A valve-like mechanism controls desensitization of functional mammalian isoforms of acid-sensing ion channels
Figures
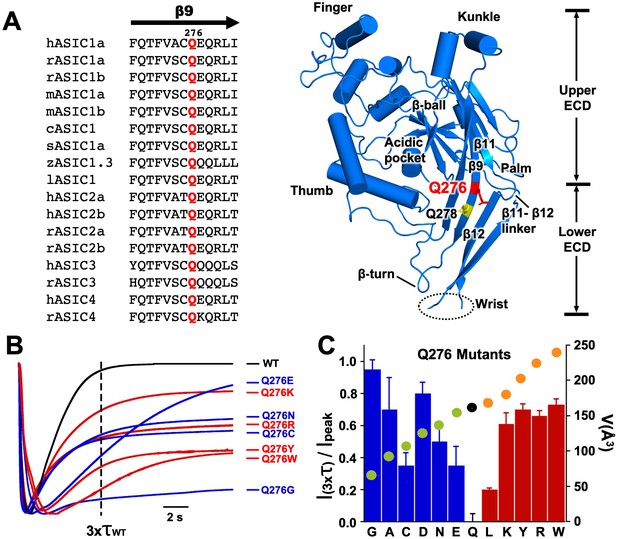
Substitutions of residue Q276 diminish or abolish desensitization.
(A) Sequence alignment of β9 strand of ASIC isoforms from various vertebrate species. Ribbon representation of the crystal structure of the extracellular domain (ECD) of a single subunit of chicken ASIC1. Positions of Q276 and Q278 in β9 are shown as sticks colored red and yellow, respectively. The β11- β12 linker near Q276 has been proposed to functionally divide the ECD into Upper ECD and Lower ECD. (B) Superimposed normalized currents of wild type hASIC1a and Q276 mutants activated by pH 6.5. The vertical dashed line marks the time corresponding to 3x the time constant of desensitization of wild-type channels (τwt). (C) Ratio of sustained/peak currents at time 3xτwt for various Q276 mutants. Columns in blue or in red represent amino acids smaller or larger than glutamine (Q). Error bars are ± SD, n = 8 to 10 cells. The filled circles superimposed on the bars indicate the volume of the corresponding amino acid in Å3 as indicated in the y-right axis: in black is Q, in green smaller and in orange larger than Q.

Representative examples of normalized current traces of various Q276 mutants and the control Q278G.
Bars above currents indicate application of pH 6.5 activating solution.
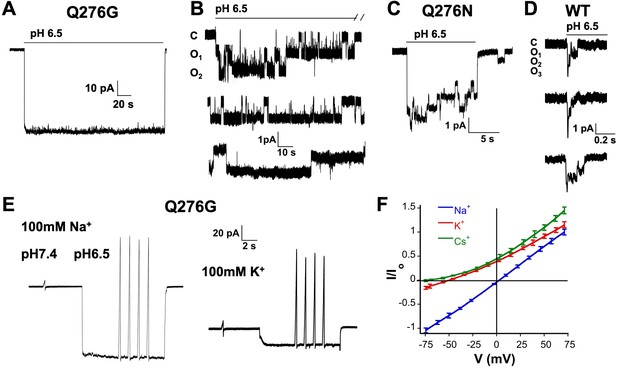
Selectivity and kinetics of Q276G and Q276N mutants.
(A) Representative example of an out-side out patch containing approximately forty Q276G channels. Currents were induced by a rapid change of external solution from pH 7.4 to 6.5 for 80 s and retuned to pH 7.4. Symmetrical 100 mM NaCl. Holding potential −60 mV. (B) Example of a patch containing two Q276G channels activated by pH 6.5 continuously for 2.4 min. Some openings last 50 s. C, O1 and O2 indicate the zero current level (closed) and one or two open channels. (C) A representative example of out-side out patch containing approximately five Q276N channels activated by pH 6.5. (D) Representative examples of unitary currents from wild-type hASIC1a channels elicited by three consecutive sweeps of pH 6.5. An expanded time scale 0.2 s is required to discern opening events. (E) Ion selectivity was calculated by changes in reversal potential according to the indicated protocol. Out-side patches expressing Q276G channels were perfused with external solutions containing 100 mM of a selected cation and 100 mM Na+ in the patch pipette. A voltage ramp from −75 to 75 mV of 250 ms duration was applied at pH 7.4. After activation with a solution containing the same external cation but buffered at pH 6.5, ramps were repeated four times for each ion. The currents obtained at pH 6.5 and 7.4 were subtracted to obtained ASIC1a specific currents and to correct for leaks. Only patches with ramps successfully measured with Na+, K+, and Cs+ were used in the calculations. (F) I-V relations of Q276G in the presence of 100 mM external Na+, K+ or Cs+. Each line represents the average of three independent patches.
