PI3K-Yap activity drives cortical gyrification and hydrocephalus in mice
Figures
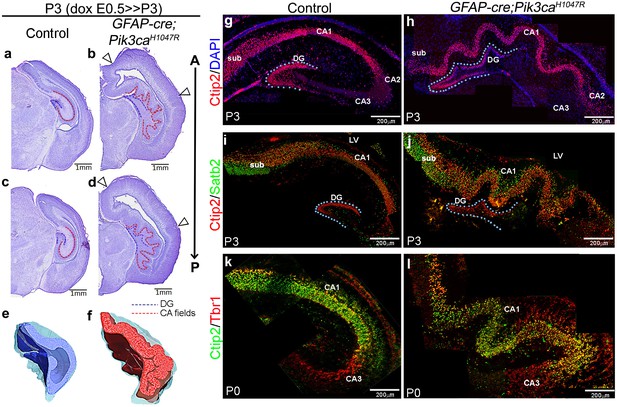
Embryonic induction of Pik3caH1047R activating mutation causes cortical gyrification in mice.
(a–d) Nissl-stained coronal sections of P3 control and GFAP-cre;Pik3caH1047R mutant hemi-forebrains revealed hippocampal (dotted lines) and neocortical gyrification (open arrowheads), when induced embryonically. (e,f) 3D models of control and mutant hippocampi. (g–l) Gross patterning of all hippocampal substructures was intact, as shown by Ctip2, Satb2 and Tbr1 expression. Medial gyrification was restricted primarily to the mutant CA1 region. CA – cornus ammonis (red dotted line, a–d); sub – subiculum; DG – dentate gyrus (blue dotted line, a-d; g–j). Scale bars: 1 mm (a–d), 100 μm (n–s). See also Figure 1—figure supplements 1–3.
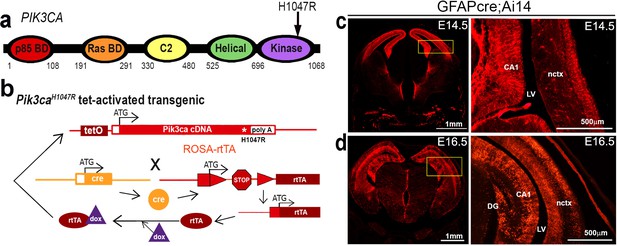
Genetic strategy for Pik3caH1047R mouse model and expression of GFAP-cre line.
(a) Schematic of PIK3CA functional domains, highlighting position of the H1047R activating mutation, modified from Roy et al. (2015). (b) Genetic strategy for tet-activated Pik3caH1047R transgenic mice (Liu et al., 2011): the human Pik3caH1047R mutation was activated in the combined presence of cre recombinase and doxycycline (dox). rtTA, reverse tetracycline-controlled trans-activator. (c,d) GFAP-cre expression, using Ai14 reporter line at E14.5 and E16.5, is stronger in medial telencephalon, compared to lateral telencephalon; boxes represent region of interest. Nctx, neocortex; LV, lateral ventricle, CA, cornus ammonis; DG, dentate gyrus. Scale bar: 500 μm, 1 mm (c).
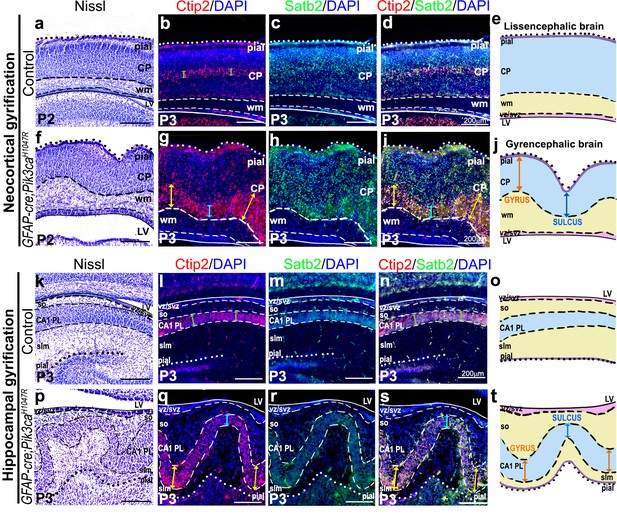
Characterization of true gyrification in Pik3caH1047R mutant neocortex and hippocampus.
(a–e, k–o) In control lissencephalic postnatal mouse brain, the ventricular (apical) and pial (basal) surfaces of both neocortex and hippocampus are smooth and parallel to each other, with no apparent difference in thickness within any intermediate cell layer or white matter (green arrows (b,d,l,n)). This is demonstrated using Nissl staining (a,k) and in schematics (e,j). Ctip2, Satb2 differentially mark the cortical plate (CP) and hippocampal pyramidal layer (CA1 PL) on either side of the lateral ventricle (LV). (f–j,p–t) Postnatal GFAP-cre;Pik3caH1047R mutant mouse brain demonstrated the criteria of true gyrification (Borrell, 2018), both in the lateral neocortex and in the hippocampal CA1, as demonstrated in schematics (j,t). Nissl staining and immunohistochemical analyses revealed folding of mutant pial surface, white matter (wm) and the neuronal layers (CP, CA1 PL) but predominantly smooth apical ventricular surfaces. The relative thickness of mutant gyri (folds distal to the ventricle; yellow/orange arrows) was greater compared to that of the adjacent mutant sulci (folds proximal to the ventricle; blue arrows) – property observed in normal gyrencephalic mammals. Point to be noted: the Pik3caH1047R mutant brain also demonstrated some extent of neuronal migration defect (this study; Roy et al., 2015). In all the panels, solid line marks ventricular edge; dashed lines distinguish progenitor zone, white matter and neuronal zone, while dotted line marks the pial edge. In schematics (e,j,o,t), pial surface is purple, neuronal plate is blue, white matter is light yellow, ventricular-subventricular zone is pink. slm – stratum lacunosum-moleculare, so – stratum oriens. Scale bars: 200 μm (a–d,f–I,k–n,p–s).
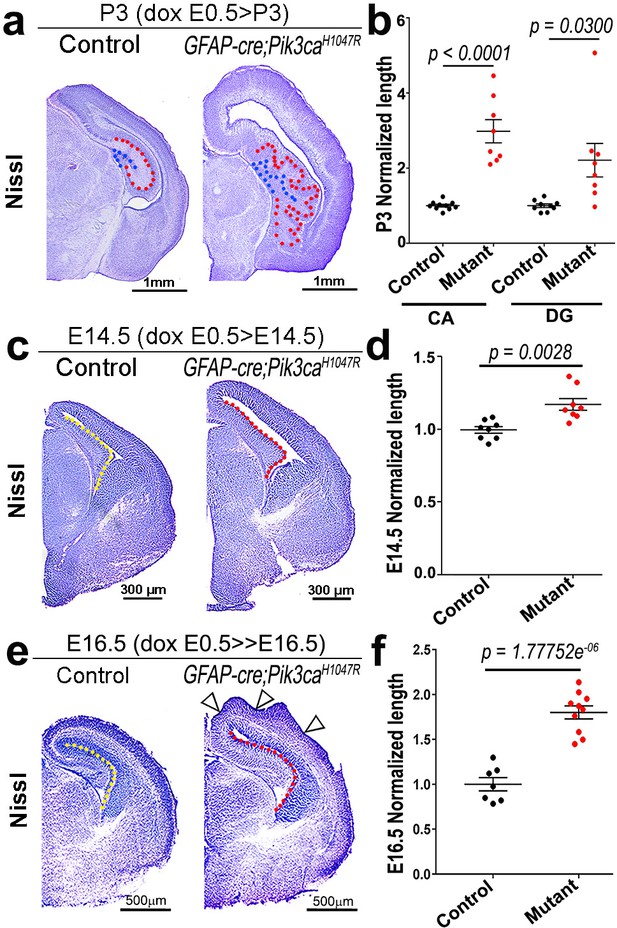
Pik3caH1047R mutant demonstrates increase in ventricular length.
