Mechanism of pharmacochaperoning in a mammalian KATP channel revealed by cryo-EM
Figures
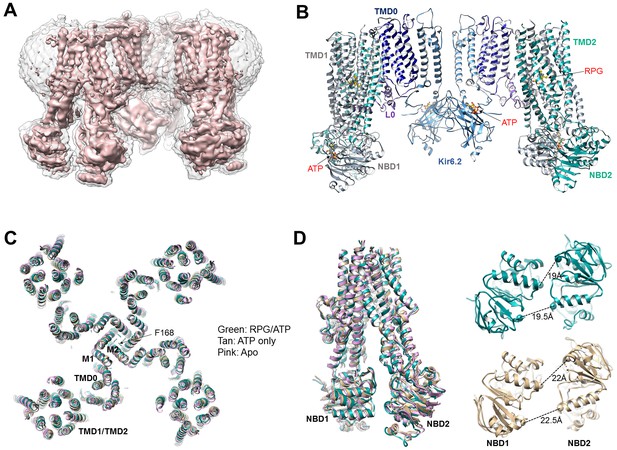
Structural determination and comparison.
(A) Unsharpened 3.9 Å C4 cryoEM reconstruction of the KATP channel bound to RPG and ATP. (B) Structural model of the channel in the RPG/ATP state. (C) Overlay of the RPG/ATP state structure, the ATP only state structure, and the apo state structure viewed from the top showing similarity of the dominant class of the ATP only and the apo state to the RPG/ATP state structure. (D) Left: Same model as (C) viewed from the side and focusing on the ABC transporter core module of SUR1 to illustrate the inward-facing conformation observed in all three structures. Right: Separation between Walker A and the signature motif in NBD1 and NBD2 (G716::S1483 and S831::G1382; Cα to Cα, indicated by the dashed line) in SUR1 bound to RPG and ATP (green) and ATP only (tan) viewed from the bottom.
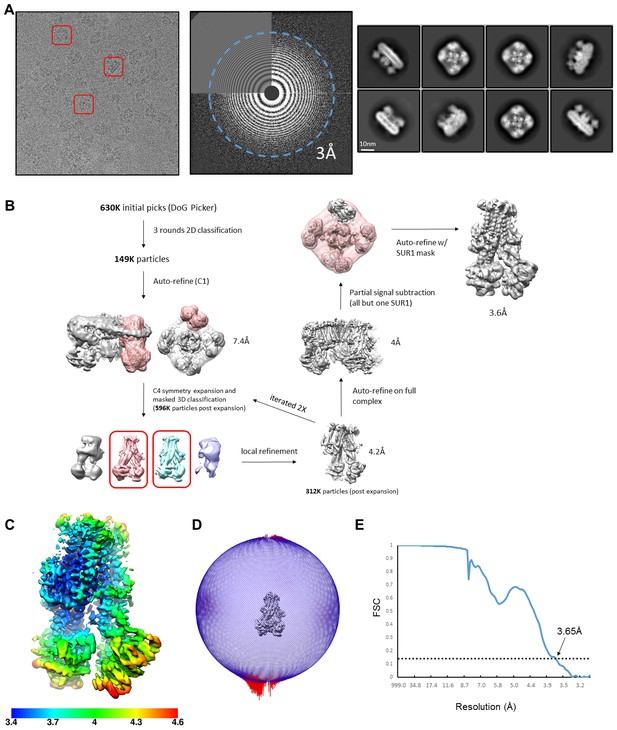
Data collection and image processing workflow for the RPG/ATP state.
(A) Left: Representative micrograph after alignment with Motioncor2. A few KATP channel complexes of various orientations are outlined by the red box. Middle: Power spectrum calculated with Ctffind4, with resolution reaching 3.0 Å. Right: Representative 2D classes. (B) Overview of data processing workflow. Particle picking was performed automatically with DoGPicker and manual inspection. All other image processing steps were performed in RELION-3. (C) Local resolution plot of focal refined SUR1 map. (D) Angular distribution plot. (E) Fourier shell correlation (FSC) of two independent half maps of focal refined SUR1.

Data processing workflow for the CBZ/ATP state.
