An explanation for origin unwinding in eukaryotes
Figures
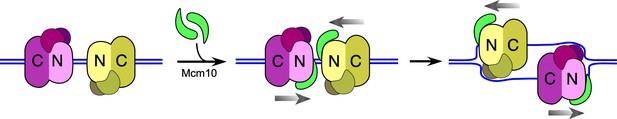
CMG action at origins.
CMG complexes, which appear as stacked rings with separate N- and C-domains, translocate in the N-first direction. Initially, two N-to-N (head-to-head) CMGs (purple and yellow) are assembled surrounding dsDNA (blue lines) at origins (left), and the tight connection of the inactive Mcm2-7 double hexamer is lost (Abid Ali and Costa, 2016; Costa et al., 2014), depicted as a separation between the CMGs (left). Upon addition of Mcm10 (green, middle) both CMGs switch to opposite strands of ssDNA and can pass one another to form bidirectional forks (right).
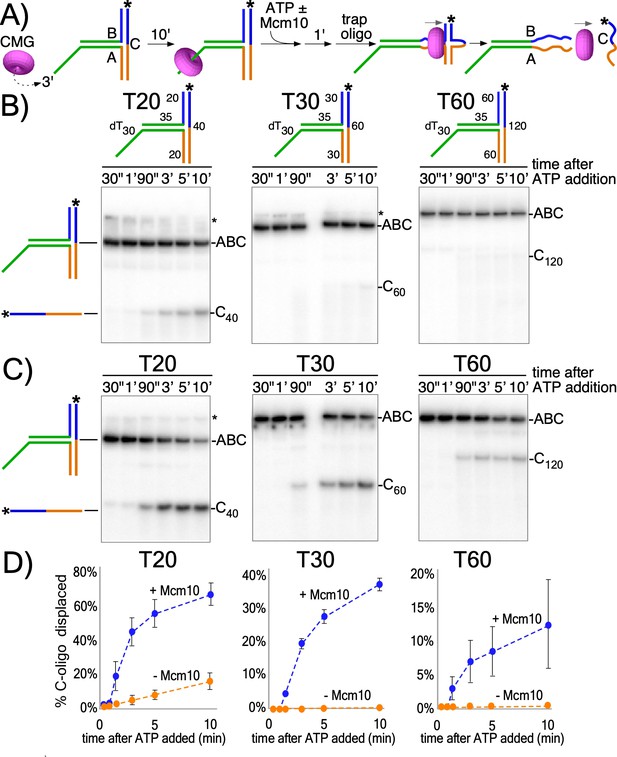
CMG translocates with force while encircling duplex DNA.
(A) Schematic of reactions using T-substrates with a 3’ dT30 ssDNA tail for CMG loading, a flush ss/ds junction and 35 bp of dsDNA (green) preceding the non-homologous arm blocks (blue and orange). See Figure 2—figure supplement 1 for details on the substrates. Unwinding the 32P-cross-bar oligo (labeled with a *) requires CMG to track with force while encircling dsDNA. (B) Native PAGE gel of time course reactions using CMG (no Mcm10) on T-substrates with different length arms. (C) Repeat of (B) with addition of Mcm10. (D) Plots of the data from (B) and (C). Values shown are the average of three independent experiments and the error bars show one standard deviation. The asterisks to the right of the gels indicate gel-shift of the substrate by CMG and Mcm10. See also Figure 2—figure supplements 2–3.
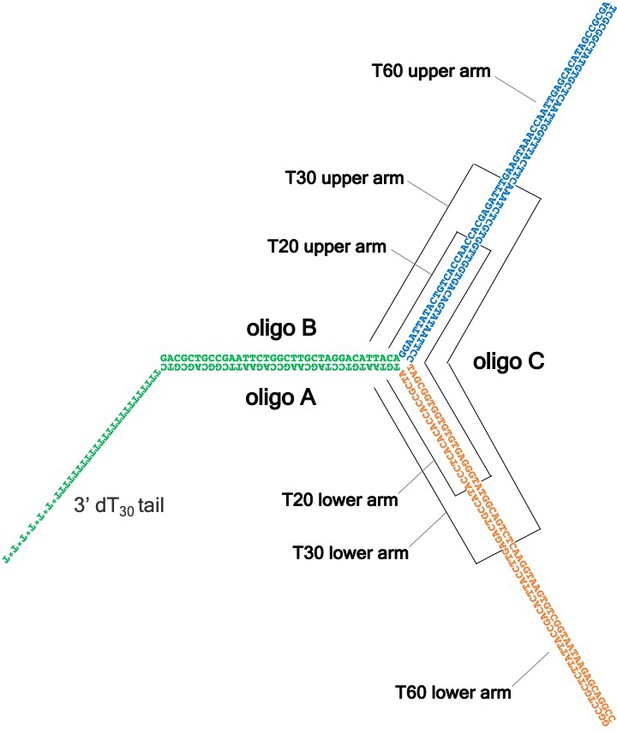
The T-shaped substrates shown in schematic form.