(a,c,e) Nissl-stained coronal sections of P3, E14.5 and E16.5 control and Pik3caH1047R mutant forebrains, induced by doxycycline from E0.5. (b) Normalized lengths of P3 CA1 and dentate gyrus (DG) measured as shown by red and blue dotted lines respectively in (a), were significantly longer in the mutant, compared to control (CA1: t = 6.372, df = 7.197; DG: t = 2.698, df = 7.165). (d,f) Normalized length of the medial ventricular lining, measured as shown in (c,e) by dotted lines in the control (yellow dotted line) and mutant (red dotted line) embryos, was significantly longer in the mutant, compared to control at both E14.5 (t = 4.065, df = 11.16) and E16.5 (t = 8.267, df = 14.4). Open arrowheads marked the occurrence of cortical folds in E16.5 mutant forebrain. Data are represented as mean ± SEM in scatter plots; 2-tailed unpaired t-tests were performed (b,d,f). Scale bars: 1 mm (a), 300 μm (c), 500 μm (e).
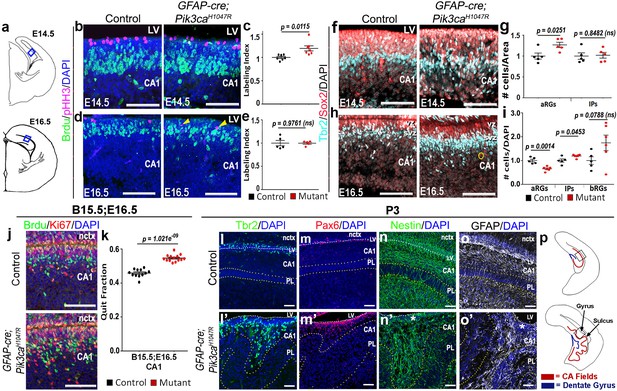
Altered neurogenesis in Pik3caH1047R mutant at embryonic and postnatal stages follows the stereotypic cortical ‘gyrification sequence’.
(a) Schematics of E14.5 and E16.5 coronal hemi-section, boxes represent region of interest (b,d,f,h,j). Images are oriented with lateral neocortex on the top, followed by lateral ventricle (LV) and then CA1 (b,d,f,h,j,l–o’). (b–e) Compared to control, E14.5 Pik3caH1047R mutants exhibited significantly higher labeling index, indicating higher proliferation rate. Though E16.5 mutant CA1 showed no overt change in labeling index, pHH3+ cells marking the M-phase appeared to be ectopic (yellow arrowheads), indicating early mis-localization of dividing cells. (f–i) Compared with respective controls, E14.5 and E16.5 GFAP-cre;Pik3caH1047R mutants, induced from E0.5, showed significant increase in primary (apical Sox2+Tbr2-, aRGs) and secondary (Tbr2+ IPs; basal Sox2+Tbr2-, bRGs) progenitor pools, in accordance with the gyrification sequence; yellow circle marks bRG. Data are normalized and represented as mean ± SEM in scatter plots; 2-tailed unpaired t-tests were performed (c,e,g,i). (j,k) Cell cycle exit during the E15.5-E16.5 period was significantly higher in the mutant CA1 than the control, as represented in mean ± SEM scatter plots (t = 10.16, degrees of freedom (df) = 21.73). (l–m’) Compared to controls, P3 mutant CA1 region showed focal increase in Pax6+ and Tbr2+ progenitors at the gyral ventricular-subventricular zone; the apical edge remained predominantly unfolded. (n–o’) Nestin+ and GFAP+ intermediate filaments in P3 were misoriented and divergent from focal points (asterisks) at the mutant gyri. (p) Schematics of P3 control and mutant hemi-sections; boxes depict regions shown in l-o’. Differences were considered significant at p<0.05; ns, not significant. nctx, neocortex; PL, pyramidal layer. Scale bars: 50 μm (b,d,f,h,j), 100 μm (l–o’). See also Figure 2—figure supplement 1.
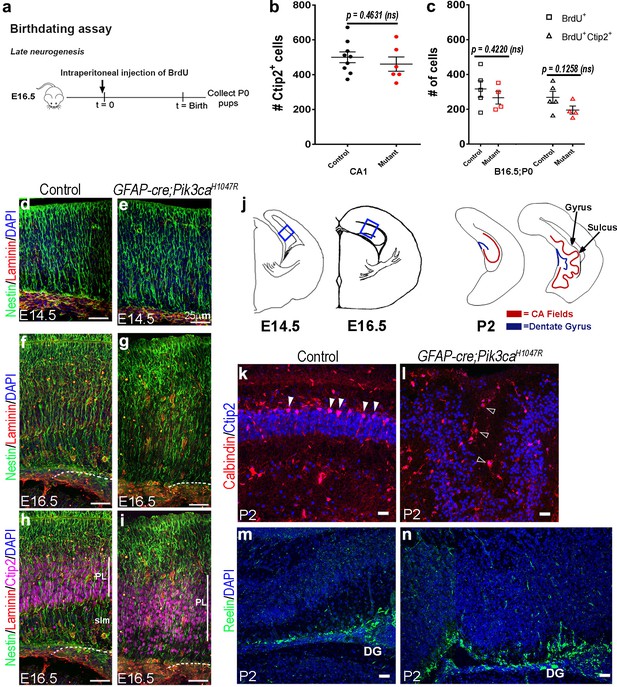
Pik3caH1047R mutant demonstrates normal cell fate specification but disrupted neural scaffold and migration.
(a) Experimental outline of birth-dating assay: BrdU was injected at E16.5 and analyzed at P0 (B16.5;P0). (b,c) At P0, total numbers of Ctip2+ cells, of cells born at E16.5 (BrdU+), and of Ctip2+ cells generated at E16.5 (Ctip2+/BrdU+; d) were not significantly different between respective controls and Pik3caH1047R mutant CA1 regions, indicating no effect of PI3K overactivation on cell fate specification in CA1. Data are represented as mean ± SEM in scatter plots; 2-tailed unpaired t-tests were performed (Ctip2+: t = 0.7623, df = 10.24; BrdU+: t = 0.8534, df = 6.91; BrdU+Ctip2+: t = 1.752, df = 6.61; b–d). Differences were considered significant at p<0.05; ns, not significant. (e–j) Nestin+ radial glial fibers appeared slightly irregular in E14.5 Pik3caH1047R mutant (f) but became progressively hyper-fasciculated and dysplastic morphology at E16.5 (d,j) compared to respective controls (e,g,i). The Laminin+ mutant basement membrane was thinner at E14.5 and thicker at E16.5, compared to respective controls. Also, E16.5 mutant PL was widely dispersed and showed severe reduction of the stratum lacunosum-moleculare (slm; j). (k) Schematics of E14.5, E16.5 (boxes marked regions shown in (e–j)) and P2 hemi-sections. (l,m) Calbindin+ cells that normally aligned along the ventricular side of P2 control CA1 PL (arrowheads), were severely dispersed (open arrowheads), clustering around the white matter of mutant gyral area. (n,o) Compared to control, P2 mutant displayed more Reelin+ cells along the gyrified neural tissue, but none were found in ectopic locations. Scale bars: 25 μm (e–j, i–o).