(A) Overview of data processing workflow. Particle picking was performed automatically with DoGPicker and manual inspection. All other image processing steps were performed in RELION-3. (B) Local resolution plot of locally refined SUR1 map. (C) Angular distribution plot. (D) Fourier shell correlation (FSC) of two independent half maps of locally refined SUR1.
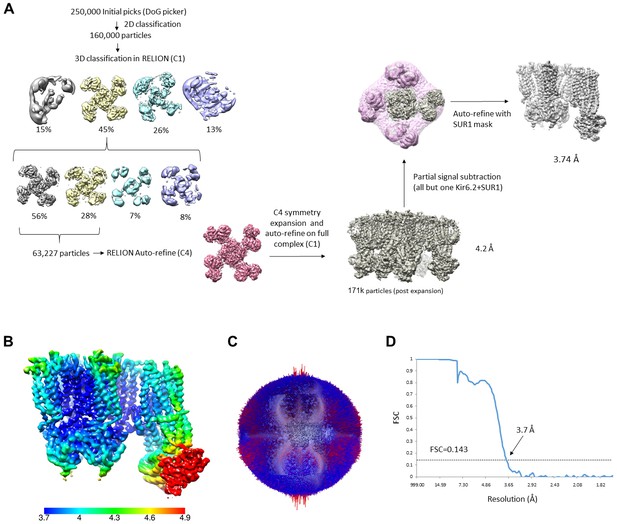
Data processing workflow for the GBC/ATP state.
(A) Overview of data processing workflow. Note the dataset used was previously published in Martin et al. (2017a) and the particles that were included in the final reconstruction (EMD-7073) were used for focused refinement in RELION-3. (B) Local resolution plot of the locally refined SUR1 map. (C) Angular distribution plot. (D) Fourier shell correlation (FSC) of two independent half maps of locally refined SUR1.
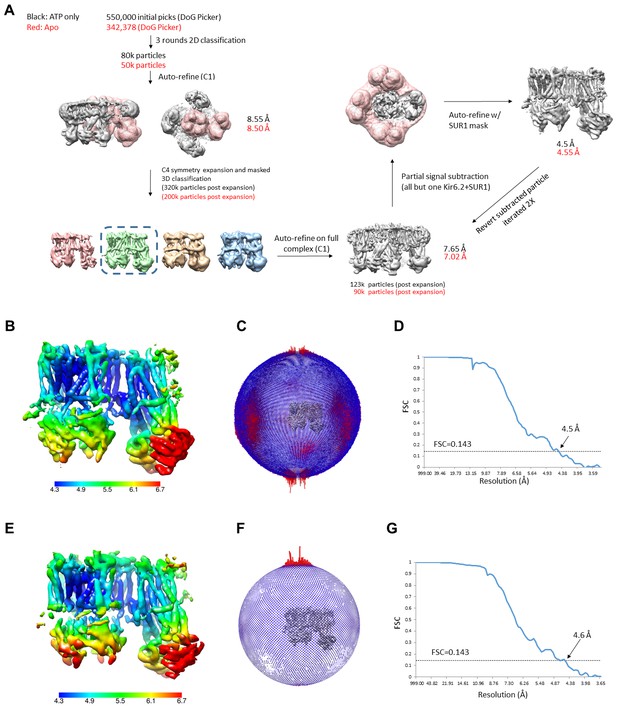
Data processing workflow for the ATP only state and the apo state.
(A) Data processing flow for the ATP only state dataset. The Apo state dataset was processed in the same manner and relevant numbers are shown in red. (B–D) Local resolution map, angular distribution plot, and FSC plot for the ATP only dataset. (E–G) Local resolution map, angular distribution plot, and FSC plot for the apo state dataset.

Comparison of the GBC/ATP state maps before and after focused refinement.
(A) Left: Cryo-EM C4 map of the SUR1-NBD1 region published in Martin et al. (EMD-7073) contoured to 2.6σ; middle: the same region from the locally refined map contoured to 8.9σ; right: same as the middle panel except the density is displayed in mesh and the structural model of the protein and bound ATP are superposed. (B) Same as (A) but showing the linker region connecting TMD2 and NBD2.
