In vivo study of gene expression with an enhanced dual-color fluorescent transcriptional timer
Figures
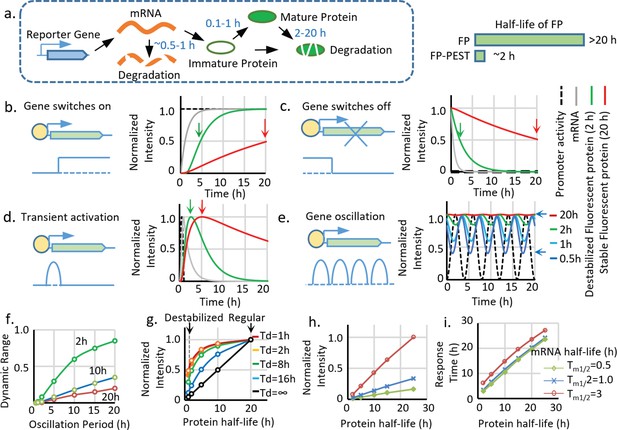
Advantages and limitations of destabilized fluorescent transcriptional reporters.
(a) Illustration of the biological processes that affect the final concentration of mature fluorescent protein, including transcription, translation, protein maturation, mRNA degradation, and protein degradation with the general half-life of mRNA, FP maturation time, and half-life of protein labeled. Destabiliation of the FP, achieved by fusion of FP with PEST domain, shortens the half-life of regular FP from over ~20 hr to ~2 hr. (b-d). Comparison between simulated signals from a destabilized fluorescent reporter (green, Tp1/2 = 2 hr) and a regular fluorescent reporter (blue, Tp1/2 = 20 hr) following switch-on, switch-off, 1 hr pulse, and oscillation with a period of 2 hr. The half-life of mRNA (Tm1/2) is set as 0.5 hr, and the protein maturation time (τm1/2) is set as 0.1 hr. The time points when the fluorescent signals reach 50% of the maximal intensity (for switch-on and switch-off) and the maximum response (for transient pulse activation) are indicated by black arrows. (e). Simulated signal intensities of fluorescent proteins with different protein half-lives to a sinusoid transcriptional oscillation with a period of 4 hr. The rest of the parameters are the same with above. The dynamic portion (the difference between the peak and valley) of the reporter (a half-life of 0.5 hr) is indicated by black arrows. (f). The dynamic range (the difference between the peak and valley of the signal compared to its average intensity as indicated in (e) of reporters with indicated half-lives generated by sinusoid transcriptional activity with different length of the period. The dynamic range of the total signal positively correlates with the oscillation period and negatively correlates with the protein half-life. (g) The intensity of the reporter signal for a constitutively active promoter (with no temporal variation) linearly depends on its protein half-life (black line). However, the maximal intensity of the reporter is less reduced by the shortened half-life for a shorter transient activation: a short-lived reporter (half-life of 2 hr) shows a 90% reduction of maximal signal compared with a stable reporter (half-life of 20 hr) for a constitutively active promoter. Nevertheless, the intensity is only reduced by 50% if the promoter is transiently activated for 1 hr. Td: the duration of a pulse transient promoter activation. (h,i) Simulated changes in maximum fluorescent intensity and response time (time to reach 50% of the maximum intensity) of the reporters with different half-lives of protein and mRNA for a promoter switch-off event.
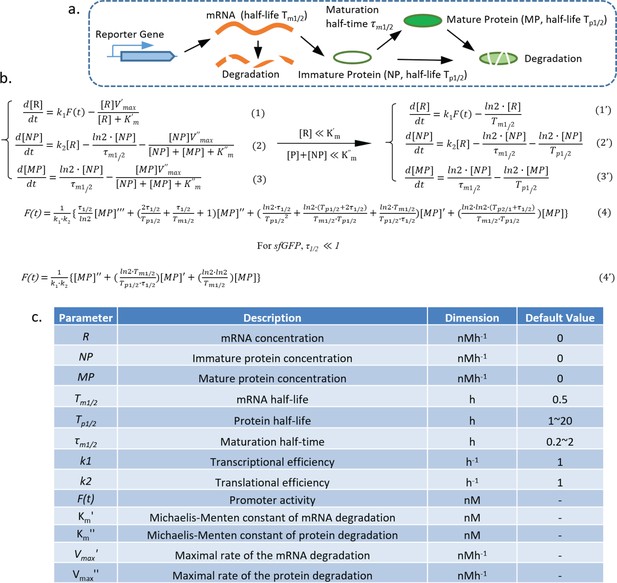
Model of the transcriptional reporter synthesis, maturation, and degradation.
(a) Illustration of the biological processes that affect the concentration of mature fluorescent protein, including transcription, translation, protein maturation, mRNA degradation, and protein degradation with key parameters labeled. (b,c) Equations describing the model generated by following the law of mass action, with the parameters shown in the table below.