These substrates, used in Figure 2 to demonstrate duplex DNA translocation with force by CMG ± Mcm10, consist of three partially complementary oligonucleotides, denoted as A, B and C. Oligo A contains a 3’ dT30 tail required for CMG loading (Figure 2—figure supplement 2) and Oligo B forms a flush duplex with oligo A (green) to allow CMG to thread onto dsDNA without unwinding it. The duplex formed by oligos A and B is followed by two regions of non-complementarity (shown in blue and orange) that vary in length depending on the substrate, as indicated. These non-complementary arms are bound by a third oligo, C, which is complementary to the upper B arm (blue) and the lower A arm (orange). Active duplex translocation by CMG ± Mcm10 is required to dislodge the C oligo by breaking the base pairs in the upper and lower arms. Oligo C is radiolabeled in the experiments of Figure 2 and complete unwinding of oligo C is monitored by its altered migration in a native PAGE gel. See Supplementary file 1 for sequences of the oligos used to construct the T20, T30 and T60 substrates.
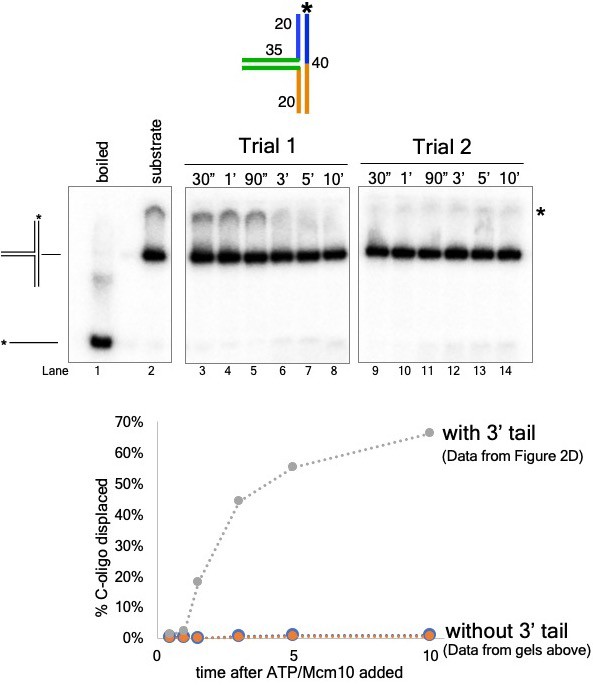
The 3’ dT30 tail is required for CMG loading onto the T-substrates.
The experiment from Figure 2C (CMG+Mcm10) was repeated using a T20 substrate with no 3’ dT30 loading tail by substituting oligo “T20A no 3’ tail” (see Supplementary file 1 for sequence) for oligo T20A. A schematic of the substrate is shown (top) as are gels from two separate trials of the experiment using this substrate (middle). The graph below shows a comparison of unwinding using the substrate with no 3’ tail to that of the 3’-tailed T20 substrate (data from Figure 2C, (D). The * next to the gels indicates gel-shift of the substrate by CMG+Mcm10.
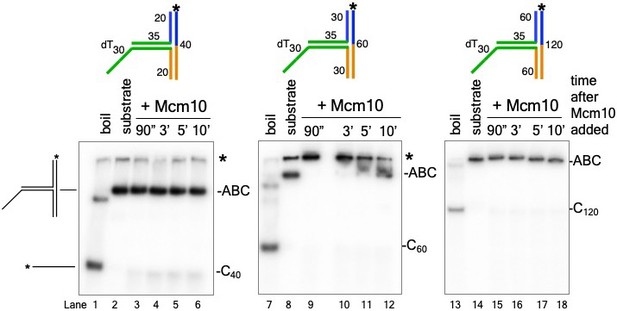
Mcm10 does not unwind the T-substrates without CMG.