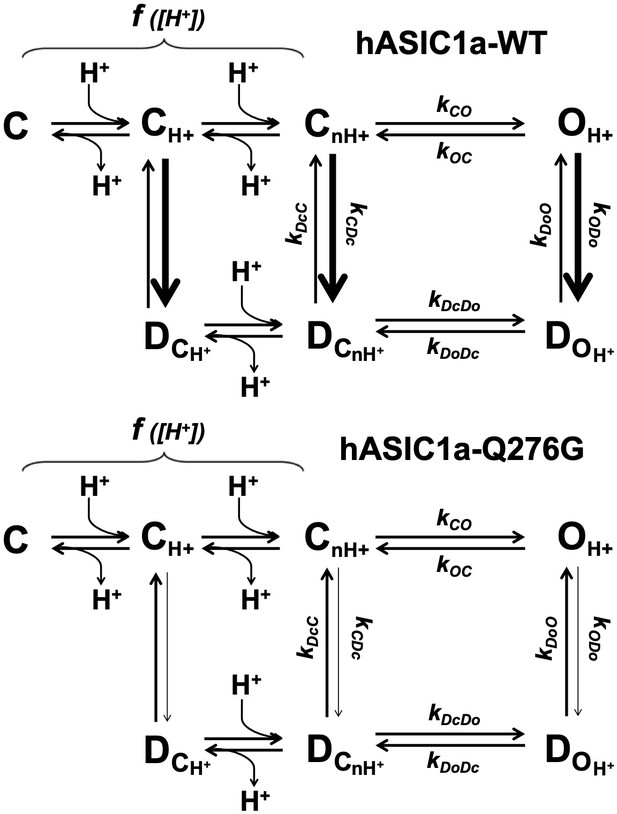
A kinetic scheme describing gating of channels that exhibit strong desensitization from the pre-open and open states.
C, O and D represent closed, open and desensitized states. C is in the resting state, CH+ are in partially activated states, and CnH+ is the opening-permissive state. In hASIC1a, the transitions between C states are H+ dependent, indicated by f[H+]. The wild-type ASIC1a open state completely desensitizes as indicated by the thick arrow that in this scheme means a high value for the rate constant kODo (transition from OH+ to DOH+). Pre-open channels (CH+) -those that make the most abundant population at low proton concentrations (pH range 7.2 to 7.0) when not all subunits within a trimer are protonated- and CnH+ undergo strong SSD (thick arrows). For Q276 mutants, specifically Q276G, the rate constants leading to desensitization –kODo and kCDc- are markedly reduced that is channels exhibit very long opening events and are resistant to SSD.
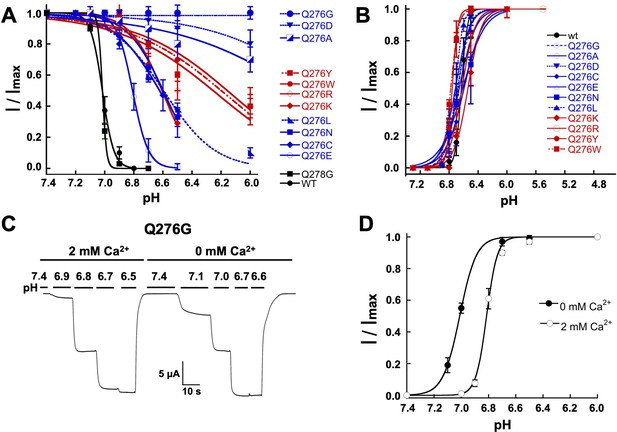
Mutations of Q276 diminish SSD.