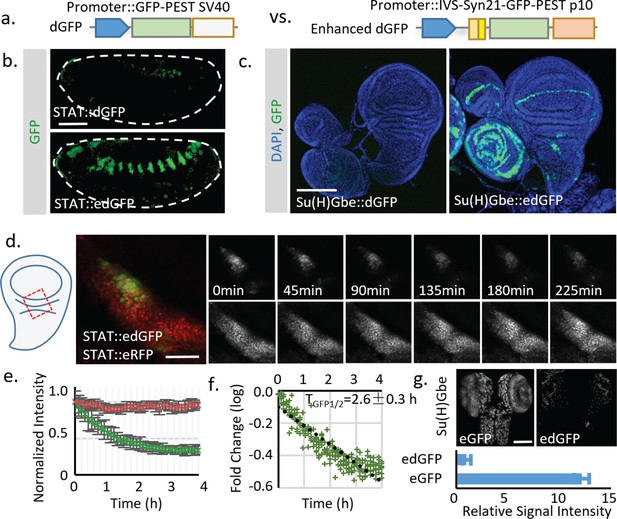
Using translational enhancers to increase the signal from destabilized fluorescent reporters for in vivo study.
(a) Illustration of the regular and destabilized GFP reporters. All the GFPs used in this study are the fast folding superfolder GFP (sfGFP). Destabilized GFP is labeled as dGFP, and dGFP reporter with translational enhancing elements is labeled as edGFP. All FPs used in this study contain the SV40 nuclear localization signal (NLS) at the N-terminus to facilitate signal segregation unless specified otherwise. (b, c) Comparison of dGFP and edGFP controlled by 6XSTAT response element in fly embryos and Su(H)Gbe Notch responding element in third instar wing imaginal discs. Images were taken with identical exposure. The contour of the embryo is outlined. (d) Dissected fly wing disc, expressing the STAT::edGFP and STAT:: eRFP, cultured ex vivo. Tissue was treated with 10 μM Actinomycin D to block transcription. STAT at the hinge region of the wing disc was imaged every 5 min for 4.5 hr. (e) The intensities of both dGFP and RFP were measured over time. Data from three independent replicates were collected and plotted. (f) The in vivo reporter half-life (TGFP1/2, representing effects of both Tm1/2 and Tp1/2) was estimated by linear regression of fluorescent intensity (in logarithmic scale). 95% confidence interval was calculated from linear regression. (g) Regular GFP and dGFP are expressed under Su(H)Gbe together with translational enhancers. Images were taken under identical parameters. The total fluorescent intensity from both reporters was plotted below with the intensity normalized to the dGFP signal. Data were collected from 10 different brains for each genotype. Scale bar: (b) 50 μm; (c, g) 100 μm; (d) 25 μm. Error bar: s.e.m.
-
Figure 2—source data 1
Source data for Figure 2e,f.
- https://doi.org/10.7554/eLife.46181.010
-
Figure 2—source data 2
Source data for Figure 2g.
- https://doi.org/10.7554/eLife.46181.011
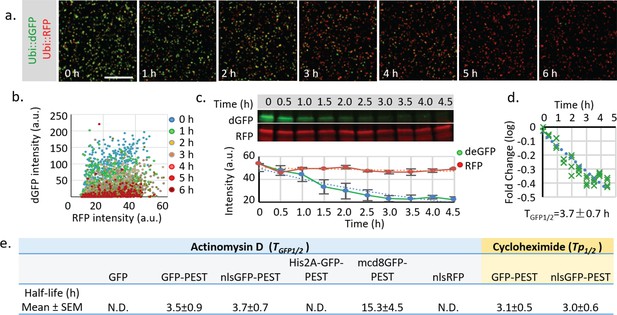
Measurement of the half-life of destabilized GFP in cultured Drosophila S2 cells.
(a) Drosophila S2 cells transiently transfected with both Ubi::nls-sfGFP-Myc-PEST and Ubi::nls-RFP-HA were treated with 10 μM Actinomycin D to block RNA synthesis for 6 hr. Images with identical exposure time were taken every hour after drug addiction. Scale bar: 100 μm. (b) The plot of fluorescence intensity of dGFP and RFP in cultured cells at different time points. (c) S2 cells treated with the transcriptional blocker Actinomycin D were harvested every 0.5 hr from time 0.0 hr to time 4.5 hr. Samples were analyzed by fluorescent western blot using rabbit anti-Myc (800 nm, Green) and mouse anti-HA (700 nm, Red). Quantification of fluorescent intensity from three independent experiments. Error bar: s.e.m. (d) Fold change of fluorescent intensity was plotted over time (logarithmic scale). The combined half-life of both mRNA and protein (TGFP1/2) was calculated by linear regression using least squares estimation. The expected value and 95% confident interval of TGFP1/2 was calculated from the linear regression. (e) Table of measured half-lives of fluorescent reporters (TGFP1/2, including both mRNA and protein half-life) measured using western blot after Actinomycin D treatment (in blue cells). Protein half-life (Tp1/2) was measured using western blot after cycloheximide (100 μg/ml) treatment to block protein synthesis (in yellow cells). Regular GFP, RFP or GFP-PEST fused with His2A is too stable to be reliably estimated within the 5 hr treatment period.