The experiments of Figure 2C were repeated using Mcm10 (no CMG) with the T20 substrate (lanes 3-6), the T30 substrate (lanes 9-12) and the T60 substrate (lanes 15-18). The unreacted substrate is shown in lanes 2, 8, and 14 and lanes 1, 7, and 13 show the migration of the unwound product by boiling. The * next to the gels indicates gel-shift of the substrates by Mcm10.
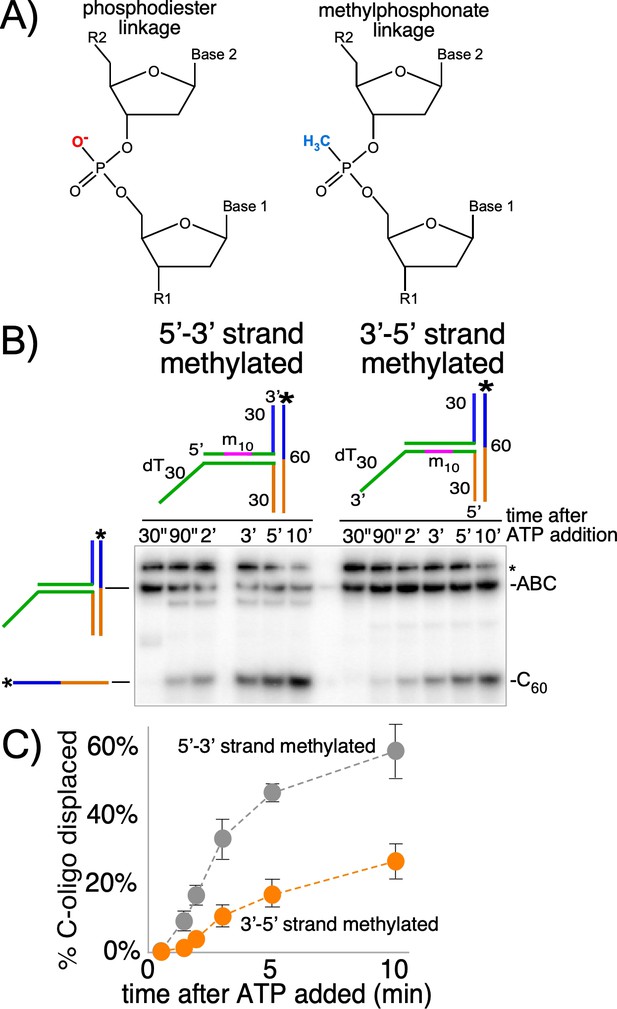
CMG+Mcm10 strand interactions during duplex translocation.
(A) Diagram of the naturally occurring negatively charged phosphodiester linkage in the phosphate backbone of a DNA chain (left) and the uncharged methylphosphonate linkages used in these experiments (right). (B) The experiment from Figure 2C (CMG+Mcm10) was repeated using a T30 substrate with 10 neutral methylphosphonate linkages (shown in pink in the schematics above the gel) in the 5’-3’ strand (left) or in the 3’-5’ strand (right) of the duplex. See Figure 3—figure supplement 2 and Supplementary file 1 for further details on the substrate. The * next to the gels indicates gel-shift of the substrate by CMG+Mcm10. (C) A plot of the data from (B) shows the averages of three independent trials using these substrates. The error bars show the standard deviations. Also see Figure 3—figure supplement 1.

Duplex strand ATPases contact only one strand while encircling duplex DNA.
(A) The E. coli clamp loader binds only one strand during action on dsDNA. (B) CMG motor domains bind ssDNA in a B-form conformation. (C) Little to no conformation change would be needed for CMG motors to bind B-form ssDNA and then slide onto dsDNA (flush end) and continue tracking on the same 3’−5’ strand.
© 2009 Elsevier. Panel A is reprinted from Simonetta et al., 2009, with permission from Elsevier. It is not covered by the CC-BY 4.0 licence and further reproduction of this panel would need permission from the copyright holder.
© 2018 Springer Nature. Panel B is reprinted from O'Donnell and Li, 2018, with permission from Springer Nature. It is not covered by the CC-BY 4.0 licence and further reproduction of this panel would need permission from the copyright holder and further reproduction of this panel would need permission from the copyright holder.