(A) Values of apparent pHSSD and (B) apparent pH50A of substitutions in position 276. Curves represent fits of data points (n = 5–8 independent measurements) to the Hill equation. (C) Current traces of Q276G activated sequentially with solutions of the indicated pH (bars above the trace) in the presence of 2 mM Ca2+ and nominal 0 mM Ca2+. Pre-conditioning protons do not induce SSD in any of the two conditions. (D) The absence of Ca2+ in the activating solution increases the apparent affinity for protons from pH50A6.8 to 7.0.
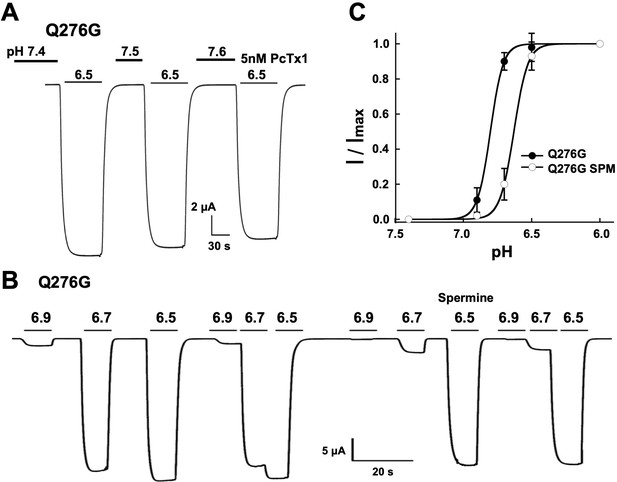
Q276G channels are insensitive to agents that modulate SSD.
(A) The tarantula toxin Pctx1 (5 nM) in preconditioning solutions of three different pH values does not change the Q276G response to pH 6.5. (B) 0.25 mM Spermine (SPM) in the bathing solution does not change the absence of SSD in Q276G but shifts the apparent pH50A from 6.8 to 6.5. (C) pH-dependence of Q276G activation in the presence of 0.25 mM spermine.
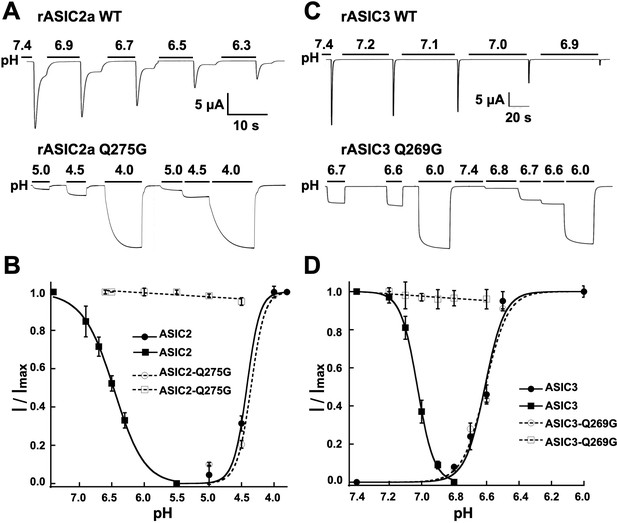
The equivalent residue Q276 in hASIC1a controls desensitization in rat ASIC2 and ASIC3.
(A) Wild-type rASIC2a currents activated by pH 4.0 undergo progressive desensitization as the preconditioning pH changes from 7.4 to 6.3, whereas mutant Q275G exhibits non-desensitizing currents that are insensitive to low preconditioning pH. (B) pH dependence of activation and SSD of wild-type ASIC2a and ASIC2a-Q275G. Error bars are ± SD, n = 5 cells. (C) and (D) similar experiments conducted with rat ASIC3 and ASIC3-Q269G except for the currents were activated with solution of pH 6.0.