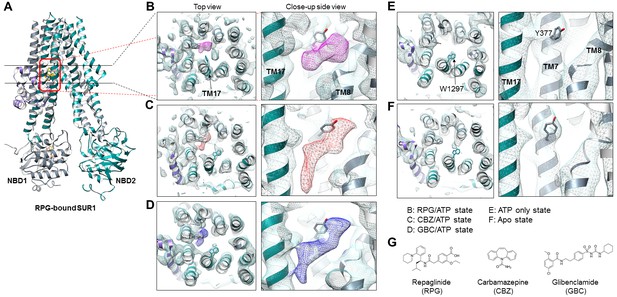
Structural comparison of the pharmacochaperone binding pocket.
(A) Structural model of the RPG-bound SUR1 ABC transporter core module viewed from the side showing the slice viewed from the top (indicated by the two black lines) and the pocket viewed from the side at higher magnification (indicated by the red box) in B-F. (B–F) The pharmacochaperone pocket viewed from the top and the side of the channel in the states indicated. To enable comparison, each map was sharpened and filtered to 4.6 Å (the resolution of the apo state reconstruction) with the Postprocessing procedure in RELION. Ligand density corresponding to RPG in (B) is shown in magenta, CBZ in (C) in red, and GBC in (D) in blue. The binding pocket is empty in both the ATP only state (E) and the apo state (F). Note the side chain of W1297 in TM17 and Y377 in TM7 are shown and labeled in panel (E) to serve as reference points. (G) Chemical structures of the three pharmacochaperones shown in B–D.

Density fitting for GBC, RPG, and CBZ.
(A) CryoEM density of GBC from the focus-refined SUR1 map contoured to 8.5σ. Left: optimal fitting of the GBC structure into the density. The electrostatic nature of the residues in the binding pocket surrounding GBC (see Figure 3B) are shown to demonstrate general electrostatic mismatch with GBC if the molecule were modeled into the density in the flipped orientation by 180° shown on the right. (B) CryoEM density of RPG from the focus-refined SUR1 map contoured to 9σ with RPG fitted into the density. (C) CryoEM density of CBZ from the locally refined SUR1 map contoured to 8.5σ. Left: A CBZ molecule is placed into one end of the density to illustrate that one stationary CBZ molecule cannot account for the full ligand density observed. Right: Two CBZ molecules are placed in the density to show sufficient density to accommodate two CBZ molecules.
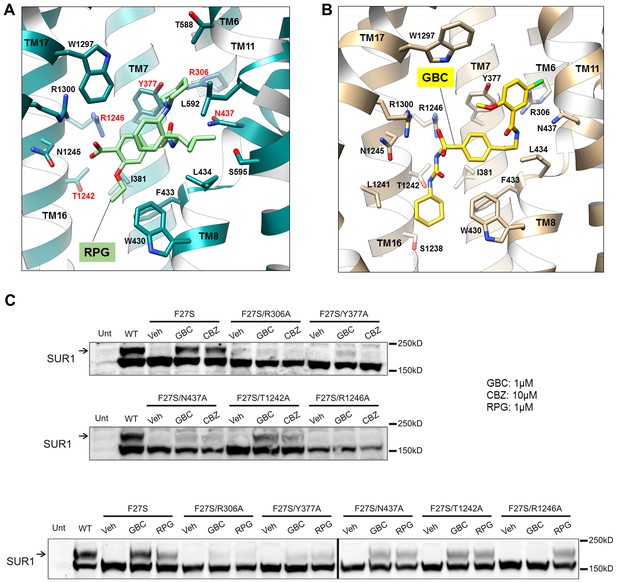
Models of the PC binding pocket.
(A) RPG binding site model, with residues mutated to alanine in C labeled in red. (B) GBC binding site model. (C) Western blots showing effects of alanine mutation of the selected residues on the ability of GBC, CBZ, and RPG to correct the processing defect caused by the F27S mutation in the TMD0 of SUR1. The arrow indicates the mature, complex-glycosylated SUR1. The lower band is the core-glycosylated immature SUR1. The thick vertical line in the bottom blot indicates samples from the same experiments run on two separate gels.