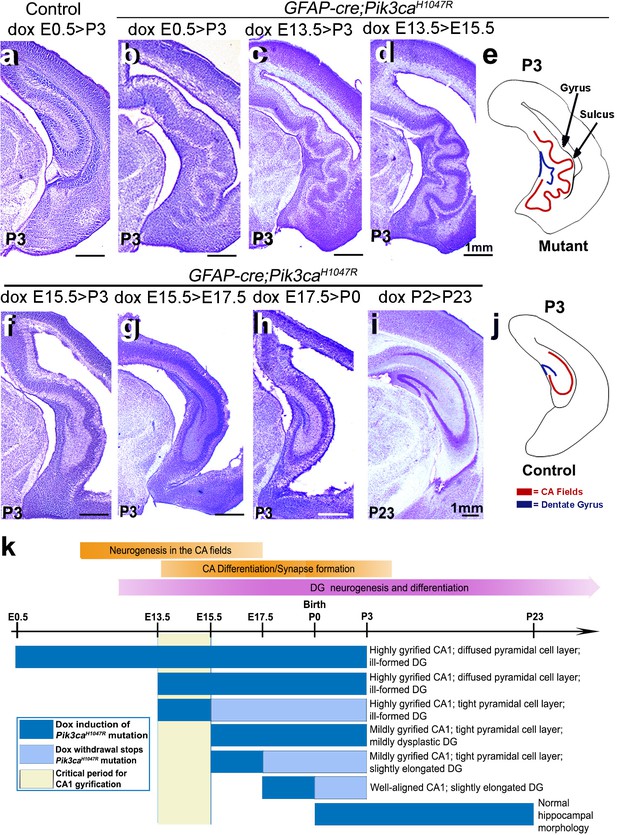
Non-random gyrification pattern in Pik3caH1047R mutant has a narrow embryonic critical period.
(a–j) Nissl-stained coronal hemi-sections depicting hippocampal morphology, alongside schematics (e,j). Compared to control (a), Pik3caH1047R mutant hippocampus showed graded severity in gyrification, depending on the time and duration of mutation induction. The critical period of the most severe folding with a non-random folding pattern ends at E15.5 (b–d). Postnatal induction of Pik3caH1047R mutation is not effective to cause cortical gyrification (i). (k) Developmental timeline of mutation induction and the corresponding hippocampal gyrification phenotype. Scale bars: 1 mm (a–d, f–i). See also Figure 3—figure supplement 1.
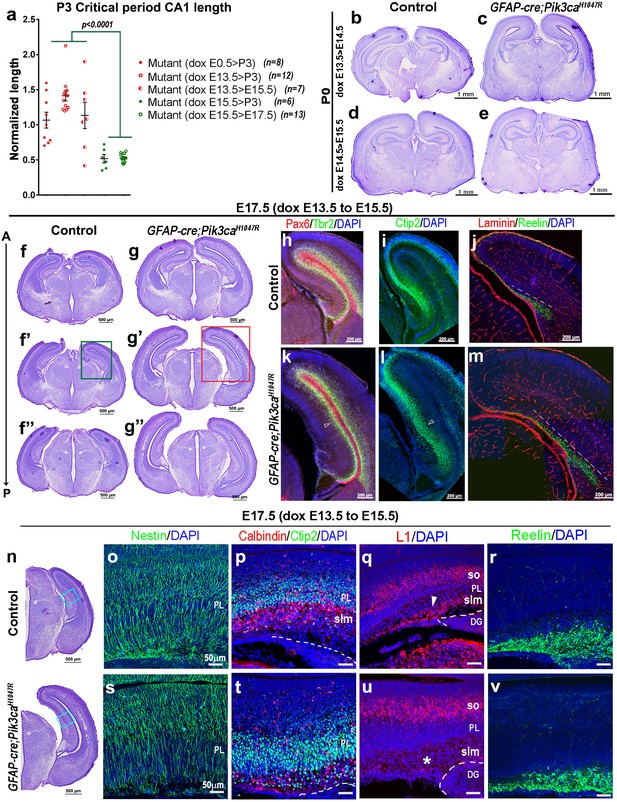
Minimal induction of PI3K overactivation is sufficient to initiate cortical folding and altered neuronal migration in E17.5 CA1.
(a) Graph showing P3 CA1 length across mutants induced in graded fashion, normalized to that of the mutant induced from E0.5. Mutants induced after E15.5 showed significantly lower CA1 lengths compared to those induced before E15.5. Data are represented as mean ± SEM in scatter plots; one-way ANOVA was followed by Tukey’s multiple comparison tests (F-value = 20.57, df = 42). (b–e) Nissl-stained coronal sections of P0 Pik3caH1047R mutants (doxycycline: E13.5 > E14.5; E14.5 > E15.5) demonstrated mild cortical dysplasia and ventriculomegaly but little gyrification, compared to respective control littermates. (f–v) E17.5 mutant, minimally induced from E13.5 to E15.5, demonstrated the first visual sign of folding, as shown in Nissl-stained coronal sections at different antero-posterior (A–P) planes (f–g”,n) and when processed for Pax6, Tbr2 (h,k), Ctip2 (i,l). Region of the first undulation is marked by open arrowheads (k,l). Reelin-Laminin immunohistochemistry indicated a slightly thicker but not overtly disrupted basement membrane and a longer Reelin+ zone (dashed lines); however, no ectopic expression of Reelin in the mutant hippocampus was observed (j,m,r,v). Compared to controls (o,p), E17.5 Pik3caH1047R mutant (dox: E13.5 > E15.5) showed hyper-fasciculated Nestin+ radial glial scaffold (s), thicker dispersed Ctip2+ PL with ectopic Calbindin+ cells (t). Mutant displayed severely reduced stratum lacunosum-moleculare (slm) (asterisk; u), marked by L1 in the control section (arrowhead; q). The mutant stratum oriens (so) appeared normal. DG – dentate gyrus. Scale bars: 1 mm (b–e), 500 μm (f–g”,n), 200 μm (h–m), 50 μm (o–v).
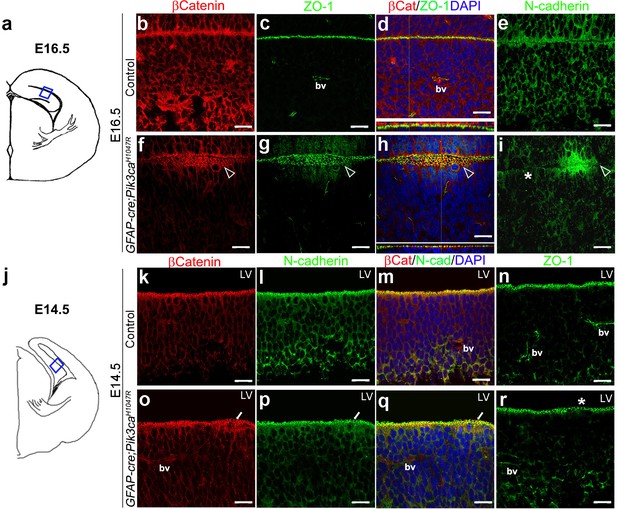
Embryonic induction of Pik3caH1047R mutation causes early disruption of apical cell adhesion.
