Neurexophilin4 is a selectively expressed α-neurexin ligand that modulates specific cerebellar synapses and motor functions
Figures
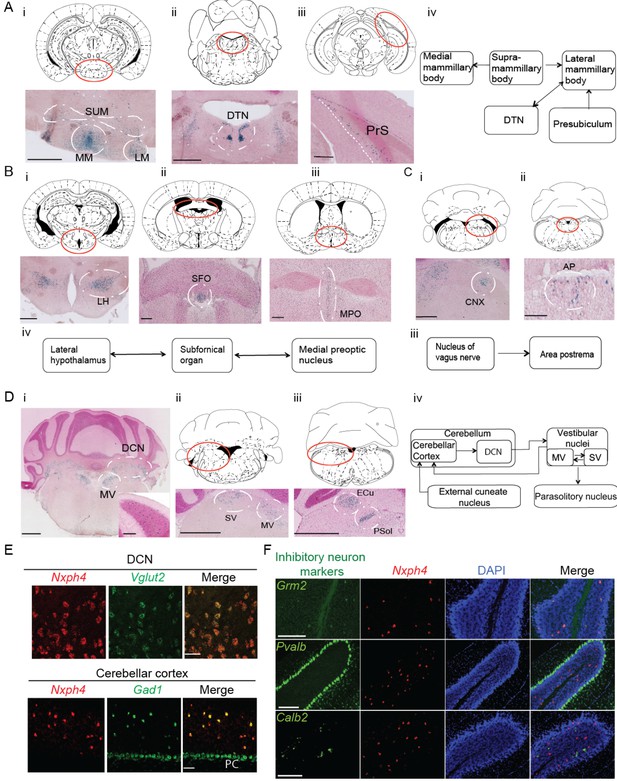
Nxph4 expression marks the components of select brain circuits.
(A–D) β-galactosidase staining of adult Nxph4βgeo/+ mice shows signals in mammillary body-related circuits (A), circumventricular organs (B–C), and cerebellar-vestibular circuits (D). Blue staining represents β-galactosidase activity. Top panels are stereotaxic maps adapted from the Paxinos and Franklin mouse brain atlas, with the red circle indicating the region for the image shown on the bottom panels. Aiv, Biv, Ciii, and Div illustrate the main connections among Nxph4+ regions in each circuit described. The inset in Di shows the blue staining in the cerebellar cortex granular layer. Scale bars: Ai, 500 μm; Aii, 100 μm; Aiii, 500 μm; Bi 500 μm; Bii, Biii 200 μm; Ci, Cii 100 μm; Di, Dii, Diii, 1 mm; Di inset, 100 μm. SUM, supramammillary body; MM, medial mammillary body; LM, lateral mammillary body; DTN, dorsal tegmental nucleus; PrS, presubiculum; LH, lateral hypothalamus; SFO, subfornical organ; MPO, median preoptic nucleus; CNX, nucleus of vagus nerve; AP, area postrema; DCN, deep cerebellar nuclei; MV, medial vestibular nucleus; SV, superior vestibular nucleus; ECu, external cuneate nucleus; Psol, parasolitary nucleus. (E) Double in situ staining of adult wild type mouse DCN and cerebellar cortex with probes against Nxph4, Vglut2 (an excitatory neuron marker), and Gad1 (an inhibitory neuron marker). PC: Purkinje cells. Scale bars: 50 μm. (F) Double in situ staining of mouse cerebellum shows that Nxph4 signals overlap with Grm2 (a Golgi cell marker) but not Pvalb or Calb2. Scale bars: 200 μm.
-
Figure 1—source data 1
Brain regions expressing Nxph4.