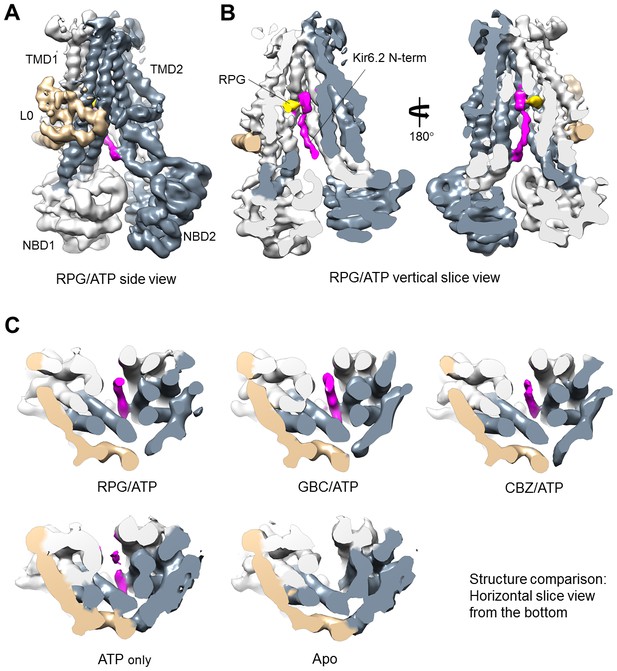
Kir6.2 N-terminus cryoEM density in SUR1.
(A) RPG-bound SUR1 from focus-refined, unsharpened map viewed from the side. The major domains are labeled in different colors. (B) Vertical slice view of the map shown in (A) that reveals the bound RPG (in gold) and the cryoEM density of Kir6.2 N-term (in magenta). (C) Comparison of the Kir6.2 N-term cryoEM density in the different structures in horizontal slices viewed from the bottom. All maps are sharpened and filtered to 6 Å. Apo and ATP-only structures are displayed at 1.8σ; RPG/ATP, GBC/ATP, and CBZ/ATP structures are displayed at 2.2σ. Note lower threshold is needed for the Kir6.2 N-term density in the ATP-only structure to become visible.
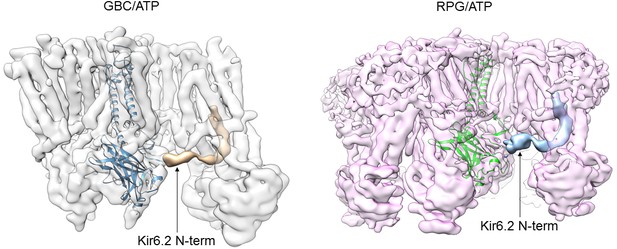
CryoEM density of Kir6.2 N-terminus in the GBC/ATP and RPG/ATP structures.
Maps were sharpened and filtered to 6 Å. The Kir6.2 N-term density shown in gold in the GBC/ATP structure and in blue in the RPG/ATP structure was obtained by subtracting the map calculated from the structure, and then low-pass filtered to 6 Å.
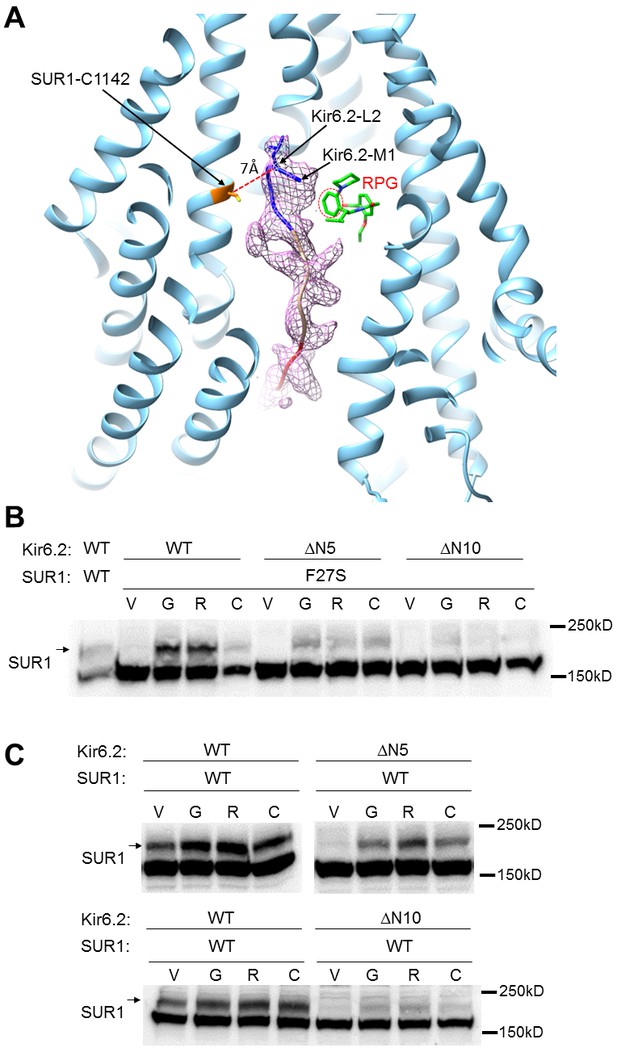
The distal N-terminus of Kir6.2 interacts with SUR1 and is required for channel biogenesis and pharmacochaperone rescue.