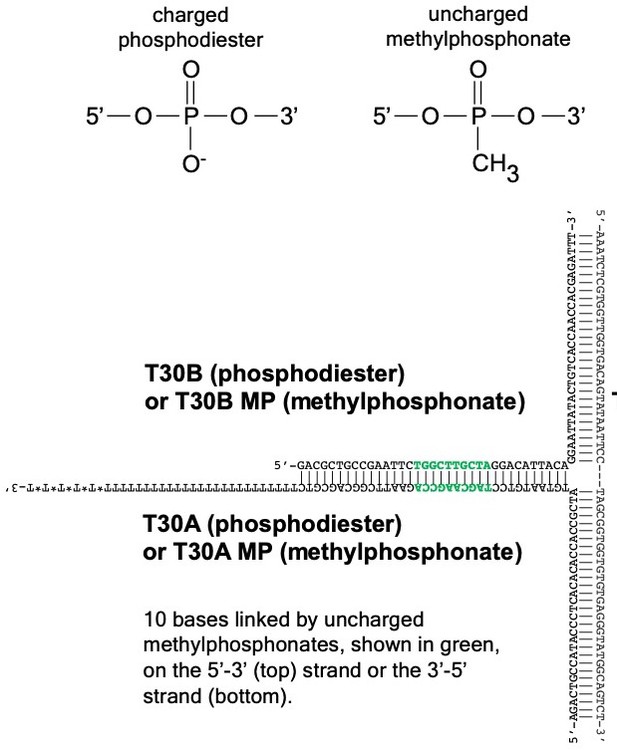
Design of substrates with methylphosphonate linkages in Figure 3.
To investigate how the CMG+Mcm10 motor interacts with the duplex DNA in its central channel, we used derivatives of oligos T30A or T30B (sequences in Supplementary file 1) in which a stretch of ten bases (shown in green in the DNA sequence) is connected by neutral methylphosphonate linkages (chemical schematic at top) instead of the normal negatively charged phosphodiester linkages on one strand or the other. The methylphosphonate derivate oligos are designated “T30A MP” and “T30B MP” (Supplementary file 1).
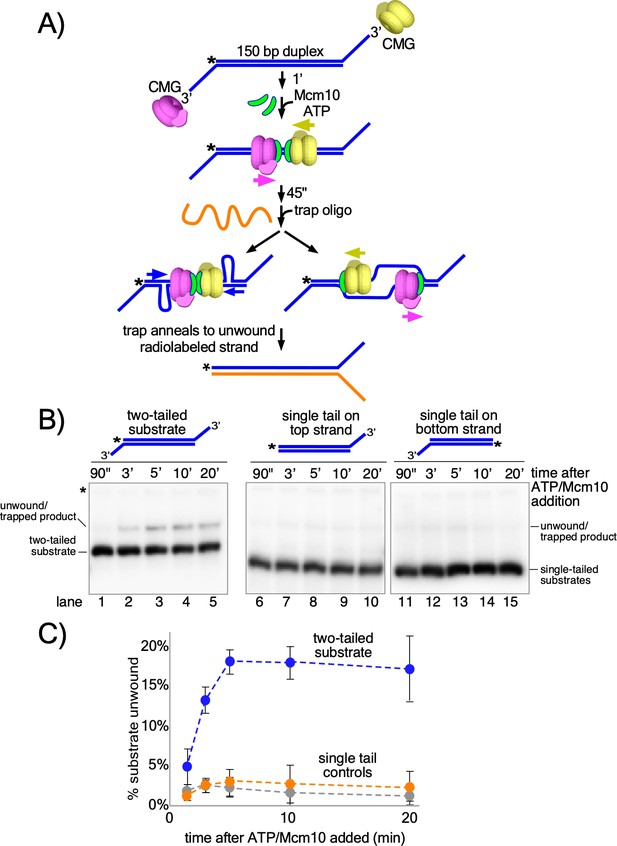
Head-to-head CMGs unwind dsDNA.
(A) Scheme of the reaction with the double tailed substrate, and two possible processes that could result in unwinding the DNA. The color scheme is the same as in Figure 1. CMG is mixed with the substrate on ice and then incubated at 30° C in the absence of ATP (top) to allow the reaction to reach temperature. 1’ later, ATP is added to allow CMGs to load onto the duplex in opposite orientations and block each other’s progress (middle). At the same time as ATP, Mcm10 is added to initiate the duplex unwinding reaction. 45’ later, an ssDNA trap oligo (orange) is added that quenches further CMG loading (Figure 4—figure supplement 1) and also anneals to the unwound radiolabeled product, creating a forked structure (bottom) that migrates at a distinct position in the gel from the substrate. Unwinding may occur either by: strand shearing (left arrow), or by strand expulsion (right arrow) to form CMGs that encircle ssDNA for conventional helicase activity. (B) Native PAGE gel time course of results using CMG + Mcm10. Lanes 1–5 show the reaction using the two-tailed substrate described in (A) while lanes 6–10 and 11–15 show control reactions using the same duplex with only a single tail at one end or the other. The migration of the substrates and unwound/trapped product are indicated to the left and right of the gel. (C) A plot of the data from (B) shows the averages of three independent trials using these substrates. The error bars show the standard deviations. See also Figure 4—figure supplements 2–3.