- https://doi.org/10.7554/eLife.46773.004
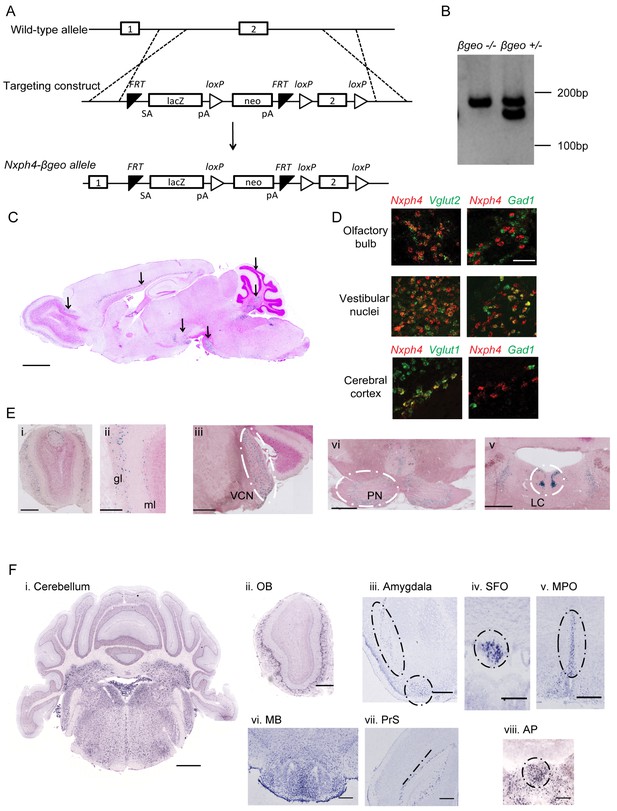
Generation of Nxph4-βgeo knock-in mouse and characterization of Nxph4 expression by β-galactosidase staining and in situ hybridization.
(A) Targeting scheme to generate Nxph4-βgeo allele. Doted lines indicate homologous recombination sites. ES cells with correct Nxph4-βgeo allele were obtained from KOMP. (B) Genotype analysis of tail DNA showed a 180 bp band from Nxph4 wild type allele and a 150 bp band from Nxph4-βgeo allele. PCR products were separated on 3% MetaPhor agarose gel. (C) A parasagittal section of an adult Nxph4βgeo/+ mouse brain processed for β-galactosidase staining, resulting in a blue reaction product where Nxph4 is expressed. Arrows indicate regions with strong Nxph4 expression. Scale bar: 1 mm. (D) Double in situ staining of adult wild type mice with probes against Nxph4, Vglut1/2 (excitatory neuron markers), and Gad1 (an inhibitory neuron marker). Nxph4 co-localized with Vglut2 in the olfactory bulb, vestibular nuclei, and cerebral cortex, but also showed overlapping signal with Gad1 in the vestibular nuclei. Scale bar, 100 μm. (E) β-galactosidase staining of adult Nxph4βgeo/+ mice shows Nxph4 expression in the glomerular layer (gl) and mitral cell layer (ml) of the main olfactory bulb, ventral cochlear nucleus (VCN), pontine nuclei (PN), and locus coeruleus (LC). Scale bars: Eii, 200 μm; others, 500 μm. (F) RNA in situ hybridization with a probe against Nxph4 detected Nxph4 in several regions of adult mouse brain. Scale bars: Cerebellum, 1 mm; OB (Olfactory Bulb), 500 μm; Amygdala, 100 μm; SFO, 200 μm; MPO, 200 μm; MB, 300 μm; PrS, 100 μm; AP, 100 μm.
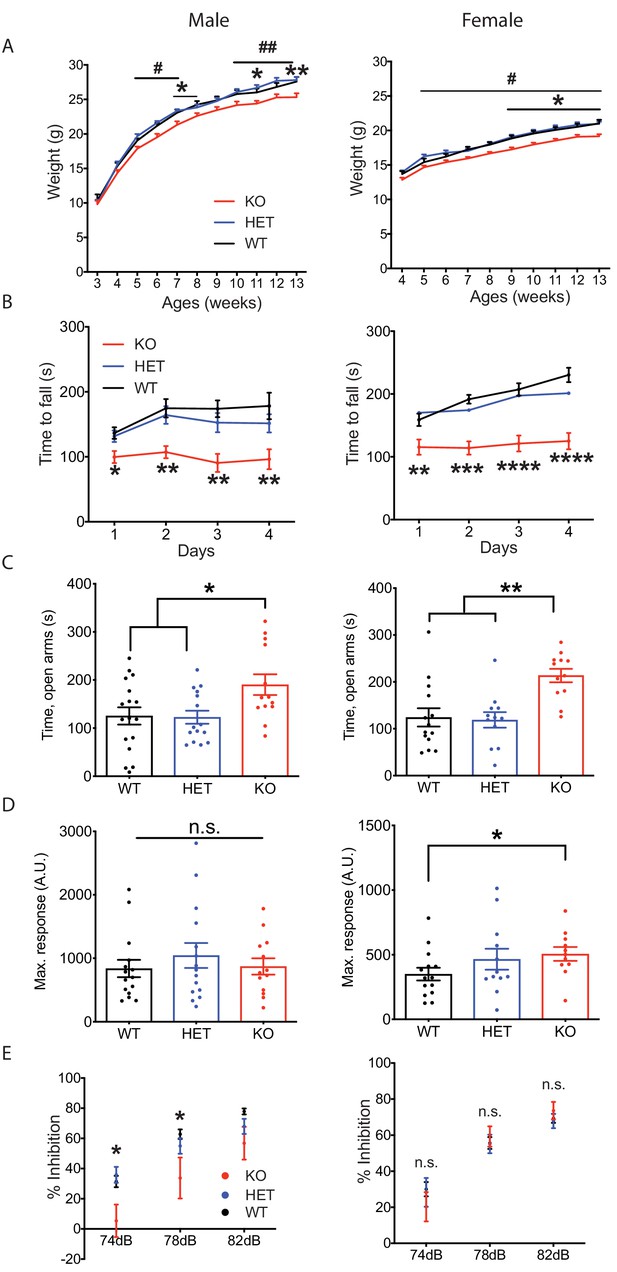
Nxph4 KO mice displayed multiple neurological deficits.