(A) Kir6.2 N-term cryoEM density (pink mesh) with superposed polyalanine model shown in the RPG (green) bound SUR1 structural model. The piperidino moiety of RPG is highlighted with dotted red line to show its close proximity to the N-terminal methionine of the modeled Kir6.2 N-terminal peptide. The relationship between SUR1 C1142 and Kir6.2 L2 is shown to illustrate their close proximity, with Cα-Cα distance of ~7 Å. The Kir6.2 N-term density map was obtained by removing densities corresponding to modeled SUR1 and RPG from the focus-refined RPG/ATP map using the Color Zone option in Chimera, contoured to 12σ. The polyalanine model of the Kir6.2 N-term corresponding to amino acids 1–5 is shown in blue, 5–10 in tan, and 10–15 in red. (B) Western blot showing that deletion of Kir6.2 amino acids 2–5 (ΔN5) or amino acids 2–10 (ΔN10) attenuated or nearly abolished, respectively, the pharmacochaperoning effect [compared to 0.1% DMSO vehicle control (V)] of GBC (G; 1 µM), RPG (R; 1 µM), and CBZ (C; 10 µM) on the SUR1-F27S mutant. (C) Western blot showing Kir6.2 ΔN5 and ΔN10 also greatly impaired maturation of WT SUR1 (V), and rendered GBC, RPG, and CBZ less and less effective in enhancing WT SUR1 maturation.

Mass spectrometric identification of a chemical crosslink between Kir6.2 peptide KGIIPEEYVLTR (5-16) and SUR1 peptide STVKALVSVQK (599-609) using CBDPS.
(A) Table listing the top seven scoring MS/MS spectra for two inter-protein (Kir6.2/SUR1; Nr. 2, 6) and four intra-protein (SUR1/SUR1; Nr. 1, 3, 4, 5, 7; Note Nrs. 3 and 4 are the same crosslink with different z values) peptide crosslinks identified by MeroX software. Q-values of zero were calculated for all identified crosslinks using a decoy database of shuffled sequences from a database of 116 common protein contaminants, indicating a false discovery rate below 1%. All precursor ions for identified crosslinks also had mass errors below two ppm. (B) Score distribution histograms for identified intra- and inter-protein crosslinks and peptide identifications (red bars = decoy matches to shuffled sequences, blue bars = matches to correct sequences). (C) Precursor ion spectrum for +four charge state crosslink between Kir6.2 (5–16) and SUR1 (599–609) showing both light and heavy forms of the crosslinked peptide resulting from the 50:50 mixture of the H8 and D8 forms of CBDPS. Only the heavy form of each 8.05 mass shifted pair was selected for MS/MS analysis. (D) Annotated MS/MS fragment ion spectrum for the +four charge state crosslink between Kir6.2 (5–16, α-peptide) and SUR1 (599–609, β-peptide). Insert shows the matched y- and b-ions along the α- and β-peptide backbones, their relative intensities and their charge states. The graph at the bottom shows the mass errors of matched fragment ions, which all had errors below 10 ppm.