(a,j) Schematic of E14.5 and E16.5 hemi-section showing area of interest. (b–i) Compared to control, E16.5 Pik3caH1047R mutant, induced from E0.5, showed abnormal zippering of forebrain apical membranes (open arrowheads, (f–h), with XZ plane indicating subtle mis-localization of cell adhesion molecules β-Catenin and ZO-1 (d,h). N-Cadherin, that is normally expressed uniformly at the juxtaposition of the apical membranes (e), appeared to be clustered around the unzippered portion and sporadically absent (asterisk) in adjacent parts of the membrane junction (i). (k–r) Compared to control, CA1 ventricular lining of E14.5 mutant showed subtle disruption in the localization of β-Catenin, N-Cadherin, ZO1, as marked by white arrows (o–q) and asterisk (r). bv – blood vessels. Scalebars: 20 μm (b–i,k–r). See also Figure 4—figure supplement 1.
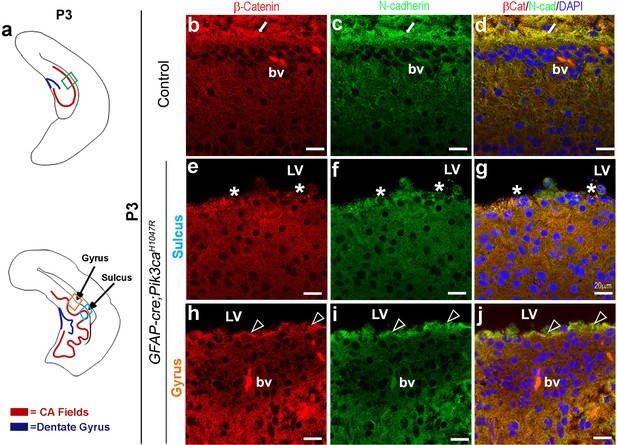
Disruption of apical cell adhesion caused by embryonic induction of PI3K overactivation persists postnatally.
(a) Schematics of P3 control and Pik3caH1047R mutant; boxed areas show areas of interest. (b–d) P3 control sections showed juxtaposition of the neocortical and hippocampal apical membranes across lateral ventricles, with uniform expression and colocalization of β-Catenin and N-Cadherin (white arrows). (e–j) Comparatively, P3 mutant sulcus showed focal absence of both β-Catenin and N-Cadherin (asterisk, (e–g), while the mutant gyrus demonstrated scattered areas of colocalization (open arrowheads) interspersed by solo expression of N-Cadherin and ectopic presence of cells at the non-zippered edge (h–j). Please note ventriculomegaly was highly pronounced in P3 mutant (e–j). Scale bars: 20 μm (b–j).
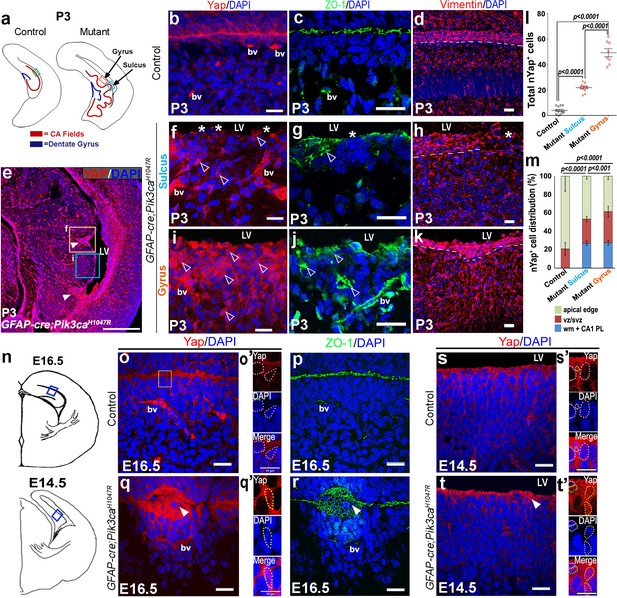
Embryonic induction of Pik3caH1047R mutation disrupts early developing ependyma and induces increase in nYap+ cells.
(a,n) Schematics of P3, E16.5 and E14.5 hemi-sections; boxed regions depict areas of interest. (b-d,o,o’p,s,s’) Ependymal markers Yap and Vimentin, and junction protein ZO-1 are localized at the apical lining of P3, E16.5 and E14.5 control CA1 respectively. Additionally, Yap and ZO-1 mark the blood vessel membranes (bv), while Vimentin mark the hippocampal radial glia. (e) P0 mutant CA1 gyral ventricular lining showed presence of focal nYap-enriched zones (arrowheads, orange box), interspersed by Yap-sparse zones at the sulci (cyan box). (f,i) Increased Yap+ nuclei in P3 mutant gyri (open arrowheads) and Yap-sparse mutant sulcus edge (asterisks) indicated disrupted ependyma. (g,j) ZO-1 was ectopically expressed in P3 mutant gyral ventricular zone, with breaks in the sulcus areas. (h,k) Vimentin+ fibers appeared to be arranged in an hour-clock pattern at the gyri; sulci demonstrated more disoriented fibers and gaps (asterisk); dashed lines mark the basal ependymal edge. (l,m) Total number and distribution of nYap+ cells in the different CA1 subzones (binned as mono-layer apical edge, ventricular-subventricular zone (vz/svz), white matter (wm) and CA1 pyramidal layer (CA1 PL)) were significantly enhanced in P3 mutant, compared to control littermates (total counts: F = 179.1, df = 28; cell distribution: F = 137.8, df = 86). The control CA1 showed minimal existence of nYap+ cells beyond the vz/svz. Data are represented as mean ± SEM in scatter plots (l) or 100% stacked columns (m); one-way and two-way ANOVA were performed respectively. (o–q’) Compared with control, E16.5 Pik3caH1047R mutant showed abnormal zippering of forebrain apical membranes, combined with focal nYap expression (arrowheads). (s–t’) Compared with control, E14.5 mutant medial apical membrane showed subtle disruption of cell adhesion causing minor buckling/unevenness. Magnified images of E14.5 and E16.5 ventricular edge demonstrated higher number of nYap+ cells in the mutant (yellow dotted lines) compared to controls (o’,q’,s’,t’); some non-nYap+ cells are marked with pink dotted lines for the purpose of comparison (o’,s’,t’). Scale bars: 20 μm (b–d, f–k, o–t), 500 μm (e), 10 μm (o’,q’,s’,t’). See also Figure 5—figure supplement 1.
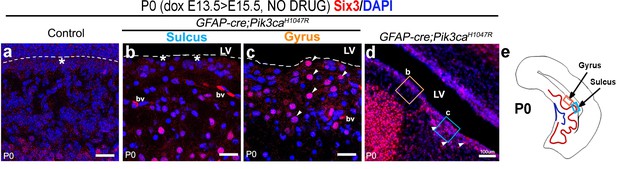
Six3+ cells ectopically concentrate at the mutant gyral ventricular zone.