(A) Plots of weight as a function of age (male n = 13, female n = 16–18; #, difference between HET and KO; * difference between WT and KO). (B) Latency to fall from the accelerating rotarod plotted as a function of training days (male n = 10–12, female n = 12). (C) Average time spent in the open arms of the elevated plus maze (male n = 13–17, female n = 12–14). (D) Mean of response to the 120 dB acoustic stimulus (male n = 13–15, female n = 12–14). (E) Pre-pulse inhibition at 74 dB, 78 dB and 82 dB pre-pulses (male n = 13–15, female n = 12–14). Data are presented as mean ± SEM. *, # p<0.05; **p<0.01; ***p<0.001; ****p<0.0001; by one-way or two-way ANOVA.
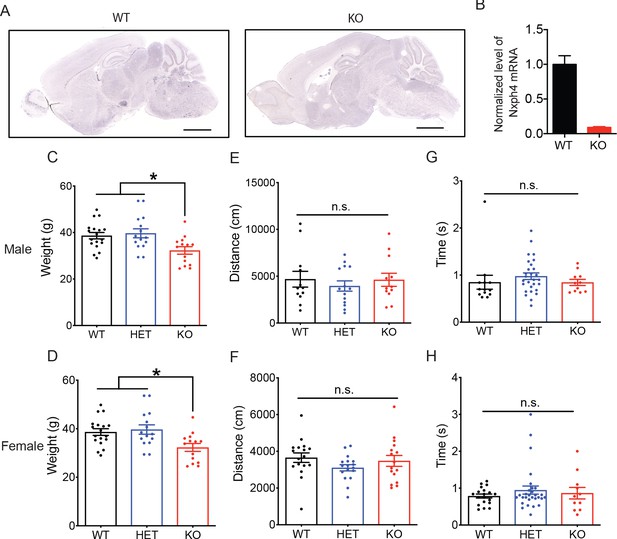
Nxph4 KO mice gained less weight but have normal locomotor and righting reflex functions.
(A) RNA in situ hybridization with a probe against Nxph4 only detected Nxph4 signal in WT but not KO mice. Scale bars: 2 mm. (B) RT-qPCR from cerebellar samples with specific primers for Nxph4 (n = 3). (C, D) Weight of male mice at 9 months of age and female mice at 13 months of age (male n = 14–18, female n = 10). (E, F) Total distance traveled in the open field assay (male n = 12–14, female n = 16–17). (G, H) Righting reflex assay was performed on postnatal day 10 mice. Y-axis shows the time that mice spent to reflex themselves after being placed on their back (male n = 11–28, female n = 11–29). Data are presented as mean ± SEM. *, p<0.05; **, p<0.01; n.s., not significant; by one-way ANOVA.
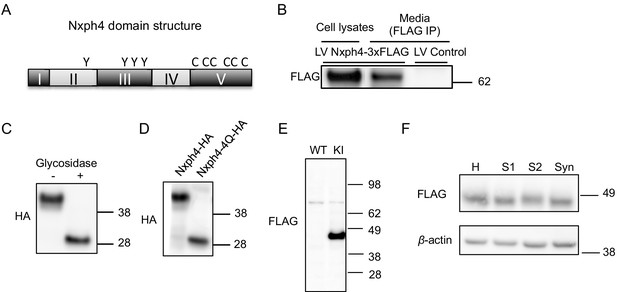
Nxph4 is a glycosylated protein that can be detected in the synaptosomes.
