | RRID:SCR_000305 |
| Other | Cryo-electron microscopy structure of rabbit TRPV2 ion channel | Zubcevic et al. (2018b) | PDB ID 5AN8 | PMID:26779611 |
| Other | Cryo-electron microscopy structure of rabbit TRPV2 ion channel | Zubcevic et al. (2018a) | EMDB ID EMD-6455 | PMID:26779611 |
| Other | Crystal structure of the TRPV2 ion channel in complex with RTx | Zubcevic et al. (2018a) | PDB ID 6BWJ | PMID:29728656 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.45779.028