(a) Control CA1 ventricular edge did not robustly express ependymal marker Six3 at P0, although normal Six3 expression in the postmitotic layer was observed (data not shown). (b–d) P0 mutant gyri showed concentration of Six3+ nuclei (arrowheads), similar to the mutant nYAP expression (see Figure 5), while they are scattered in the sulci lacking in the ventricular zone (asterisks). (e) Schematic of P0 mutant showing location of gyrus and sulcus. bv – blood vessels; LV – lateral ventricle. Scale bars: 20 μm (a–c), 100 μm (d).
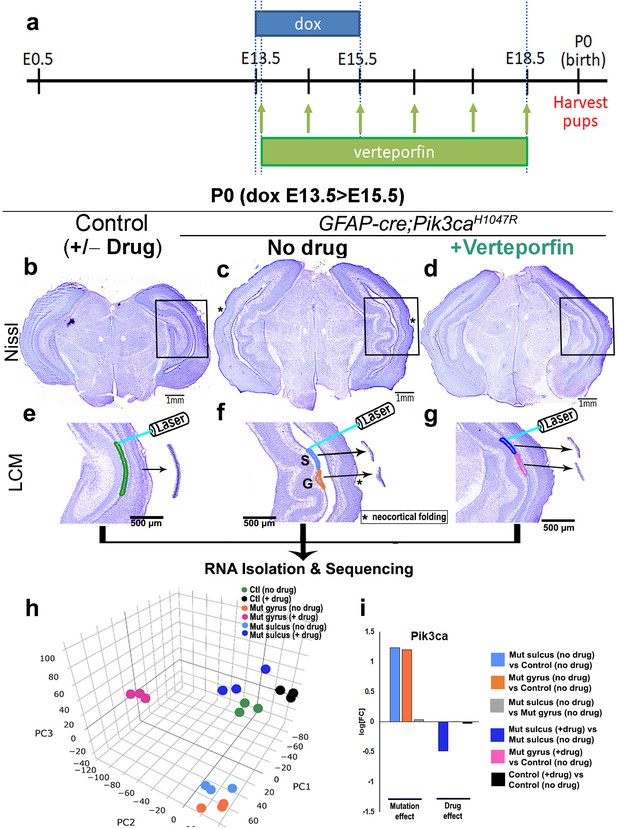
Translocation of Yap from nucleus to cytoplasm by verteporfin attenuates gyrification and ventriculomegaly in Pik3caH1047R mutant.
(a) Developmental timeline of verteporfin administration (E13.5 > E18.5). (b–d) Nissl-staining revealed attenuation of gyrification and absence of ventriculomegaly in P0 Pik3caH1047R mutant (induced E13.5 > E15.5) post-verteporfin administration. Boxes mark the respective areas of interest for (e–g). (e–g) Flowchart of laser capture microdissection (LCM) of marked ventricular-subventricular zone tissue samples, and RNA sequencing. Asterisks (c,f) mark neocortical gyrification in the untreated mutant brains, that were also eventually attenuated by verteporfin (d,g). (h) Principal component (PC) analysis revealed that verteporfin administration drives ventricular Pik3ca mutant gene expression towards that of control tissue. PC1 is largely explained by PI3K signaling, PC2 by Hippo-Yap signaling. (i) Graphs showing the effect of PI3K overactivation and of verteporfin treatment in Pik3ca. Scale bars: 1 mm (b–d), 500 μm (e–g). See also Figure 6—figure supplements 1–3, Figure 6—source datas 1 and 2.
-
Figure 6—source data 1
Significant gene list and differential expression analysis from P0 Pik3caH1047R mouse RNA-seq data.
A truncated gene list is shown based on selected parameters (logFC >0.263, FDR < 0.05, true for at least one biological pair comparison).
- https://doi.org/10.7554/eLife.45961.020
-
Figure 6—source data 2
Gene lists used in gene set enrichment analyses.
- https://doi.org/10.7554/eLife.45961.021
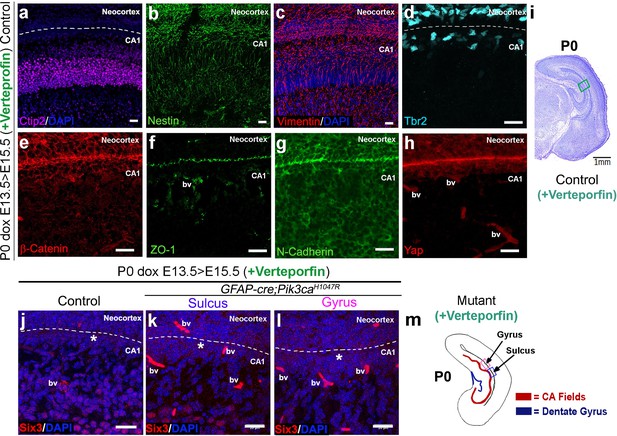
Verteporfin administration has no overt effect on control littermates.
(a–j). Verteporfin treatment did not overtly affect the normal expression of Ctip2, radial glial markers Nestin and Vimentin, intermediate progenitor marker Tbr2, cell adhesion molecules like β-Catenin, ZO-1, N-Cadherin, developing ependymal markers Yap and Six3, as well as the overall brain morphology. Box in (i) depicts area of interest. (k,l) Verteporfin treatment also did not overtly affect the Six3 expression in the developing ependyma of P0 control CA1 and mutant sulcus; however, it considerably reduced the focal concentration of Six3+ cells at the mutant gyral ventricular zone (asterisks). (m) Schematic of P0 mutant (verteporfin treated) showing location of gyrus and sulcus. bv – blood vessels. Scale bars: 20 μm (a–h, j–l), 1 mm (i).
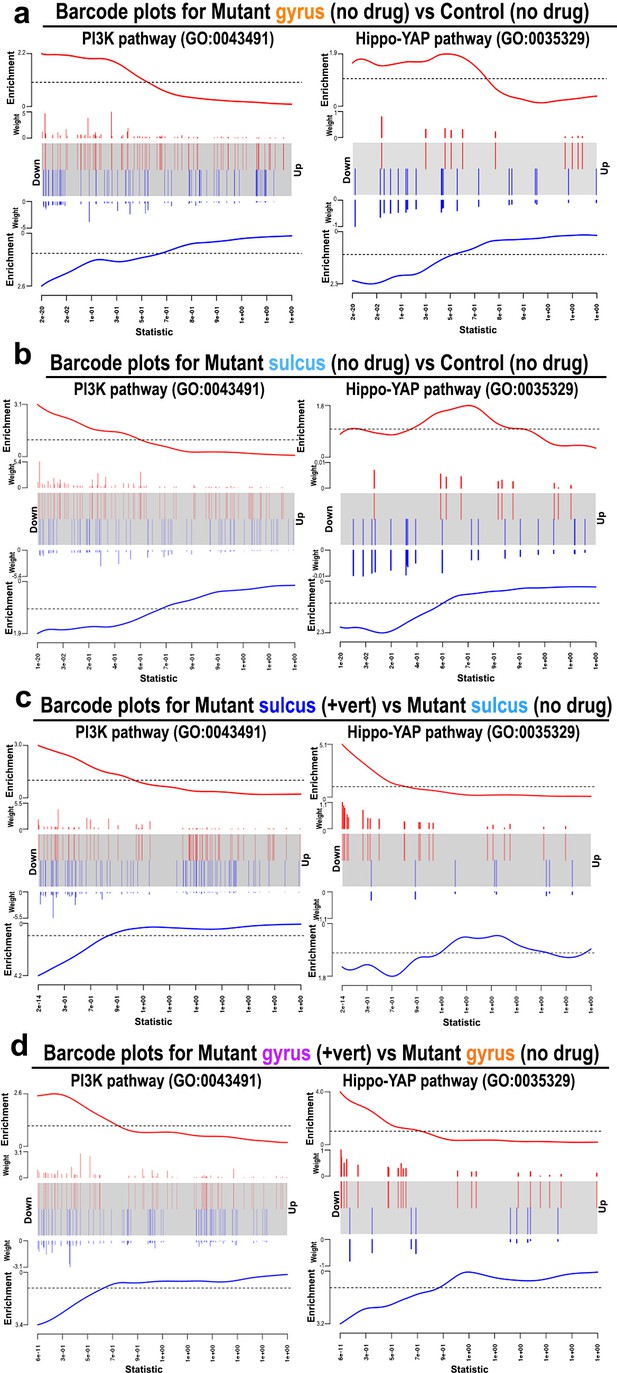
Barcode plots across groups showing gene enrichment.