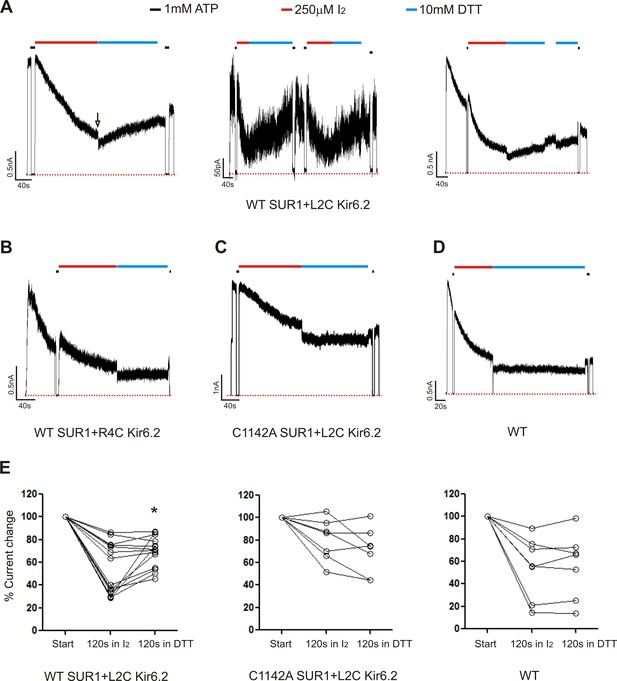
Patch-clamp recording of cysteine mutants to probe location of the Kir6.2 N-terminus.
(A) Three examples of traces of channels formed by WT(C1142) SUR1 and L2C Kir6.2 showing DTT-induced current recovery following I2 exposure. Channels were exposed to 250 µM I2 or 10 mM DTT as indicated by the bars above the traces. Baseline was obtained by exposing channels to 1 mM ATP. The arrow in the first trace indicates the blocking effect of the DTT that was readily reversed upon return to K-INT. Note as we have documented before (Lin et al., 2003; Pratt et al., 2012), DTT alone did not cause significant current run-up or run-down aside from the reversible blocking effect (not shown). (B, C, D) Representative traces of control channels formed by WT SUR1 and R4C Kir6.2, C1142A SUR1 and L2C Kir6.2, or WT SUR1 and Kir6.2. (E) Quantification of current changes at the end of 120 s exposure to I2, and at the end of subsequent 120 s exposure to DTT (expressed as % of currents at the start of I2 exposure). The asterisk indicates statistical significance comparing currents at the end of I2 exposure and the end of DTT exposure using paired student’s t-test (p<0.05).
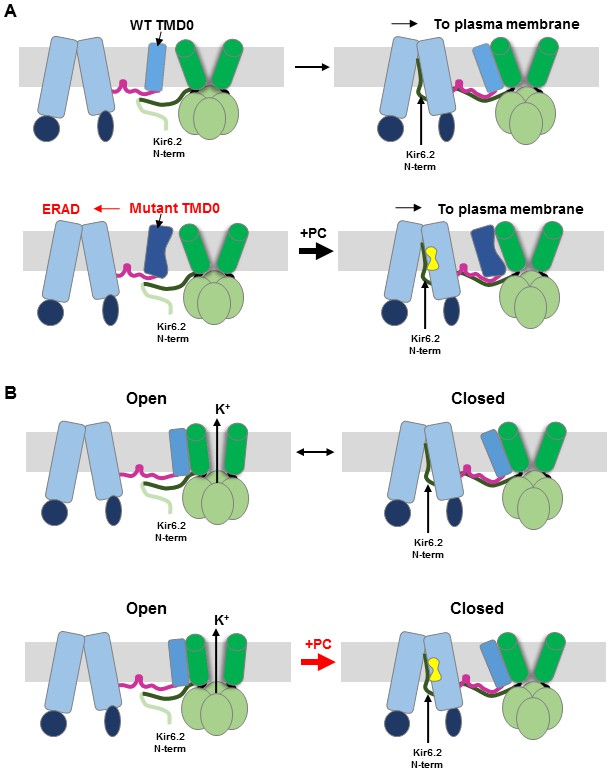
Cartoon of pharmacochaperoning and channel inhibition mechanism.