-
Figure 2—figure supplement 1—source data 1
Source data for Figure 2—figure supplement 1b.
- https://doi.org/10.7554/eLife.46181.007
-
Figure 2—figure supplement 1—source data 2
Source data for Figure 2—figure supplement 1c.
- https://doi.org/10.7554/eLife.46181.008
-
Figure 2—figure supplement 1—source data 3
Source data for Figure 2—figure supplement 1d.
- https://doi.org/10.7554/eLife.46181.009
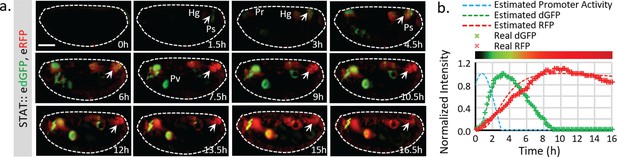
Live imaging of both destabilized sfGFP and stable RFP to reveal endogenous STAT dynamics.
(a) Time-lapse imaging of developing fly embryos expressing both STAT::edGFP and STAT::eRFP from stages 12 to 17 when STAT activity starts to increase. Maximum intensity z-projections of the mid-section (120 μm) are shown. Arrowheads indicate the posterior spiracles and hindgut region. The signal from the same structure turns from green to red over time. (b) The total dGFP and RFP fluorescent signals within the posterior spiracles and hindgut region (indicated by arrows in a) are plotted as colored crosses. The estimated promoter activity was calculated using equations (4) (Figure 1—figure supplement 1b), and plotted as a dashed blue line. The simulated responses of dGFP and RFP from the estimated transcriptional signal using equations (1)-(3) (Figure 1—figure supplement 1b) are plotted as dashed green and red lines respectively. The merged intensity from estimated dGFP and RFP signals is illustrated as a color bar at the top of the plot. Scale bar: 100 μm.
-
Figure 3—source data 1
Source data for Figure 3b.
- https://doi.org/10.7554/eLife.46181.016
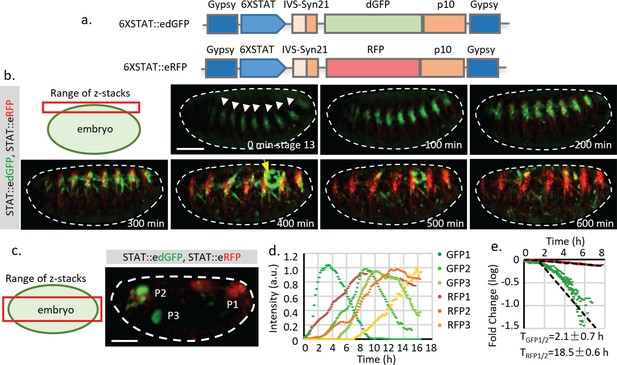
Quantification of STAT reporter dynamics.
(a) The STAT reporter consists of gypsy insulators at each end to prevent potential influence of nearby promoters, a 6XSTAT response element from STAT effector Socs36E, a minimal promoter from the hsp70 heat shock gene (HSPmin), an intervening sequence (IVS), a Syn21 sequence, a fly codon-optimized fluorescent protein with nuclear localization signal (NLS), a destabilized GFP (dGFP) which is sfGFP fused with a PEST degradation sequence from mouse ornithine decarboxylase (MODC), and p10 polyA signal. The dGFP together with the translational enhancing elements were called edGFP. (b) Frames from live imaging of STAT::edGFP and STAT::eRFP reporter in developing embryos (from developmental stage 13 to 17). Maximum intensity projections of z-stack images that capture the surface of the embryo (25 μm) are shown (Video 2). The surface signal was projected separately from the mid-section because it is much weaker than the signal from internal organs. STAT activity is initially detected at the tracheal pits (indicated by white arrowheads). As embryonic development continues, the dGFP signal disappears whereas the RFP signal remains. Meanwhile, STAT activity, revealed by STAT::edGFP in the gonad (indicated by yellow arrow), also increases and decreases during the time of imaging. (c, d) z-projection of the internal organs of a developing fly embryo expressing both STAT::edGFP and STAT::eRFP reporters (Video 1). Fluorescent intensities of both GFP and RFP from three different positions, P1 (posterior spiracles and hindgut), P2 (pharynx), and P3 (proventriculus), are measured. Original and normalized total fluorescent signals from P1 (GFP1 and RFP1), P2 (GFP2 and RFP2), and P3 (GFP3 and RFP3) are plotted in (d) respectively. (e) Estimation of dGFP and RFP half-life (TGFP1/2 = 2.1 hr, TRFP1/2 = 18.5 hr, which represent the effects from both Tm1/2 and Tp1/2) using P1 (posterior spiracles and hindgut) signals from six different embryos. The degradation of dGFP is moderately deviated from the simplified model, suggesting that the substrate concentration may be comparable with the Michaelis-Menten (MM) constant. Therefore, the estimated half-life parameter is an average over time. Scale bar: (b.c) 100 μm.