(a–d) Barcode plots showing gene enrichment of PI3K pathway and Hippo-YAP pathway for the following comparisons: mutant sulcus (no drug) versus control (no drug; pPI3K=2.02e−20, pYap = 2.35e−18), mutant gyrus (no drug) versus control (no drug; pPI3K=4.92e−20, pYap = 3.33e−18), mutant sulcus (+verteporfin) versus mutant sulcus (no drug; pPI3K=7.74e−19, pYap = 2.05e−17), and mutant gyrus (+verteporfin) versus mutant gyrus (no drug; pPI3K=1.96e−19, pYap = 5.67e−18). The central portion of the plot shows the ranked list of all genes, ranked by p-value, with the smallest p-values to the left and the largest to the right. Genes in the set are highlighted with colored bars: red and blue bars mark upregulated and downregulated genes respectively. The ‘weights’ show the logarithmic fold change between the two groups, giving a measure of the effect size, and the outer plots (labeled ‘Enrichment’) show the relative enrichment of the genes in the set. Thus, the enrichment plot highlights those genes that provide evidence that the whole gene set is being differentially expressed.
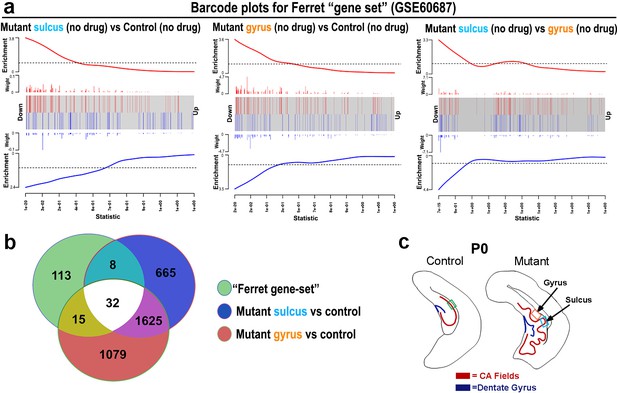
Pik3ca mutant gyrification is physiologically relevant.
(a) Barcode plots of a significant set of gyrification-related ferret genes (GSE60687), mapped to mouse, for the following comparisons: mutant sulcus versus control (no drug; p=9.65e−21); mutant gyrus versus control (no drug; p=1.57e−21), mutant sulcus versus gyrus (no drug; p=1.64e−19). The plots indicate that top genes mapped between the ferret data and our mouse data are highly affected in our dataset. (b) Venn diagram indicates that genes relevant to normal ferret gyrification have significant overlap with those differentially expressed in the mouse mutant gyrification zones. (c) Schematics of P0 control and mutant hemi-sections, indicating regions of interest.
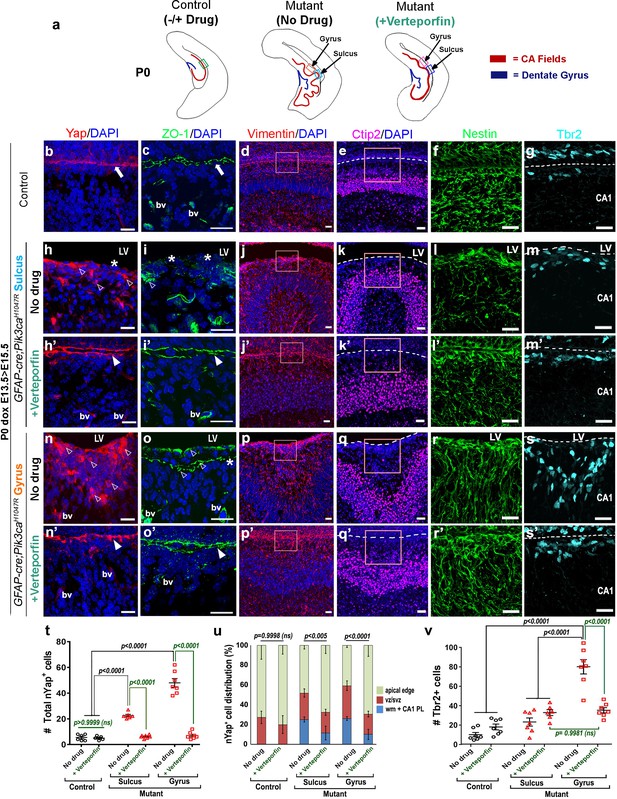
Reduction of nYap by verteporfin re-establishes developing apical junctions and suppresses focal proliferation in Pik3caH1047R mutant.
(a) Schematics of P0 control and mutant (±verteporfin) forebrain hemi-sections; boxes depict respective areas of interest. (b–g) P0 control untreated brain expressed cYap, adherens junction protein ZO-1 and radial glial markers Vimentin and Nestin, uniformly along the juxtaposed apical membranes. Ctip2 and Tbr2 were expressed in pyramidal neurons and IPs respectively. (h–s’) Verteporfin treatment significantly normalized Yap localization, from nucleus to cytoplasm, restored apical junctions and streamlined disrupted radial glial scaffold, as compared to the untreated P0 (dox E13.5 > E15.5) mutant gyri and sulci. (t,u) The treatment significantly reduced the total nYap+ cell number in the mutant hippocampal gyri and sulci (F = 70.47, df = 32) as well as normalized the nYap+ cell distribution to the control levels (F = 134.4, df = 96). This also significantly decreased the Tbr2+ progenitor population (F = 31.45, df = 35), especially in the mutant gyral zones, as quantitated in (v). This resulted in attenuation of the extent of gyrification and re-zippering of the apical membranes to halt the progressive ventriculomegaly. Data are represented as mean ± SEM in scatter plots (t); two-way ANOVA was followed by Tukey’s post-tests. ns, not significant. Open arrowheads, disrupted/ectopic junctional and ependymal proteins in mutant; asterisks, absence of adhesion/ependymal molecules at the mutant apical edge; arrowheads, rescued apical ventricular lining in the mutant post-verteporfin treatment; bv, blood vessels; LV, lateral ventricle. Scalebars: 20 μm (b–s’). See also Figure 7—figure supplements 1–2.