(A) A domain model of Nxph4 (adapted from Missler and Südhof, 1998). I: signal peptide; II: a variable domain; III: a conserved domain; IV: a linker region; V: C-terminal domain. Positions of N-glycosylation sequences are marked by letter Y, and the conserved cysteine residues are identified by the letter C. (B) Immunoblotting of samples from cultured cortical neurons that are infected with lentivirus expressing Nxph4-3xFLAG or the control lentivirus. Nxph4-3xFLAG was detected in the cell lysates as well as the media. (C) Treatment with glycosidase altered the electrophoretic motility of recombinant Nxph4-HA. (D) Nxph4-HA-4Q mutant has a smaller molecular mass compared with the wild type recombinant Nxph4-HA. (E) Immunoblotting analysis detects Nxph4-3xFLAG expression in the KI mouse synaptosomes. (F) Immunoblotting analysis of fractions derived from cerebellar synaptosomal preparation detects Nxph4-3xFLAG in the synaptosomes. β-actin was used as loading control. H: homogenate. S1 and S2 are successive supernatants in the synaptosomal preparation protocol. S2 is also the cytosolic fraction. Syn: synaptosomes.
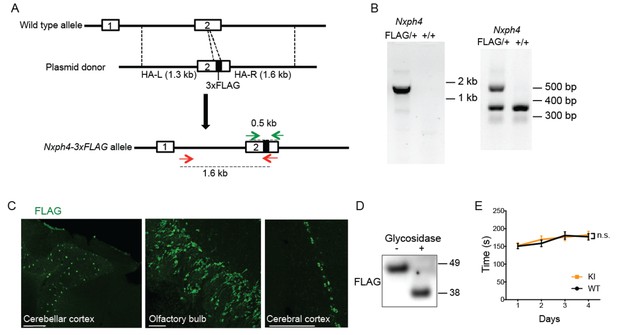
Generation and characterization of Nxph4-3xFLAG knock-in mice.
(A) A schematic diagram of the strategy to generate an Nxph4-3xFLAG knock-in allele. An sgRNA is designed to excise the coding region corresponding to the C-terminal region of Nxph4 third domain. In the donor vector, the 3xFLAG sequence is inserted after the sequence of Nxph4 third domain. The homologous arms of the donor vector are indicated as HA-L and HA-R. Two pairs of primers designed for genotyping are shown with green and red arrows. (B) Representative genotyping results using two different pairs of primers to distinguish Nxph4FLAG/+ from Nxph4+/+ mice. Left, the forward primer is outside of the HA-L and the reverse primer in within 3xFLAG region (red arrows in A). Right: the forward and the reverse primers flank the 3xFLAG sequence (green arrows in A). (C) Representative immunofluorescence images illustrate the expression of Nxph4-3xFLAG in specific brain regions. Scale bars: 200 μm. (D) Treatment with glycosidase altered the electrophoretic motility of Nxph4-3xFLAG. (E) Latency to fall from the accelerating rod (mixed male and female mice, n = 17–19). Data are presented as mean ± SEM. n.s., not significant; by two-way ANOVA.
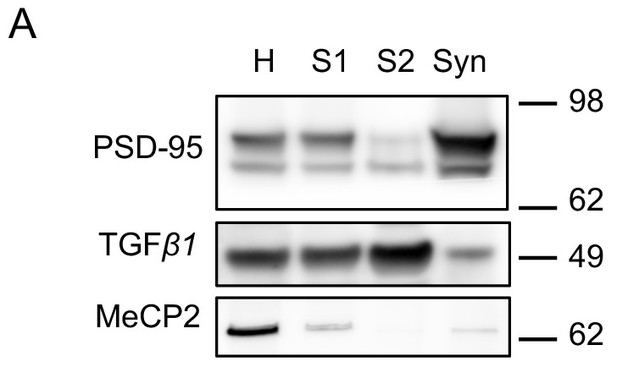
Validation of synaptosomes preparation.
(A) Immunoblot analysis of fractions derived from different steps of synaptosomal preparation. Synapse specific protein PSD-95 showed enriched signal in purified synaptosomes as expected. Somatic protein TGF-β1 is enriched in the cytosolic fraction S2. Nuclear protein MeCP2 is barely detectable in S1, S2 or Syn. H: homogenate. S1 and S2 are successive supernatants in the synaptosomal preparation protocol. S2 is also the cytosolic fraction. Syn: synaptosomes.