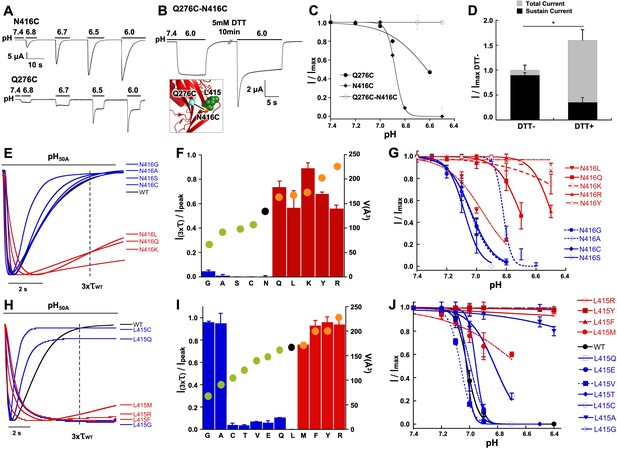
Functional and physical interactions of Q276 and N416 residues.
(A) Current traces of single cysteine mutants N416C and Q276C activated by increasing concentrations of protons. N416C channels completely desensitize, whereas Q276C exhibit a fraction of sustained current. (B) The double mutant Q276C-N416C exhibits only sustained currents. After treatment with DTT a desensitizing current appeared. Inset shows predicted locations of the two cysteines in the closed conformation and a putative disulfide bond as a line linking the residues. (C) Comparison of the pH dependence of SSD of single and double mutants shows the double mutant is insensitive to SSD. (D) Columns represent the fractions of sustained and peak currents before and after DTT treatment. Currents were normalized to the values prior to DTT; n = 15 cells. Asterisks indicate p values < 0.005. (E) Normalized currents of substitutions of N416 superimposed on the wild type (black trace). (F) Ratio of sustained/peak currents at time 3xτwt of N416 mutants ordered from low to large side chain volume indicated by the filled circles on the bars. Error bars are SD, n = 8 to 10 cells. (G) pH dependence of SSD of N416 mutants. Lines are the fit of data points (n = 5–8 independent measurements) to the Hill equation ± SD. (H) Normalized currents of substitutions of L415 superimposed on the wild type (black trace). (I) Ratio of sustained/peak currents at time 3xτwt of L415 mutants ordered from low to large side chain indicated by the filled circles on the bars. Error bars are SD, n = 8 to 10 cells. (J) pH dependence of SSD of L415 mutants. E data point is the mean ± SD of 5 to 8 independent cells.
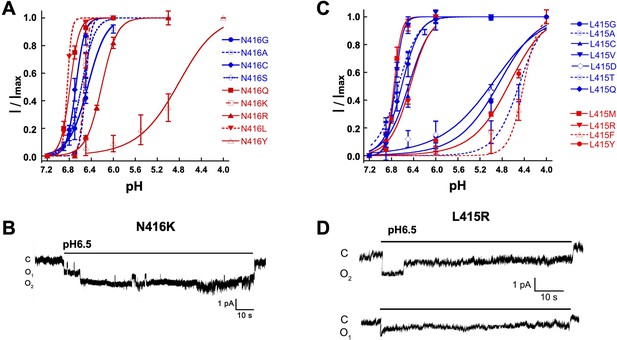
Mutations in the b11-b12 linker decrease sensitivity to protons and prolonged duration of openings.
(A) pH dependence of activation of N416 mutants. Some substitutions markedly decrease the apparent pH50A. (B) Representative example of an out-side patch expressing two N416R channels activated with external solution of pH 6.5. The recording shows uninterrupted long opening events of 10 to 30 s duration. (C) pH dependence of activation of L415 mutants. Similar as for N415 mutants, some substitutions in position L415 also display higher values of apparent pH50A than wild type channels. (D) Representative example of an out-side patch expressing a single L415R channel activated with external solution of pH 6.5. The recording shows few but very long opening events.
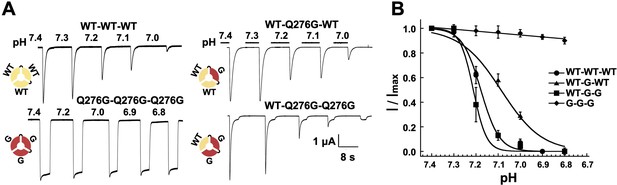
One WT subunit in ASIC1a trimer is sufficient to induce SSD.