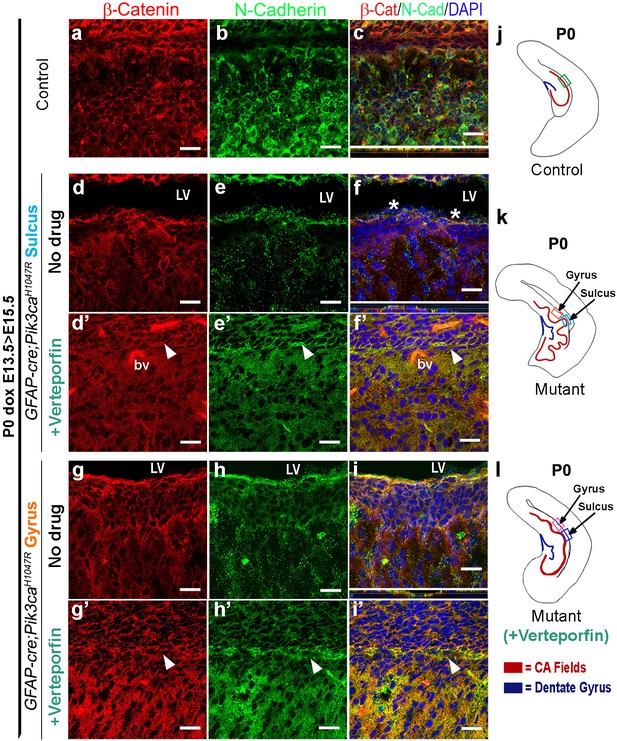
Reduction of nYap by verteporfin re-establishes cell adhesion in Pik3caH1047R mutant.
(a–c) Cell adhesion molecules, namely β-Catenin, N-Cadherin, were uniformly expressed specifically along the juxtaposed apical membranes in the P0 (dox E13.5 > E15.5) control brains. (d–i’) Verteporfin treatment restored the disrupted apical cell adhesion, observed in the untreated P0 (dox E13.5 > E15.5) mutant gyri and sulci. This resulted in attenuation of the extent of gyrification and re-zippering of the apical membranes to halt the progressive ventriculomegaly. (j–l) Schematics of P0 control and mutant (±Verteporfin) forebrain hemi-sections; boxes depict respective areas of interest. Asterisks, absence of adhesion/ependymal molecules at the mutant apical edge; arrowheads, rescued apical ventricular lining in the mutant post-verteporfin treatment; bv, blood vessels; LV, lateral ventricle. Scale bars: 20 μm (a–i’).
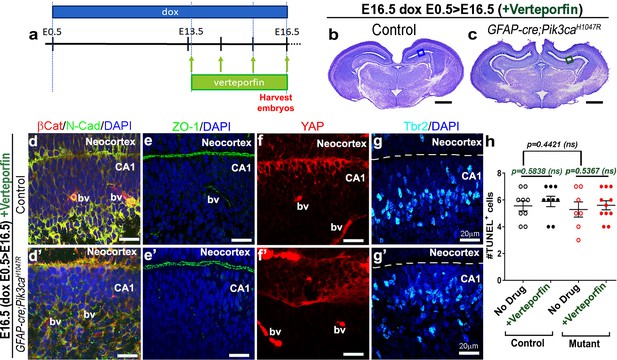
Effect of verteporfin on apical junctions initiates early.
(a) Developmental timeline of verteporfin administration (E13.5 > E16.5). (b,c) Nissl-stained coronal sections of E16.5 Pik3caH1047R mutant and control (induced E13.5 > E15.5), post-verteporfin administration, revealed similar morphology. Boxes mark the respective areas of interest. (d–g’) Verteporfin-treated mutants demonstrated increased alignment of junctional proteins, normalized progenitor pool and reduced occurrence of unzippered apical membranes (compare with Figure 4). Dashed lines mark the apical ventricular lining. (h) Study of apoptosis revealed no significant differences (F = 0.3318, df = 32) in the number of TUNEL+ cells between E16.5 control and mutant (±Verteporfin) CA1 regions. Data is represented as mean ± SEM in scatter plots; one-way ANOVA was performed followed by Tukey post-test. bv, blood vessels; dashed line, lateral ventricle. Scalebars: 500 μm (b,c), 20 μm (d–g’).
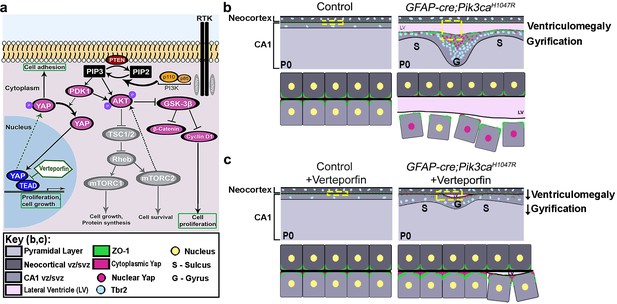
Summary: PI3K-Yap gyrification model.
(a) Schematic of PI3K-AKT-MTOR pathway, its connection with Yap signaling and mode of verteporfin action. The activity of the PI3K enzyme (dimer of catalytic and regulatory subunits) initiates the PI3K-AKT-MTOR pathway cascade. In parallel, downstream of the Hippo pathway, Yap binds to transcriptional regulator proteins in the nucleus, to facilitate cell growth and proliferation. Activation of Hippo pathway results in translocation of nuclear Yap to cytoplasm through phosphorylation. Cytoplasmic Yap, in turn, promotes cell adhesion. The activating Pik3ca mutation results in Yap translocation from cytoplasm to nucleus, possibly via Pdk1. In this study, verteporfin, a nuclear Yap inhibitor, restored cytoplasmic Yap. (b,c) Schematics summarizing main findings of the paper: PI3K over-activation resulted in disruption of cell adhesion at the neural-ependymal transition zone causing ventriculomegaly, and also in differential proliferation of progenitors, thus triggering the gyrification sequence cascade (effect of genotype). In mutant mice, verteporfin attenuated these anomalies (effect of drug), leading to reduction in gyrification severity as well as ventriculomegaly. Area within the yellow dashed boxes in the top rows of (b) and (c) is magnified in the respective bottom rows. Color coding for elements in (b) and (c) is explained in the figure ‘key’. vz/svz, ventricular/subventricular zone.