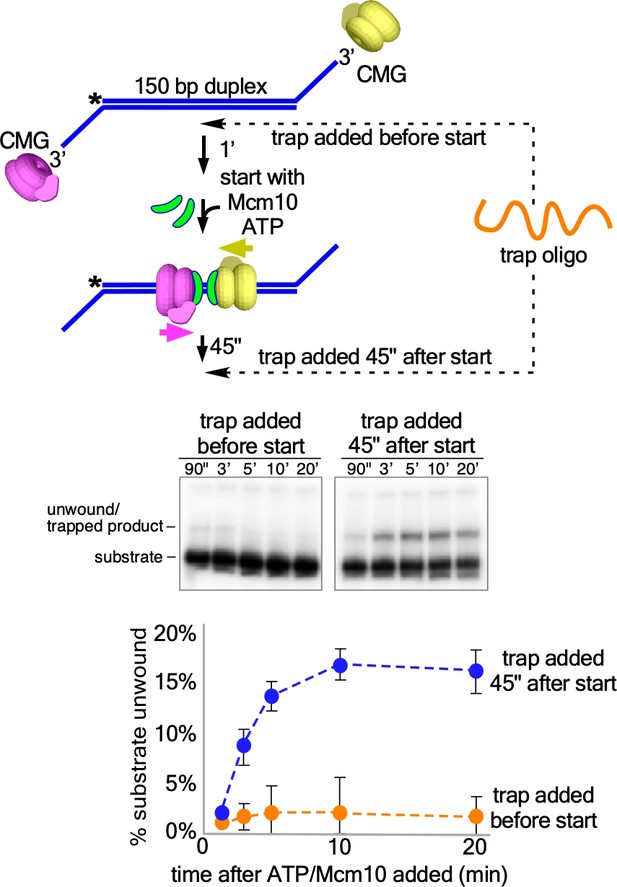
Addition of oligo trap prevents further loading of CMGM onto the two-tailed substrate.
The experiment in Figure 4B was repeated with addition of the oligo trap prior to addition of ATP/Mcm10 (left gel) or 45” after addition of ATP/Mcm10 (right gel) as in Figure 4B. The reaction scheme is shown above the gels, indicating the two different points of trap addition, and the data are plotted below. The values shown are the average of three independent experiments and the error bars show one standard deviation.
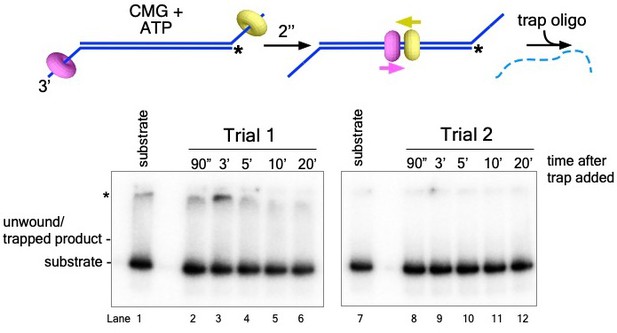
CMG alone does not unwind the origin-duplex substrate.
The experiment in Figure 4B, lanes 1-5, was repeated with CMG alone (no Mcm10). Because Mcm10 was not added, the experimental design was streamlined to an initial 2’ incubation with CMG, ATP, and the substrate (for CMG loading) after which the trap oligo was added to monitor product formation (scheme at top). Shown are the native PAGE gels from two separate experiments.
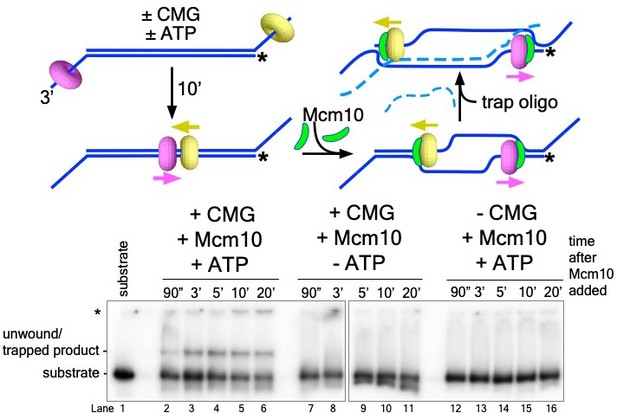
Unwinding of the origin-duplex substrate in Figure 4 requires ATP and CMG.