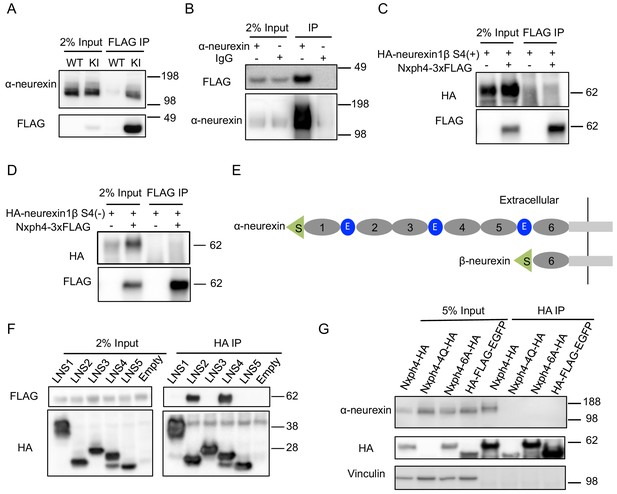
Nxph4 forms a complex with α-neurexin in vivo.
(A) Synaptosomes (tissue used: olfactory bulb, hypothalamus, midbrain, hindbrain, and the cerebellum) from Nxph4-3xFLAG KI or WT (negative control) mice were precipitated with an antibody against FLAG. Bound proteins as well as 2% input were analyzed by immunoblotting with anti-FLAG and anti-α-neurexin antibodies as indicated. (B) Brain lysates from Nxph4-3xFLAG KI mice were precipitated with an anti-α-neurexin antibody. Elution and 2% input were analyzed by immunoblotting with anti-α-neurexin and anti-FLAG antibodies. IgG was used as negative control. (C, D) Nxph4-3xFLAG and HA-neurexin1β S4(+) (with the insertion of splicing site 4, C) or HA-neurexin1β S4(-) (without the insertion of splicing site 4, D) were co-expressed in HEK293T cells. Cell lysates were precipitated with an anti-FLAG antibody. Bound proteins were analyzed by immunoblot showing pulling down of Nxph4-3xFLAG but not HA-neurexin1β. Cells transfected with HA-neurexin1β alone were used as negative control. (E) Schematic drawing of the extracellular domain structure of α- and β-neurexins. α-neurexin contains 6 LNS domains interspersed by 3 EGF-like repeats. β-neurexin has a single LNS6 domain. S: signal peptide; 1–6: LNS1-6; E: EGF-like domain. (F) Nxph4-3xFLAG was co-expressed with individual α-neurexin specific LNS domains in HEK293T cells. Culture media was precipitated by an anti-FLAG antibody. LNS2 and LNS4 were co-precipitated with Nxph4-3xFLAG. (G) Cultured primary cortical neurons overexpressing wild type or mutant Nxph4-HA were subjected to co-IP with an anti-HA antibody. Elution and 5% input were analyzed by immunoblotting with anti-HA, anti-α-neurexin, and anti-vinculin antibodies. Wild type and Nxph4 mutants were fused with mCherry. Nxph4-4Q-HA was not detectable in 5% input and was only detected as a faint band in the IP sample.
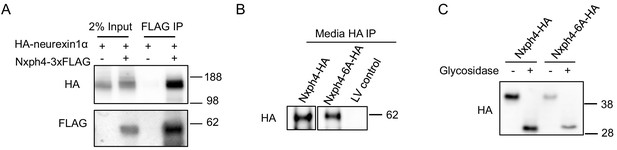
Nxph4 interacts with neurexin1α in vitro and the Nxph4-6A is secreted and glycosylated.
(A) HA-neurexin1α and Nxph4-3xFLAG were co-expressed in HEK293T cells. Cell lysates were precipitated with beads conjugated with an anti-FLAG antibody. Bound proteins as well as input were analyzed by immunoblotting. (B) Media from primary cultured neurons over-expressing wild type and mutant Nxph4 was collected. After precipitation by beads conjugated with an anti-HA antibody, bound proteins were analyzed by immunoblotting. (C) Both wild type Nxph4 and Nxph4-6A showed similar reduction of molecular mass after glycosidase treatment.
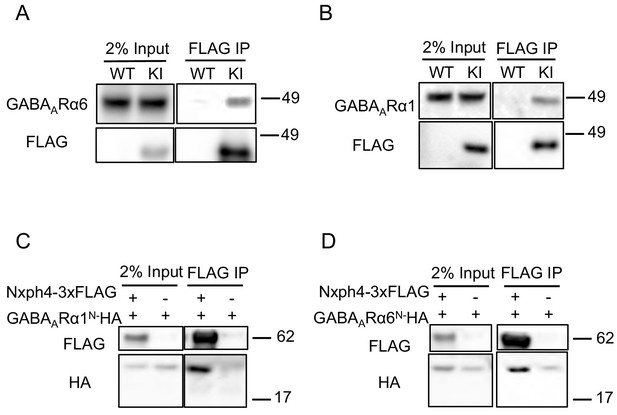
Nxph4 interacts with GABAARs.