Videos
3D model of P3 control hippocampus.
https://doi.org/10.7554/eLife.45961.0063D model of P3 GFAP-cre;Pik3caH1047R hippocampus.
https://doi.org/10.7554/eLife.45961.007Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Mus musculus) | GFAP-cre | gift (JJ Zhao); PMID: 11668683 | (IMSR Cat# JAX:004600, RRID:IMSR_JAX:004600) | gifted by Dr. Jean J Zhao (Dana Farber Cancer Inst., Boston, USA) |
| Genetic reagent (Mus musculus) | Pik3caH1047R | gift (JJ Zhao); PMID: 21822287 | MGI: 5526971 | gifted by Dr. Jean J Zhao (Dana Farber Cancer Inst., Boston, USA) |
| Genetic reagent (Mus musculus) | Ai14/+; Ai14 | Jax labs (stock #007914), PMID:20023653 | (IMSR Cat# JAX:007914, RRID:IMSR_JAX:007914) | PMID: 20023653 |
| Genetic reagent (Mus musculus) | Rosa26-rtTA; Rosa | PMID: 15784609 | (IMSR Cat# JAX:005670, RRID:IMSR_JAX:005670) | gifted by Dr. Jean J Zhao (Dana Farber Cancer Inst., Boston, USA) |
| Antibody | Anti-BrdU antibody [BU1/75 (ICR1)] | Abcam | (Abcam Cat# ab6326, RRID:AB_305426) | IHC (1:100) |
| Antibody | Anti-BrdU-Fluorescein antibody Formalin grade | Roche | (Roche Cat# 11202693001, RRID:AB_514484) | IHC (1:100) |
| Antibody | Rabbit anti Calbindin D-28k | Swant | (Swant Cat# CB38, RRID:AB_2721225) | IHC (1:1500) |
| Antibody | Anti-Ctip2 antibody [25B6] - ChIP Grade | Abcam | Abcam Cat# ab18465, RRID:AB_2064130) | IHC (1:250) |
| Antibody | Rabbit GFAP antibody | Dako (now Agilent) | (Agilent Cat# Z0334, RRID:AB_10013382) | IHC (1:2500) |
| Antibody | Anti-Neural Cell Adhesion Molecule L1 Antibody, clone 324 | Millipore | (Millipore Cat# MAB5272, RRID:AB_2133200) | IHC (1:200) |
| Antibody | Rabbit Anti-Laminin | Sigma | (Sigma-Aldrich Cat# L9393, RRID:AB_477163) | IHC (1:25) |
| Antibody | Purified Mouse Anti-N-Cadherin | BD Biosciences | (BD Biosciences Cat# 610921, RRID:AB_398236) | IHC (1:150) |
| Antibody | Mouse Anti-Nestin Antibody, clone rat-401 | Millipore | (Millipore Cat# MAB353, RRID:AB_94911) | IHC (1:200) |
| Antibody | Rabbit Purified anti-Pax-6 Antibody | Biolegend | (BioLegend Cat# 901301, RRID:AB_2565003) | IHC (1:300) |
| Antibody | Mouse anti-Pax6 antibody | DSHB | N/A | IHC (1:300; deposited in DSHB by Kawakami A.) |
| Antibody | Phospho-Histone H3 (Ser10) (6G3) Mouse mAb | Cell Signaling | (Cell Signaling Technology Cat# 9706, RRID:AB_331748) | IHC (1:200) |
| Antibody | Anti-Reelin Antibody, a.a. 164–496 mouse reelin, clone G10 | Millipore | (Millipore Cat# MAB5364, RRID:AB_2179313) | IHC (1:1200) |
| Antibody | Anti-SATB2 antibody [SATBA4B10] - C-terminal | Abcam | (Abcam Cat# ab51502, RRID:AB_882455) | IHC (1:400) |
| Antibody | Rabbit anti-Six3 polyclonal antibody; | Rockland Antibodies | (Rockland Cat# 600–401-A26S, RRID:AB_11181864) | IHC (1:200) |
| Antibody | Rabbit anti-Sox2 antibody | Thermo Fisher Scientific | (Thermo Fisher Scientific Cat# PA1-094, RRID:AB_2539862) | IHC (1:400) |
| Antibody | Rabbit anti-Tbr1 antibody | Millipore | (Millipore Cat# AB10554, RRID:AB_ 10806888) | IHC (1:400) |
| Antibody | EOMES Monoclonal Antibody (Dan11mag) Tbr2 antibody | eBioscience | (Thermo Fisher Scientific Cat# 14-4875-82, RRID:AB_11042577) | IHC (1:200) |
| Antibody | Anti-Vimentin antibody [EPR3776] - Cytoskeleton Marker | Abcam | (Abcam Cat# ab92547, RRID:AB_10562134) | IHC (1:200) |
| Antibody | YAP (D8H1X) XP Rabbit mAb | Cell Signaling | (Cell Signaling Technology Cat# 14074, RRID:AB_2650491) | IHC (1:100) |
| Antibody | ZO-1 Monoclonal Antibody (ZO1-1A12), Alexa Fluor 488 | Thermo Fisher Scientific | (Thermo Fisher Scientific Cat# 339188, RRID:AB_2532187) | IHC (1:500) |
| Antibody | Purified Mouse Anti-β-Catenin Clone 14 | BD Biosciences | (BD Biosciences Cat# 610154, RRID:AB_397555) | IHC (1:200) |
| Antibody | Rabbit β-Catenin antibody | Abcam | (Abcam Cat# ab2365, RRID:AB_303014) | IHC (1:100) |
| Antibody | Goat anti-Rat IgG (H + L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 | Thermo Fisher Scientific | (Thermo Fisher Scientific Cat# A-21247, RRID:AB_141778) | IHC (1:400) |
| Antibody | Goat anti-Mouse IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | (Thermo Fisher Scientific Cat# A-11029, RRID:AB_2534088) | IHC (1:400) |
| Antibody | Goat anti-Rabbit IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 568 | Thermo Fisher Scientific | (Thermo Fisher Scientific Cat# A-11036, RRID:AB_10563566) | IHC (1:400) |
| other | DAPI stain | Molecular Probes | (Thermo Fisher Scientific Cat# D1306, RRID:AB_2629482) | 1:10000 |
| Commercial assay or kit | ApopTag Plus Fluorescein In situ Apoptosis Detection Kit | Chemicon/Millipore | Cat# S7111 | |
| Commercial assay or kit | SMART-Seq v4 Ultra Low Input RNA Kit | Takara | Cat# 634889 | |
| Commercial assay or kit | Nextera XT kit | Illumina | Cat# FC-131–1024 | |
| Commercial assay or kit | KAPA qPCR complete kit | Biorad | KK4844 Complete kit 500 × 20 ul reactions | |
| Commercial assay or kit | RNeasy Micro Kit | Qiagen | Cat# 74004 | |
| Commercial assay or kit | Bioanalyzer 6000 Pico Kit | Agilent | Cat# 5067–1513 | |
| Chemical compound, drug | Verteporfin | US Pharmacopeial Convention | Cat# 1711461 | |
| Chemical compound, drug | BrdU Labeling Reagent | Life Technologies | Cat# 000103 | |
| Chemical compound, drug | Doxycycline (doxycycline hyclate) | Sigma | Cat# D9891-25G | |
| Software, algorithm | BioViz3D version 3.1 | BioViz3D | BioViz3D, RRID:SCR_017162 | |
| Software, algorithm | salmon aligner v0.11.3 for R | PMID: 28263959 | ||
| Software, algorithm | Bioconductor tximport package, v1.8.0 for R | PMID: 26925227 | ||
| Software, algorithm | Bioconductor edgeR package v3.22.3 | PMID: 19910308 | (Bioconductor, RRID:SCR_006442) | Software |
| algorithm | glmTreat function in edgeR | PMID: 19176553 | ||
| Software, algorithm | Bioconductor GEOquery v2.48.0 | PMID: 17496320 | (GEOquery, RRID:SCR_000146) | |
| Software, algorithm | limma packages, v3.36.2 for R | PMID: 25605792 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.45961.026
-
Reporting standard 1
The ARRIVE guidelines checklist.
- https://doi.org/10.7554/eLife.45961.027