-
Figure 3—figure supplement 1—source data 1
Source data for Figure 3—figure supplement 1d.
- https://doi.org/10.7554/eLife.46181.014
-
Figure 3—figure supplement 1—source data 2
Source data for Figure 3—figure supplement 1e.
- https://doi.org/10.7554/eLife.46181.015
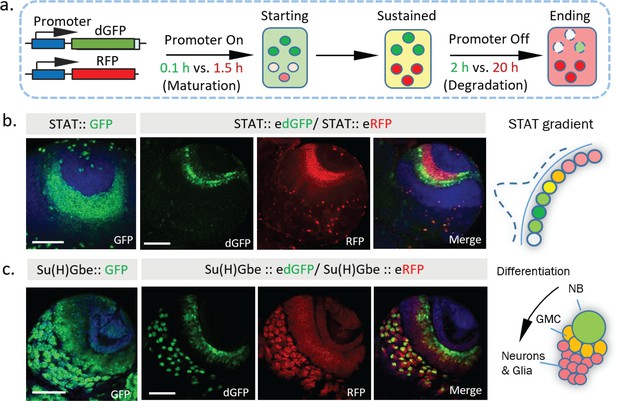
Dynamics of STAT and Notch activity revealed in fixed tissues.
(a) The destabilized GFP (dGFP) combined with stable RFP functions as a fluorescent timer to reveal transcriptional dynamics. The maturation and degradation half-lives of dGFP and RFP are indicated. (b. c) A comparison between the regular GFP reporter with a combination between dGFP and RFP reporter under control of either STAT response elements or Notch response element Su(H)Gbe. Both reporters are visualized in third instar larval brains. NB, neuroblast; GMC, Ganglion mother cells. Scale bar: (b, c) 50 μm.
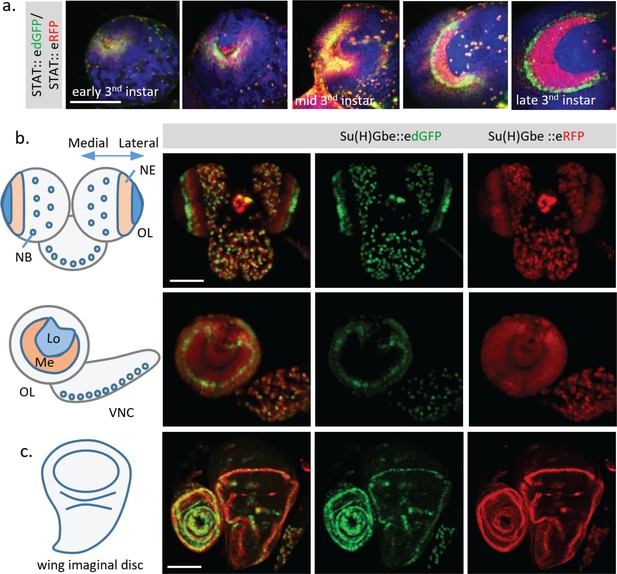
Additional dynamics of Notch and STAT activity revealed in different fixed tissues and stages.
(a) Change of STAT activity in third instar larval optic lobe (OL) at different developing stages. (b) Activities of Notch signaling revealed by Su(H)Gbe::edGFP and Su(H)Gbe::eRFP in third instar larval brain. In the optic lobe (OL), the Notch signal is down-regulated by a ‘proneural wave’ that sweeps across the neuroepithelium (NE) from the medial to the lateral regions, to trigger the NE to neuroblast (NB) transition. Consistent with this, a gradient change of Notch activity from red to green was observed in the NE region. Red signal (Notch low) located medially, and a band of green signal (Notch high) located at the lateral side of the OL, were observed, revealing the site of Notch pathway activation and progression of the proneural wave. Lo, lobula; Me, medulla; VNC, ventral nerve cord. (c) Both dGFP and RFP reporters show activity in the wing margin region of the third instar wing disc, with the GFP signal concentrated in a more defined cell population. Scale bar: (a,b,c) 100 μm.
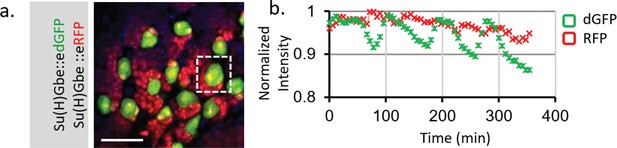
Live imaging of Notch activity in dividing Type I neuroblasts (NBs) in larval brain.