The experiment in Figure 4B, lanes 1-5, was repeated to demonstrate that unwinding of the two-tailed substrate by the complete system (lanes 2-6, with CMG, Mcm10 and ATP) is dependent upon both CMG (lanes 12-16, no CMG) and ATP (lanes 7-11, no ATP). This experiment also demonstrates that Mcm10 alone does not unwind the substrate (lanes 12-16). For these experiments, CMG was pre-incubated with the substrate in the presence of ATP (where indicated) for 10’ before the addition of Mcm10.
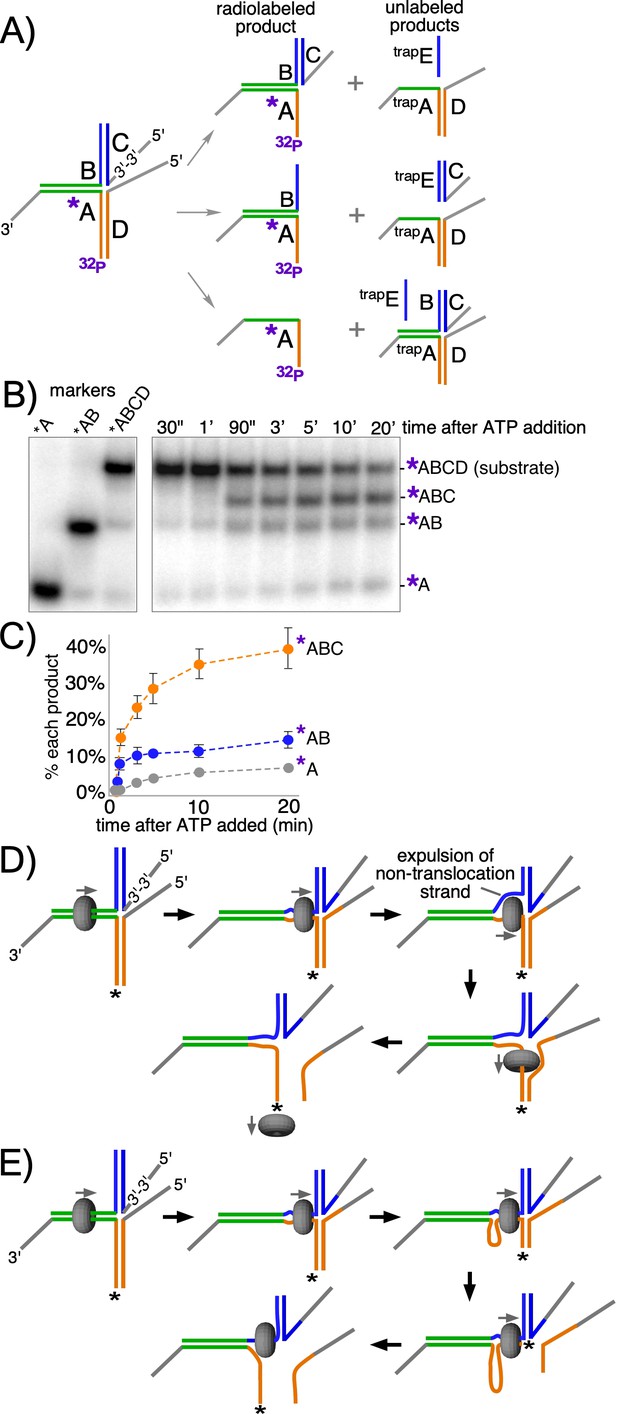
CMG + Mcm10 preferentially unwinds the tracking arm of a T-substrate.