(A) Current traces of four trimers containing three wild-type subunits (WT-WT-WT), one (WT-Q276G-WT) or two (WT-Q276G-Q276G) mutant subunits elicited by pH 6.5 exhibit SSD but not channels with three (Q276G-Q276G-Q276G). WT-Q276G-Q276G trimers exhibit a small component of current resistant to desensitization. (B) pH response to SSD of the four trimers shown in (A).
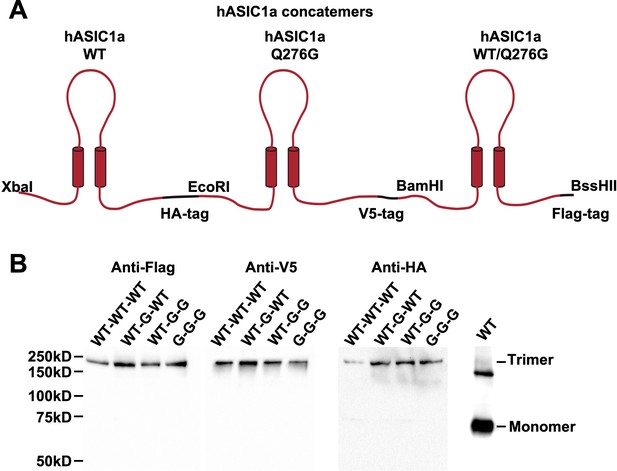
Schematic and expression of trimers containing wild type or mutant monomers (Q276G).
(A) Cartoon representation of hASIC1a trimers constructed by ligation of three subunits using unique restriction sites. Each subunit has the indicated tag at the carboxyteminus. (B) Western blot analysis of the four trimers using anti-Flag, anti-V5 and anti-HA monoclonal antibodies. WT monomer (~68 kDa) detected with anti-HA monoclonal. The upper band ~130 kDa corresponds to aggregated subunits owing to overloading the lane with protein.
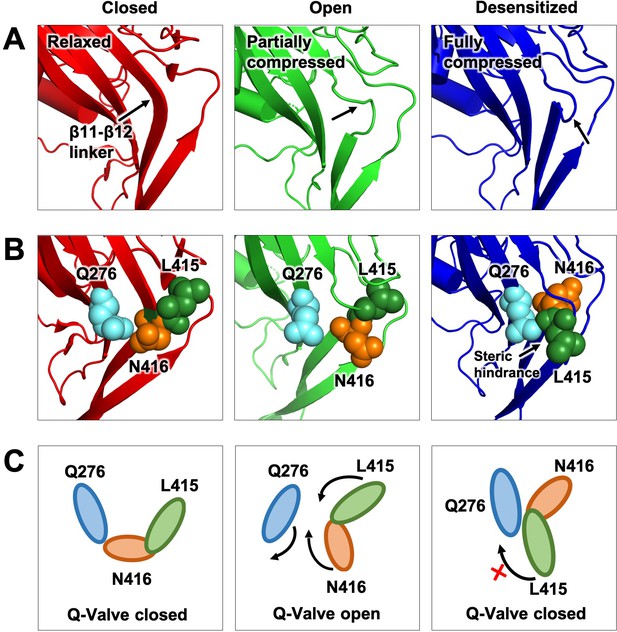
Proposed Q-valve mechanism to control of hASIC1a desensitization.
(A) Ribbon representation of the area containing the Q-valve: β9 and β11- β12 linker in the ECD of cASIC1 in Closed, Open and Desensitized conformations. The linker is compressed upon proton activation inducing 180o rotation of L415 and N416. Slight upward movement of β12 further increases the compression in the desensitized channel. (B) Side chains of Q276, L415 and N416 are shown as colored spheres. (C) Cartoon depicting proposed mechanism of the Q-valve: in the Closed state Q276 and L415 close the valve. In the Open state, the valve opens as protons induce a conformational change of the upper ECD that compresses the upper vestibule formed by β strands of the palm and the β11- β12 linker. Compression of the linker promotes an 180o rotation of N416 and L415 in opposite directions and facing Q276: trajectory indicated by the arrows. Red cross shows the valve closed and how Q276 could prevent backward rotation of L415, which in turn would relax the linker preventing full desensitization. Predicted intermediate states and direction of rotation are derived from functional analysis of substitutions in the three key residues making the valve.