(a) Notch activity is monitored by both Su(H)Gbe::edGFP and Su(H)Gbe::eRFP reporters. Dissected third instar larval brain was cultured together with the larval fat body in Schneider's medium and imaged every 5 min. A z-stack covering the entire NB cell body was taken. In the central brain region, the large neuroblasts (NBs) (green) undergo asymmetric cell division and create new NBs and much smaller progeny cells (red). (b) The total signal from a single NB was measured using Bitplane Imaris software. The dGFP channel showed an oscillation with amplitude ~8% of the average signal, whereas the RFP channel is generally flat. The change of dGFP is consistent with an asymmetric NB cell division event (~1.5 hr) that produces a small progeny cell marked by RFP, suggesting that Notch activity may be affected during cell division. Scale bar: 25 μm.
-
Figure 4—figure supplement 2—source data 1
Source data for Figure 4—figure supplement 2b.
- https://doi.org/10.7554/eLife.46181.022
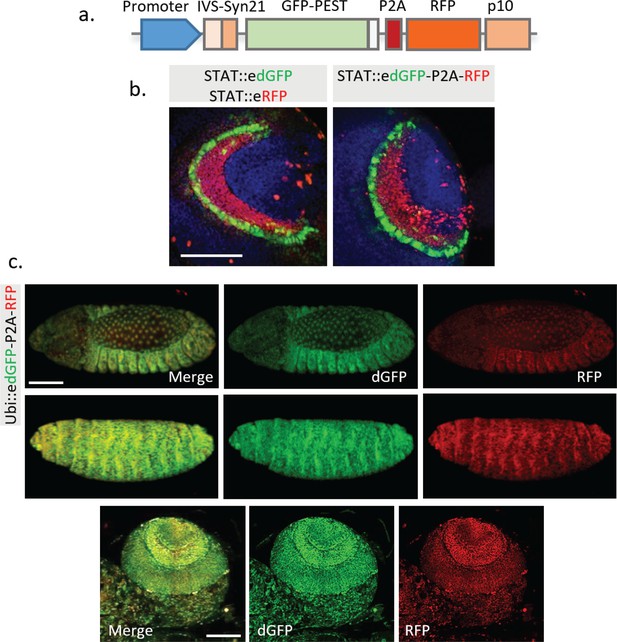
Combination of edGFP and RFP into a single construct using 2A peptide.
(a) Structure of the multicistronic reporter containing edGFP and RFP. (b) Comparison between STAT::edGFP, STAT::eRFP and STAT::edGFP-P2A-RFP in third instar larval brain. (c) No significant change of ratio between dGFP compared to RFP was observed in the embryo and larval brain when both proteins are controlled by a constitutively active Ubiquitin promoter. Representative images from the stage 14 (top panel) and 16 (middle panel) fly embryos, and third instar larval optic lobe (bottom panel) are shown. Scale bar: (b) 50 μm; (c) 100 μm.
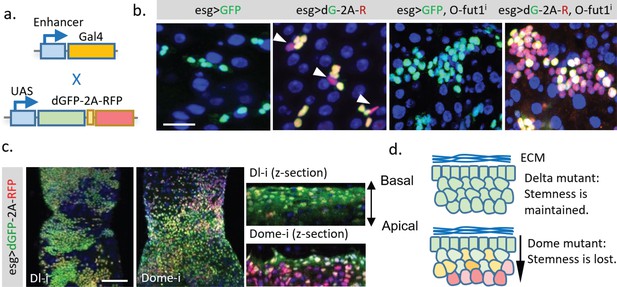
Application of the dGFP-P2A-RFP dynamic reporter to study gene expression dynamics of fly intestine stem cells.
(a) UAS-TransTimer, a UAS controlled multicistronic reporter containing dGFP and RFP connected by the P2A peptide was generated and crossed with the intestine stem cell driver esg-Gal4. (b) Compared with the regular GFP reporter, TransTimer (dGFP-P2A-RFP/dG-2A-R) reveals the differentiating cells, which reduce the expression of the escargot (esg) stem cell marker (arrowheads). Knocking-down O-fut1 inhibits Notch activity and causes ISC tumors. Compared to regular GFP, significant heterogeneity is revealed in the over-proliferating cell clusters using TransTimer. (c,d) Intestine tumors are generated by knocking down the Notch ligand Dl or the cytokine receptor Domeless (Dome). The double-headed arrow indicates the z-direction with the basal side of intestine epithelium facing up and apical side facing down. Scale Bar: (b) 25 μm; (c) 100 μm.
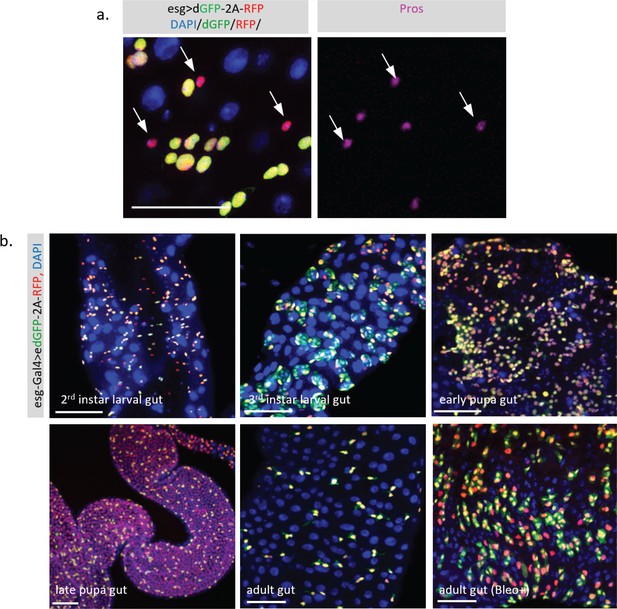
Dynamic activity of TransTimer in the fly intestine.