(A) Top: During WT channel biogenesis, KNt insertion into the SUR1 central cavity facilitates SUR1-TMD0 interaction with Kir6.2 for successful channel assembly. For SUR1 containing TMD0 trafficking mutations, it is unable to assemble with Kir6.2 in the absence of a pharmacochaperone (PC), and is targeted for ER-associated degradation (ERAD). Upon binding of PC to the binding pocket in SUR1, the Kir6.2 N-term becomes stabilized in the central cavity of the SUR1 ABC core, allowing assembly of the mutant TMD0 with Kir6.2 and trafficking of the complex to the plasma membrane. (B) Insertion of KNt into SUR1’s central cavity prevents Kir6.2 from adopting an open conformation (top) to close the channel. However, this interaction is labile and reversible (indicated by the black arrow going both directions). Binding of PC in the SUR1 pocket stabilizes Kir6.2 N-term in the central cavity and pushes channel equilibrium towards a closed state (indicated by the thick red arrow pointing to the closed state). In addition, PC binding stabilizes SUR1 in an inward-facing conformation, unable to be stimulated by Mg-nucleotides. Crosslinking of SUR1’s endogenous C1142 with engineered Kir6.2-L2C also traps the Kir6.2 N-term in the central cavity, closing the channel (see Figure 5).
Tables
Statistics of cryo-EM data collection, 3D reconstruction and model building.
https://doi.org/10.7554/eLife.46417.008| Data collection | Rpg/ATP | Cbz/ATP | Gbc/ATP | ATP only | Apo |
|---|---|---|---|---|---|
| Microscope | Krios | Krios | Krios | Krios | Arctica |
| Voltage (kV) | 300 | 300 | 300 | 300 | 200 |
| Camera | Falcon III | Gatan K2 | Gatan K2 | Gatan K2 | Gatan K2 |
| Camera mode | Counting | Super-resolution | Super-resolution | Super-resolution | Super-resolution |
| Defocus range (µm) | −1.0 ~ −2.6 | −1.4 ~ −3.0 | −1.4 ~ −3.0 | −1.4 ~ −3.0 | −1.4 ~ −3.0 |
| Movies | 5765 | 4413 | 2180 | 2344 | 2047 |
| Frames/movie | 240 | 60 | 60 | 60 | 60 |
| Exposure time (s) | 60 | 15 | 15 | 15 | 15 |
| Dose rate (e-/pixel/s) | ~0.7 | 8 | 8 | 8 | 8 |
| Magnified pixel size (Å) | 1.045 | 1.72* | 1.72* | 1.71* | 1.826** |
| Total Dose (e-/Å^2) | ~40 | ~40 | ~40 | ~40 | ~40 |
| Reconstruction | |||||
| Whole channel | |||||
| Software | Relion 3.0 | Relion 2 | Relion 2 | Relion 2 | Relion 2 |
| Symmetry | C4 | C4 | C4 | C4 | C4 |
| Particles refined | 24,747 | 138,000 | 63,227 | 80,304 | 34,527 |
| Resolution (masked) | 3.9 Å | 4.4 Å | 4.07 Å | 4.88 Å | 5.31 Å |
| SUR1 focused refinement | |||||
| Software | Relion 3.0 | Relion 3.0 | Relion 3.0 | Relion 3.0 | Relion 3.0 |
| Symmetry | C1 | C1 | C1 | C1 | C1 |
| Particles refined | 312,000 | 499,095 | 171,420 | 123,757 | 90,058 |
| Resolution (masked) | 3.65 Å | 4.34 Å | 3.74 Å | 4.5 Å | 4.55 Å |
| Model Statistics | (Includes KNt) | (Includes KNt) | |||
| Map CC (masked) | 0.6559 | 0.7771 | 0.7065 | 0.8155 | 0.7677 |
| Clash score | 3.37 | 7.52 | 3.22 | 2.60 | 3.11 |
| Molprobity score | 1.58 | 1.95 | 1.60 | 1.46 | 1.5 |
| Cβ deviations | 0 | 0 | 0 | 0 | 0 |
| Ramachandran | |||||
| Outliers | 0% | 0% | 0% | 0% | 0% |