From analysis of the functional data of mutant channels and cASIC1 structures it emerges a possible mechanism for desensitization wherein the central elements are three residues: Q276 located in the middle of β9 strand and L415 and N416 in the β11- β12 linker.
In the closed conformation, the β11- β12 linker is relaxed and the side chains of Q276 and N416 are in proximity keeping the Q-valve closed. Proton-induced activation separates the β strands of the lower palm and compresses the β11- β12 linker leading to opening of the Q-valve and enabling 180o rotation of L415 and N416 in opposite directions with their side chains facing Q276. We speculate that the conformational change produced by the full rotation of L415 and N416 propagates downward through the β12 strand to TM2 to close the gate of the pore. In the desensitized conformation Q276 blocks L415 from flipping back, it remains trapped in the desensitized state. This proposed mechanism explains why Q276G cannot close the valve because L415 can rotate back preventing desensitization. Conversely, when bulky side chains substitute Q276, rotation of N416 and L415 are hindered also preventing or slowing desensitization. Without full and sustained compression of the β11- β12 linker the gate remains open explaining the long mean open times of the mutants and absence of desensitization. This mechanism is also consistent with cross-linking of C276 with C416 or C415 both prevent rotation of residues in the β11- β12 linker leading to inhibition of desensitization.
Tables
Summary of calculated values of apparent pH50A of activation, pH50SSD steady-state desensitization and corresponding Hill coefficients: n Overlap of activation and desensitization currents in mutants with decreased desensitization prevented to determine pH50SSD values, they are reported as <pH 6.0 or ND.
Numbers are the mean ± SD of five to eight independent measurements.
| pH50a | N | pH50ssd | N | ||
|---|---|---|---|---|---|
| WT | 6.69 ± 0.21 | 3.5 | 7.09 ± 0.05 | 10 | |
| Q278G | 6.68 ± 0.18 | 3.4 | 7.07 ± 0.02 | 14 | |
| Q276 Mutants | |||||
| pH50a | n | pH50ssd | n | ||
| Q276G | 6.67 ± 0.15 | 3.5 | <6.00 | ND | |
| Q276A | 6.71 ± 0.12 | 4.2 | <6.00 | ND | |
| Q276D | 6.68 ± 0.31 | 4.7 | <6.00 | ND | |
| Q276C | 6.61 ± 0.21 | 3.9 | <6.60 | ND | |
| Q276E | 6.66 ± 0.18 | 4.2 | 6.80 ± 0.11 | 8 | |
| Q276N | 6.64 ± 0.23 | 3.4 | <6.60 | ND | |
| Q276L | 6.71 ± 0.13 | 3.8 | 6.6 ± 0.20 | 3 | |
| Q276K | 6.55 ± 0.32 | 3.7 | <6.60 | ND | |
| Q276R | 6.62 ± 0.23 | 4.1 | <6.30 | ND | |
| Q276Y | 6.75 ± 0.11 | 3.8 | <6.20 | ND | |
| Q276W | 6.72 ± 0.09 | 3.4 | <6.00 | ND | |
| L415 Mutants | |||||
| pH50a | n | pH50ssd | n | ||
| L415G | 4.88 ± 0.11 | 1.3 | <6.00 | ND | |
| L415A | 4.50 ± 0.20 | 2 | <6.00 | ND | |
| L415D | 5.00 ± 0.11 | 1 | <6.00 | ND | |
| L415C | 6.00 ± 0.10 | 2.1 | 7.02 ± 0.05 | 18 | |
| L415V | 6.70 ± 0.11 | 6 | 7.02 ± 0.07 | 15 | |