(a) The small cells with high RFP and low dGFP signals are positive for the enteroendocrine (EE) cell marker Pros, suggesting that these cells are differentiated cells that turn off esg. (b) Patterns of the dynamic reporter driven by esg-Gal4 in the Drosophila intestine at different developmental stages. In the second instar, the adult midgut progenitors (AMPs) exist as individual cells and disperse throughout the intestine. Using TransTimer, we observed considerable variation in the dGFP/RFP ratio, indicating an unexpected heterogeneity in this cell population. In the third instar intestine, the AMPs proliferate and form clusters called midgut imaginal islands (MIIs). Some AMPs with high Notch activity start to differentiate into peripheral cells (PCs), which surround the AMPs to create a transient stem cell niche. Consistent with this model, cells at the periphery of MII showed more red color, suggesting that these cells started to differentiate and lost their stemness. During the early pupal stage, the MIIs break down, while PCs and a large portion of AMPs differentiate into pupal intestine cells, which will be degraded at the end of the pupal stage. Consistent with this, many esg +AMPs turn red as these cells start to differentiate. During metamorphosis, the larval intestine is completely turned over, and the late pupal intestine, which will become the future adult intestine, is newly generated from AMPs. Therefore, except for the ISCs that still maintain the dGFP signal, differentiated gut cells all show the RFP signal, suggesting that they were recently derived from esg +AMPs. During the adult stage, esg +ISCs and enteroblast (EBs) are mitotically inactive and predominately show a yellow color. However, in the presence of the damaging reagent Bleomycin, which triggers strong cell proliferation and differentiation, a significant separation between dGFP and RFP signal is observed. Scale bar: (a, b) 50 μm.
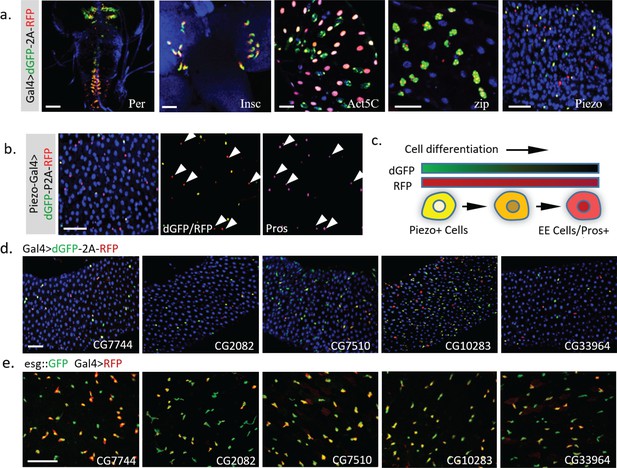
Dynamic pattern of TransTimer driven by different Gal4 drivers.
(a) Examples of Gal4 lines that show clear dynamic patterns with TransTimer (UAS-dGFP-P2A-RFP) in various organs: Per-Gal4, controlled by the circadian rhythm regulator Period (larval brain and ventral ganglion); insc-Gal4, expressed in Type II neuroblasts (larval brain); Act5C-Gal4, controlled by the act5C enhancer (larval intestine); GMR51F08-Gal4, controlled by the enhancer from the fly myosin heavy chain Zipper (larval intestine, adult intestine precursor cells); Piezo-Gal4 (BL58771), controlled by the cloned enhancer from mechanical sensitive ion channel Piezo (adult intestine). (b) Dynamic reporter under control of Piezo-Gal4[KI], a Gal4 knock-in after the first ATG of Piezo. Piezo + cells with high RFP and low GFP are positive for the EE cell marker Prospero (Pros). (c) Illustration of the differentiation process from Piezo + EP (enteroendocrine precursor) to Pros + EE cells. (d) The expression dynamics of UAS-TransTimer driven by different Gal4 lines in the fly intestine. (e) Gal4 activities are detected in a subpopulation of the esg +stem cells. Stem cells are marked by esg::GFP and the expression of Gal4s are revealed by UAS-RFP. Scale bar: (a-e) 50 μm.