| Allowed | 7.03% | 9.66% | 7.98% | 6.29% | 5.93% |
| Favored | 92.97% | 90.34% | 92.02% | 93.71% | 94.07% |
| RMS deviations | |||||
| Bond length | 0.009 | 0.008 | 0.007 | 0.005 | 0.009 |
| Bond angles | 1.166 | 1.121 | 0.992 | 1.085 | 1.233 |
-
*Super-resolution pixel size 0.86; **Super-resolution pixel size 0.913.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Cricetus cricetus) | ABCC8 (SUR1) | UniProt database | Q09427 | |
| Gene (Rattus norvegicus) | KCNJ11 (Kir6.2) | UniProt database | P70673 | |
| Recombinant adenovirus (Cricetus cricetus) | FLAG-tagged hamster SUR1 | PMID: 28092267 | FLAG-epitope inserted at the N-terminus of SUR1 and cloned into AdEasy vector for production of adenovirus | |
| Recombinant adenovirus (Rattus norvegicus) | Rat Kir6.2 | PMID: 28092267 | N/A | Constructed using the AdEasy vector for production of adenovirus |
| Recombinant adenovirus | tTA | PMID: 28092267 | N/A | Adenovirus for over-expression of Tetracycline-controlled transactivator (tTA) under CMV promoter used for Tet-Off system |
| Recombinant DNA reagent (Cricetus cricetus) | FLAG-tagged ham SUR1 in pECE | PMID: 11226335 | N/A | FLAG-epitope inserted at the N-terminus of SUR1 |
| Recombinant DNA reagent (Rattus norvegicus) | Rat Kir6.2 in pcDNA3 | PMID: 14707124 | N/A | |
| Cell line (Rattus norvegicus) | INS-1 clone 832/13 | PMID: 10868964 | RRID:CVCL_7226 | |
| Cell line (Chlorocebus aethiops) | COSm6 | PMID: 11226335 | RRID:CVCL_8561 | |
| Chemical compound, drug | Digitonin | Calbiochem | CAS 11024-24-1 | |
| Chemical compound, drug | ATP | Sigma-Aldrich | A7699 | |
| Chemical compound, drug | Glibenclamide | Sigma-Aldrich | G0639 | |
| Chemical compound, drug | Repaglinide | Sigma-Aldrich | R9028 | |
| Chemical compound, drug | Carbamazepine | Sigma-Aldrich | C4024 | |
| Chemical compound, drug | CBDPS-H8/D8 | Creative Molecules, Inc | Cat. Number: 014S | |
| Chemical compound, drug | FuGENE6 | Promega | E2691 | |
| Peptide | FLAG-peptide | Sigma-Aldrich | F3290 | |
| Antibody | Anti-FLAG M2 affinity gel | Sigma-Aldrich | A2220 | |
| Antibody | Anti-SUR1 (rabbit polyclonal | PMID: 17575084 | N/A | (1:100) |
| Antibody | Horseradish Peroxidase conjugated goat anti-rabbit secondary | Jackson ImmunoResearch | Code: 111-035-144 | (1/1000) |
| Software, algorithm | Serial EM | PMID: 16182563 | http://bio3d.colorado.edu/SerialEM | |
| Software, algorithm | MOTIONCOR2 | PMID: 28250466 | http://msg.ucsf.edu/em/software/motioncor2 | |
| Software, algorithm | CTFFIND4 | PMID: 26278980 | http://grigoriefflab.janelia.org/ctffind4 | |
| Software, algorithm | DoGPicker | PMID: 19374019 | https://sbgrid.org/software/titles/dogpicker | |
| Software, algorithm | Relion-3 | PMID: 30412051 | https://www2.mrc-lmb.cam.ac.uk/relion | |
| Software, algorithm | COOT | PMID: 20383002 | http://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot | |
| Software, algorithm | UCSF Chimera | PMID: 15046863 | http://www.cgl.ucsf.edu/chimera | |
| Software, algorithm | Pymol | Schrödinger | https://pymol.org/2 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.46417.018