(A) Scheme of the substrate and possible reaction products depending on the pathway of unwinding. Oligo A is radiolabeled at the 5’ end (32P) and oligo C contains a reverse polarity (3’−3’) linkage to create a 5’ flap that prevents CMG loading onto oligo C (Figure 5—figure supplement 2). Two traps are used in the reaction, each in 40-fold excess over the substrate: one is unlabeled oligo A (trapA, green and orange), which prevents unwound oligos D or B from re-annealing to unwound 32P-oligo A; the other is oligo E (trapE, blue), which is complementary to oligo C and thus prevents unwound oligo C from re-annealing to oligo B. These traps are shown annealed to the possible products (right). (B) Representative native PAGE gel of the assay time course using CMG + Mcm10. The migration positions of the indicated DNA makers are shown at the left and the time course of the reaction is on the right. (C) Plot of the data from (B). The values shown are the average of three independent experiments and the error bars show one standard deviation. (D) Scheme of the strand expulsion pathway. CMG partially unwinds the two heterologous arms, creating ssDNA (top middle) and then the non-tracking strand B is expelled to the exterior of the ring (top right), followed by CMG encircling ssDNA (bottom right) and finishing unwinding of the lower flapped arm (bottom left). (E) Schematic of the DNA shearing pathway. CMG melts both arms (top middle) but at unequal rates (top right) such that the lower arm with the tracking strand is fully unwound before the upper arm (bottom right), leaving CMG encircling the non-tracking ssDNA (bottom left). See also Figure 5—figure supplement 1.
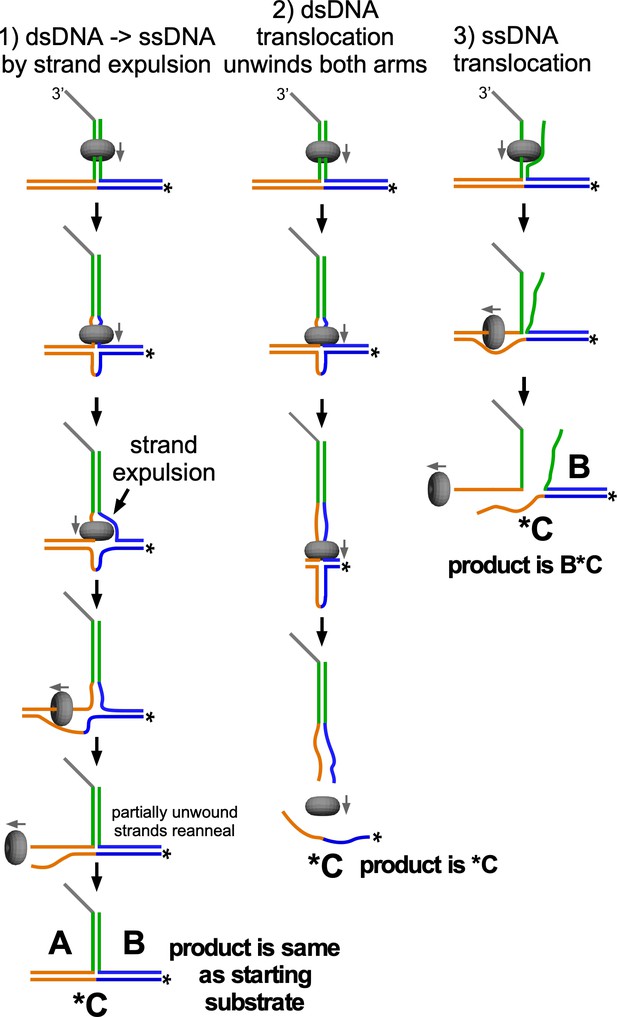
Description of why strand expulsion would not be observed using the substrate in Figure 2.
If CMG loads onto the 3’ end, threads onto the duplex (green), partially unwinds the arms, and then expels the non-tracking strand (1, left), the two non-homologous arms would be only partially unwound and could re-anneal after passage of CMG, re-forming the starting substrate. Duplex translocation without strand expulsion (2, middle) results in unwinding of both arms, displacing the radiolabeled C oligo. This is the only unwinding product observed in Figure 2. If CMG were to “lift up” the 5’ end of the green duplex and translocate exclusively along ssDNA (3, right), this would unwind the lower/left arm (orange) but not the upper/right arm (blue), and the radiolabeled C oligo would remain bound to the B oligo. This product was not observed in Figure 2. The trap used in Figure 2 is an excess of unlabeled oligo C, which prevents fully unwound oligo A or B from re-annealing to the unwound radiolabeled oligo C.