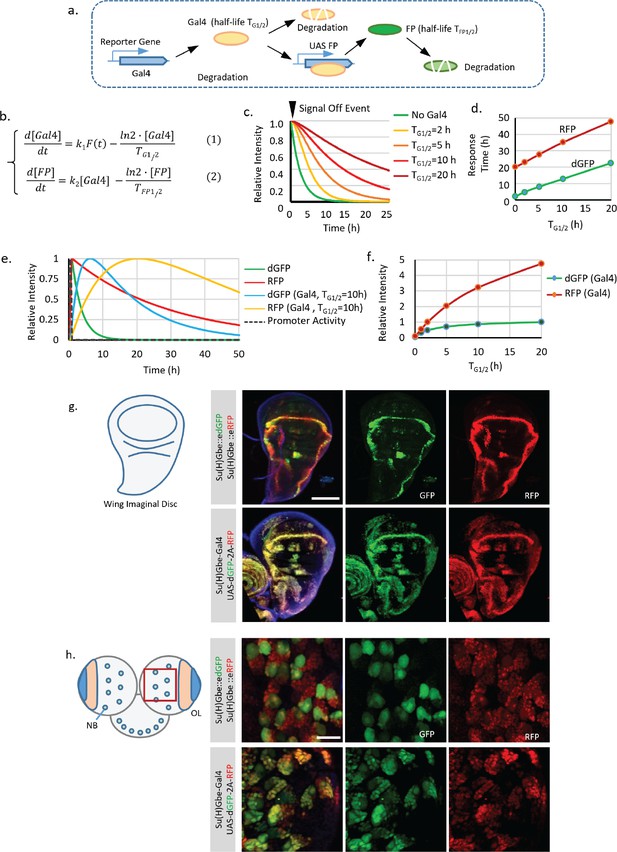
Effect of using Gal4/UAS system on the transcriptional timer.
(a) Illustration of the synthesis and degradation processes of fluorescent protein (FP) controlled by the Gal4/UAS system. (b) The degradation of Gal4 and FP was modeled to follow the first order kinetics with half-life TG1/2 for Gal4 and half-life TFP1/2 for FP. The production and degradation of mRNA were not explicitly expressed to simplify the model as it is not the major contributor to the system dynamics. (c,d) An ‘off’ event of the signal was modeled. The response time (time to reduce the signal to half of the original) is generally proportional to the sum of TG1/2 and TFP1/2. (e,f) A pulse of signal activation (duration of 1 hr) was modeled. Signals from dGFP and RFP controlled directly by the promoter was compared with signals from dGFP and RFP driven by the promoter controlled Gal4 (half-life of 10 hr). Including Gal4 as an intermediate significantly delays the response of the FPs. (f) Presence of Gal4 also increases the maximum intensity of the FPs in response to the pulse signal. This amplification effect increases as the half-life of Gal4 grows longer and gradually saturates as the TG1/2 surpasses the TFP1/2. (g,h) dGFP and RFP were driven either directly by the Notch responding element Su(H)Gbe or indirectly by Su(H)Gbe-Gal4. Wing imaginal discs and brain from third instar larval were analyzed. Images were all captured with full dynamic range (with the brightest pixel near the 100% saturation). Scale bar: (g) 100 μm; (h) 25 μm.
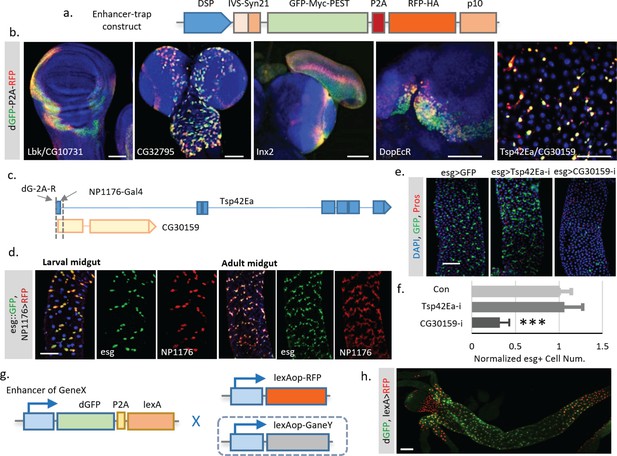
Enhancer trap screen for endogenous transcription dynamics.