| L415E | 6.60 ± 0.10 | 3.1 | 6.90 ± 0.05 | 8.9 | |
| L415T | 6.68 ± 0.18 | 2.3 | 6.95 ± 0.04 | 15 | |
| L415Q | 6.61 ± 0.11 | 2.6 | 6.82 ± 0.03 | 4.8 | |
| L415M | 6.70 ± 0.14 | 6.3 | <6.00 | ND | |
| L415R | 6.46 ± 0.21 | 2.3 | <6.00 | ND | |
| L415F | 4.41 ± 0.12 | 3.7 | <6.00 | ND | |
| L415Y | 4.69 ± 0.22 | 1.5 | <6.00 | ND | |
| N416 Mutants | |||||
| pH50a | n | pH50ssd | n | ||
| N416G | 6.50 ± 0.02 | 3.6 | 7.04 ± 0.03 | 7 | |
| N416A | 6.50 ± 0.06 | 8 | 6.80 ± 0.01 | 6 | |
| N416C | 6.68 ± 0.05 | 5.3 | 7.03 ± 0.05 | 5.6 | |
| N416S | 6.58 ± 0.04 | 3.5 | 7.08 ± 0.01 | 7 | |
| N416Q | 6.79 ± 0.02 | 7.8 | 6.71 ± 0.06 | 5.6 | |
| N416L | 6.85 ± 0.03 | 5.2 | 6.96 ± 0.01 | 4 | |
| N416K | 6.60 ± 0.05 | 8 | >6.00 | ND | |
| N416R | 6.22 ± 0.04 | 5 | >6.00 | ND | |
| N416Y | 4.85 ± 0.03 | 1.8 | >6.00 | ND | |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Anti-V5 HRP Mouse monoclonal | ThermoFisher Scientific | 46–0708 | Dilution 1:2000 |
| Antibody | Anti FLAG M2 HRP Mouse monoclonal | Sigma | A8592-.2MG | Dilution 1:2000 |
| Antibody | HA-probe (F-7) Mouse monoclonal | Santa Cruz Biotechnology | sc-7392 | Dilution 1:2000 |
| Antibody | Mouse IgG Secondary Antibody HRP conjugate | ThermoFisher Scientific | A16072 | Dilution 1:10000 |
| Recombinant DNA reagent (DNA plasmid) | pcDNA3.1-hASIC1a | PMID: 23048040 | Cloned from HEK293T | |
| Recombinant DNA reagent (DNA plasmid) | pcDNA3.1-ratASIC2a | PMID: 11382806 | Cloned from rat dorsal root ganglia | |
| Recombinant DNA reagent (DNA plasmid) | pcDNA3.1-ratASIC3 | PMID: 11382806 | ||
| Recombinant DNA reagent (DNA plasmid) | pcDNA3.1- hASIC1aHA-hAS IC1aV5-hASIC1aFl | This paper | Expression plasmid wt hASICa trimer | |
| Recombinant DNA reagent (DNA plasmid) | pcDNA3.1- hASIC1aQ276GHA-hASIC1aQ276GV5-hASIC1aQ276GFl | This paper | Expression plasmid hASIC1aQ276G trimer | |
| Recombinant DNA reagent (DNA plasmid) | pcDNA3.1-hASIC1aHA-hASICQ276GV5-hASIC1aFl | This paper | Expression plasmid one mutant subunit Q276G | |
| Recombinant DNA reagent (DNA plasmid) | pcDNA3.1-hASIC1aHA-hASICQ276GV5-hASIC1aQ276GFl | This paper | Expression plasmid two mutant subunits Q276G | |
| Peptide, | Psalmotoxin-1 | Alomone labs | STP-200 | five nM |
| Commercial assay or kit | QuickChange Site-Directed Mutagenesis Kit | Agilent | 200523 | Mutagenesis |
| Commercial assay or kit | mMESSAGEmMACHINE T7 | ThermoFisher Scientific | AM1344 | Synthesis of cRNA |
| Commercial assay or kit | Pierce BCA Protein Assay Kit | ThermoFisher Scientific | CN: 23227 | Protein quantification |
| Chemical compound, drug | Spermine | SIGMA-ALDRICH | S3256-1G | 0.25 mM |
| Chemical compound, drug | Tris(2-carboxyethyl)phosphine hydrochloride | SIGMA-ALDRICH | C4706 | 5 mM |
| Software, algorithm | PyMOL | RRID:SCR_000305 | ||
| Software, algorithm | KaleidaGraph | RRID:SCR_014980 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.45851.016