(A,B) Cerebellar synaptosomes from three Nxph4-3xFLAG KI or WT (negative control) mice were precipitated with an antibody against FLAG. Bound proteins as well as 2% input were analyzed by immunoblotting with anti-FLAG and anti-GABAARα6 (A), or anti-GABAARα1 (B) antibodies as indicated. (C) Nxph4-3xFLAG and the N-terminal extracellular domain of GABAARα1 were co-expressed in HEK293T cells. Cell lysates were precipitated with an anti-FLAG antibody and elution was analyzed by immunoblot showing the precipitation of Nxph4-GABAARα1N complex. Cells transfected with GABAARα1N-HA alone were used as negative control. (D) Nxph4-3xFLAG binds to the N-terminal extracellular domain of GABAARα6.
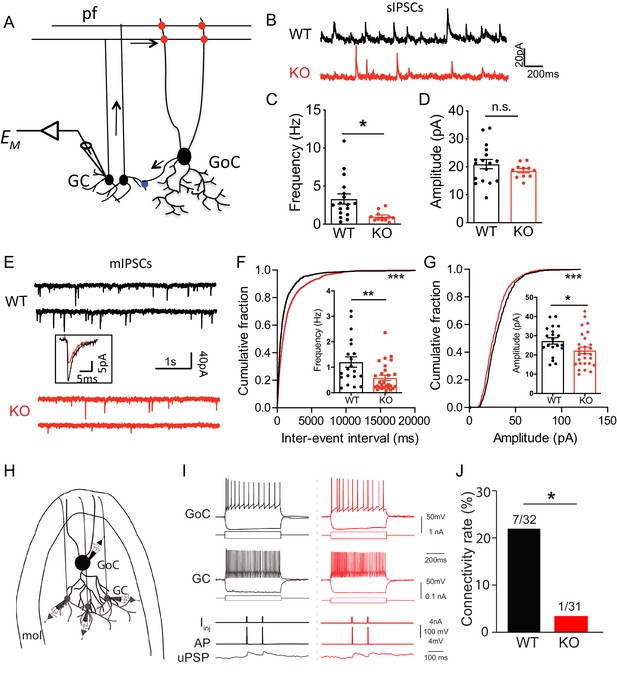
Loss of Nxph4 reduced inhibition onto cerebellar granule cells.
(A) A simplified diagram illustrating the inhibitory circuit of cerebellar granular layer and single-cell recording experimental design. GC: granule cells; GoC: Golgi Cells; pf: parallel fibers. Arrows indicate the direction of information flow. (B) Representative traces of spontaneous IPSC recorded from the WT and KO cerebellar granule cells. (C–D) Statistical analysis of sIPSC frequency (C, Mann-Whitney U test) and amplitude (D, t-test) (n = 11–17 cells from 4 WT and 4 KO mice). (E) Representative traces of miniature IPSCs recorded from the WT and KO cerebellar granule cells. Insect is the averaged traces. (F–G) Statistical analysis of mIPSC frequency (F, Mann-Whitney U test) and amplitude (G, t-test). Cumulative probability plots were analyzed by Kolmogorov-Smirnov test (n = 20–28 cells; 6–7 mice). (H) Multi-channel recording configuration. A GoC was first identified and recorded in the granular layer, and then nearby GCs were sequentially recorded to test the connectivity in GOC->GC while inducing action potentials in the GoC. mol: molecular layer. (I) Samples of connected GoC->GC pairs in WT and KO, showing their firing patterns and unitary GABAergic postsynaptic poetntials (uPSP). The recordings were performed in the presence of AMPA and NMDA receptor antagonists to only detect GABAergic synaptic transmission. Given the high-chloride internal solution being used, GABAergic synaptic potentials were depolarized at the resting membrane potentials. Iinj: injected current; AP: action potential. (J) The connectivity rate (GoC->GC) was significantly lower in KO compared with WT (4 WT and 3 KO mice). Chi-square test. Data are presented as mean ± SEM. n.s., not significant; *p<0.05; ***p<0.001.