(a) A P-element containing Drosophila synthetic core promoter (DSCP), translational enhancing elements, and dGFP-P2A-RFP (TransTimer) was randomly mobilized in the fly genome to identify enhancers with dynamic expression. edGFP and RFP are tagged by Myc and HA epitopes, respectively, to allow signal enhancement by immunohistochemistry. (b) Examples of enhancer trap lines that show dynamic patterns in different organs: Lbk, an Immunoglobulin-like protein, or CG10731, the subunit S of mitochondrial ATP synthase complex (wing disc); CG32795, a novel membrane protein (larval brain); inx2, a gap junction protein (larval brain and eye disc); DopEcR, Dopamine and Ecdysteroid receptor (larval brain); and Tsp42E, a tetraspanin protein; and CG30159, a novel gene with unknown function (adult intestine). (c) The dGFP-P2A-RFP and the NP1176-Gal4 enhancer trap insertions at the promoter region of Tsp42Ea and CG30159. Exons are shown in the diagram (blue for Tsp42Ea, yellow for CG30159). Tsp42Ea and CG30159 share the same promoter region. (d) NP1176-Gal4 (marked by UAS-RFP) shows stem cell expression like esg (marked by esg::GFP). (e, f) Knocking down CG30159 significantly reduces stem cell numbers (esg+ cells) in the intestine, while RNAi against Tsp42Ea does not have a significant phenotype. Number of esg +cells were quantified within 100 μm^two regions from n = 8 (GFP control), n = 7 (Tsp42Ea-i), and n = 9 (GC30159-i) adult fly intestines. p-value<0.001. Cell numbers are normalized according to the control. (g) Schematic of the TransTimerLex for enhancer trap with RFP replaced by the bacterial transcriptional repressor LexA. This reporter allows both two-color contrast and further genetic manipulation of the target cell population using the LexA/lexAop binary expression system. (h) An enhancer trap line, under potential control of Larp enhancer, was crossed with lexAop::RFP. The anterior region of the larval intestine is shown, revealing a decrease of transcriptional activity in the proventriculus. Scale bar: Scale bar: (b, h) 50 μm; (d, e) 100 μm.
-
Figure 7—source data 1
Source data for Figure 7f.
- https://doi.org/10.7554/eLife.46181.030
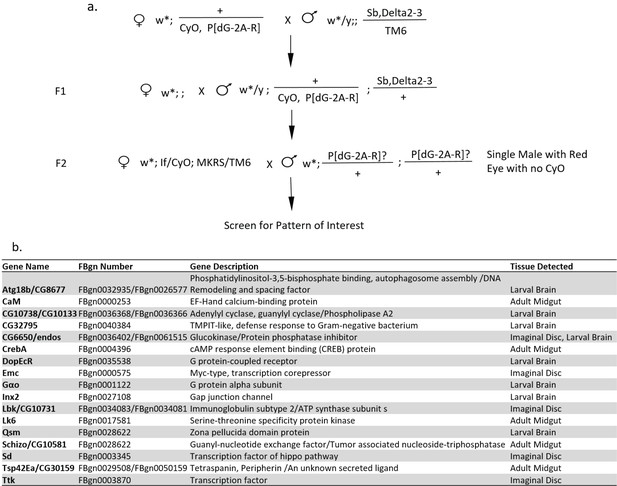
Enhancer trap screen for gene expression dynamics.
(a) Genetic crossing scheme for random insertions of a P-transposable element with TransTimer (P[dG-2A-R]) on the second and third chromosomes. Fly lines with interesting expression patterns were balanced and mapped using splinkerette PCR. (b) List of identified enhancer trap lines with interesting dynamic expression patterns. The names of the genes with promoter regions located within 3 kb range of the reporter insertion sites are listed in the table. In most cases, the insertions showed a clear association with the promoter region of a specific gene. Representative images of the expression patterns are shown in Supplementary file 3.
Videos
Live imaging of developing fly embryo (mid projection).
Embryo was imaged at room temperature using a Zeiss Lightsheet Z1 microscope with a 20X (N.A. 1.0) lens. Z-stack images (2 μm between each slice) were acquired every 10 min. A maximal intensity z-projection of mid-section (120 μm) is shown in the video. Video was taken from stage 13 to stage 17 when the somatic muscles contract. Genotype of the sample is: w; STAT::edGFP/STAT::eRFP.
Live imaging of developing fly embryo (surface projection).
Embryo was imaged at room temperature using a Zeiss Lightsheet Z1 microscope with a 20X (N.A. 1.0) lens. Z-stack images (2 μm between each slice) were acquired with 10 min time intervals. A maximal intensity z-projection of surface (25 μm) is shown. Video was taken from stage 13 to stage 17 before the somatic muscles contraction. Genotype of the sample is: w; STAT::edGFP/STAT::eRFP.
Additional files
-
Supplementary file 1
Gal4s with dynamic expression patterns.
(Gal4s analyzed in the main figures were not listed.)
- https://doi.org/10.7554/eLife.46181.031
-
Supplementary file 2
Gal4s without significant expression dynamics in different fly organs.
- https://doi.org/10.7554/eLife.46181.032
-
Supplementary file 3
Enhancer trap lines of Transcriptional Timer (HSPmini-IVS-p21-nlsGFP-Myc-PEST-P2A-nlsRFP-HA-p10) with expression dynamics in different fly organs.
- https://doi.org/10.7554/eLife.46181.033
-
Supplementary file 4
Cloning information and sequences of constructs.
- https://doi.org/10.7554/eLife.46181.034
-
Supplementary file 5
Mapping Results of enhancer trap lines by splinkerette PCR.
- https://doi.org/10.7554/eLife.46181.035
-
Supplementary file 6
Key resources table.
- https://doi.org/10.7554/eLife.46181.036
-
Transparent reporting form
- https://doi.org/10.7554/eLife.46181.037