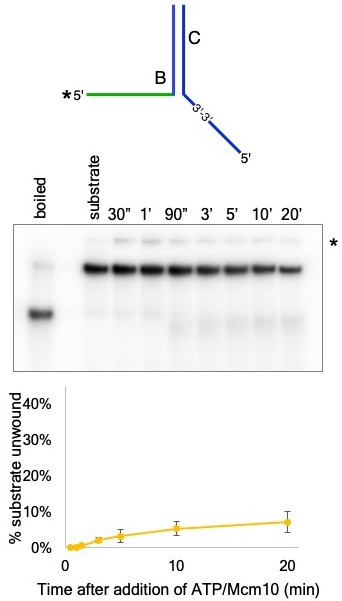
CMG+Mcm10 does not load onto the reverse polarity 5’ flap.
The oligo used to form a 5’ dT flap on the upper arm of the substrate used for Figure 5 is examined for its ability to prevent CMG+Mcm10 loading. The substrate used here was formed by annealing oligo T50C to radiolabeled T50B (schematic at top; See Supplementary file 1 for oligo sequences) and the reaction was performed exactly as described for the experiment of Figure 5. The * next to the gel indicates gel-shift of the substrate by CMG+Mcm10. The graph (bottom) shows the averages of three separate trials using this substrate. The error bars show the standard deviations.
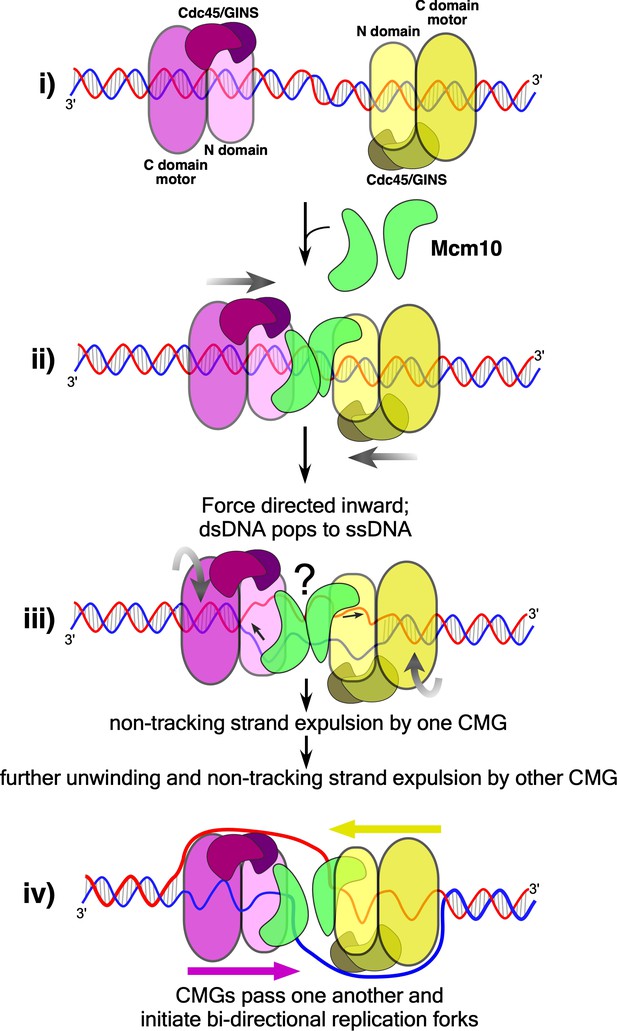
Model of CMG action at an origin.
(i) Two head-to-head CMGs (purple and yellow) are formed at an origin surrounding dsDNA (blue and red). (ii) Addition of Mcm10 (green) promotes movement of the two CMGs toward one another (gray arrows). (iii) Upon colliding, the two CMG motors continue to exert force on their respective tracking strands and may continue to rotate (curved gray arrows), placing tension on the dsDNA between them and causing it to unwind. The presence of Mcm10 promotes significant shearing and melting of dsDNA. Small black arrows show the direction of DNA strand movement through the respective CMGs. While ssDNA is illustrated both inside and between CMGMs, it remains possible that DNA unwinding occurs within the N-and C-tiers of individual CMGs or some other process. The question mark reflects this uncertainty. (iv) The unwinding of dsDNA by CMGM observed in the current study provides a sufficient length of ssDNA for CMGs to expel the non-tracking strand and thus transition from encircling dsDNA to ssDNA and track 3’−5’ on their respective ssDNA past one another to form bidirectional forks (see also Figure 1).
Additional files
-
Supplementary file 1
DNA oligonucleotides used in this study.
- https://doi.org/10.7554/eLife.46515.018
-
Transparent reporting form
- https://doi.org/10.7554/eLife.46515.019