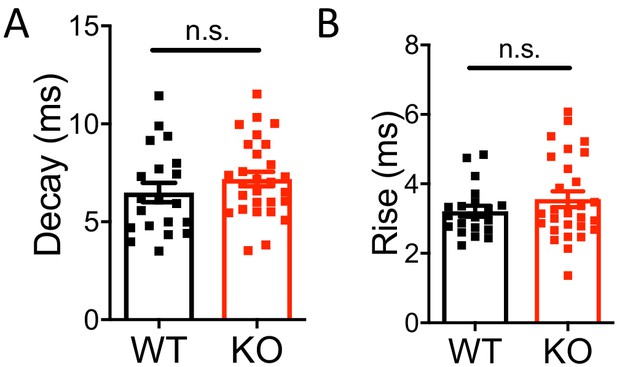
Miniature IPSC decay and rise time were not affected in Nxph4 KO mice.
(A–B) Statistical analysis of mIPSC decay (A, t-test) and rise time (B, Mann-Whitney U test). Data are presented as mean ± SEM. n.s., not significant.
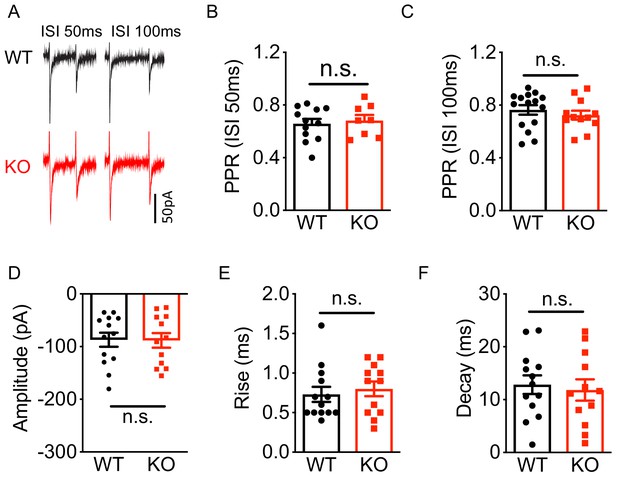
Deletion of Nxph4 KO did not affect mossy fibers-granule cell EPSC.
(A) Sample traces of granule cell EPSC, evoked by stimulating mossy fibers, exhibited paired-pulse depression at ISI (inter-stimulus interval) 50 ms and ISI 100 ms from WT and KO mice. (B–F) Summary of eEPSC paired-pulse ratio (EPSC2/EPSC1, PPR) at ISI 50 ms and ISI 100 ms, amplitude, rise time, and decay (n = 5–7 mice). Data are presented as mean ± SEM. n.s., not significant; by Mann-Whitney U test or t-test.
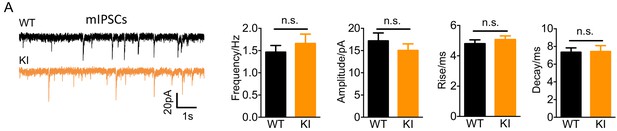
Nxph4-3xFLAG KI mice showed normal mIPSC.
(A) Miniature IPSCs recorded from the KI and WT cerebellar granule cells. From left to right: representative mIPSC traces, mean mIPSC frequency, amplitude, rise, and decay (n = 11–14 cells; 5mice). Data are presented as mean ± SEM. n.s., not significant; by t test.
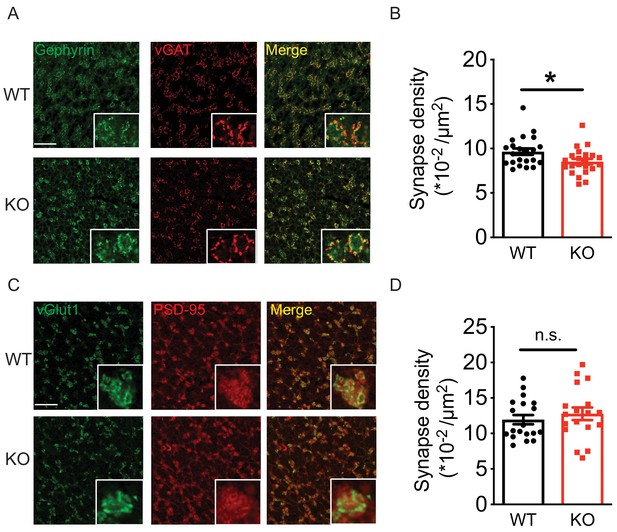
Loss of Nxph4 reduced Golgi-granule inhibitory synapse number.
(A) Gephyrin and vGAT staining in the cerebellar granular layer indicates Golgi-granule inhibitory synapses. (B) Quantification of puncta co-expressing gephyrin and vGAT as an indicator of inhibitory synapse number. (C) vGlut1 and PSD-95 staining in the cerebellar granular layer indicates mossy fiber-granule cell excitatory synapses. (D) Quantification of puncta co-expressing vGlut1 and PSD-95 as an indicator of excitatory synapse number. Data are presented as mean ± SEM. n.s., not significant; *p<0.05; by t test (B) or Mann-Whitney U test (D).
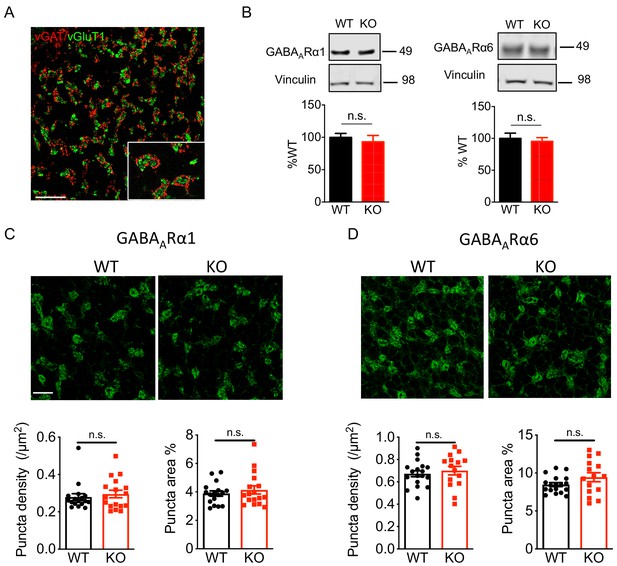
Nxph4 KO mice showed normal expression and localization of GABAARs.
(A) In the cerebellar granular layer, vGAT marks the Golgi cell inhibitory terminals and vGlut1 marks the mossy fiber endings. Scale bar: 20 μm. (B) Immunoblotting analysis of GABAARα1/6 in the cerebellar synaptosomes of the WT and Nxph4 KO mice (n = 6–8). (C–D) Top: Immunofluorescence staining of GABAARα1 (C) and GABAARα6 (D) in the cerebellar granular layer. Scale bar: 20 μm. Bottom: Quantification of GABAAR cluster density and size (n = 17 sections; 3 mice). Data are presented as mean ± SEM. n.s., not significant; by t test.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Mouse monoclonal anti-FLAG M2 | Sigma-Aldrich | Cat# F1804 RRID:AB_262044 | (1:1000) |
| Antibody | Rabbit polyclonal anti-pan Neurexin-1 | Millipore | Cat# ABN161 RRID:AB_10917110 | (1:1000) |
| Antibody | Mouse monoclonal anti-HA1.1 | Bioledgend | Cat# 901513, RRID:AB_2565335 | (1:5000) |
| Antibody | Mouse monoclonal anti-Vinculin | Sigma-Aldrich | Cat# V9131, RRID:AB_477629 | (1:10000) |
| Antibody | Rabbit polyclonal anti-GABAARα1 | Synaptic System | Cat# 224 203, RRID:AB_2232180 | (1:1000) |
| Antibody | Rabbit polyclonal anti-GABAARα6 | Synaptic System | Cat# 224 603, RRID:AB_2619945 | (1:1000) |
| Antibody | Mouse monoclonal anti-Vglut1 | Millipore | Cat# MAB5502, RRID:AB_262185 | (1:1000) |
| Antibody | Guinea pig polyclonal anti-vGAT | Frontier Institute co. Ltd | Cat#: VGAT-GP-Af1000, RRID: AB_2571624 | (1:1000) |
| Antibody | Mouse monoclonal anti-beta Actin | Abcam | Cat#: ab20272 RRID:AB_445482 | (1:2000) |
| Antibody | Rabbit polyclonal anti-TGF beta 1 | Abcam | Cat#: ab92486, RRID:AB_10562492 | (1:1000) |
| Antibody | Rabbit polyclonal anti-MeCP2 | Zoghbi Lab | #0535 | (1:1000) |
| Antibody | Mouse monoclonal anti-Gephyrin | Synaptic system | Cat#: 14701, RRID:AB_887717 | (1:1000) |
| Antibody | Rabbit polyclonal anti- PSD-95 | Cell signaling | Cat # 2507, RRID:AB_561221 | (1:1000) |
| Recombinant DNA reagent | pLenti-mCherry | Addgene | Cat# 36084, RRID:Addgene_36084 | |
| Recombinant DNA reagent | pLenti-Nxph4-3xFLAG-mCherry | This paper | N/A | Results subsection‘Nxph4 is a secreted glycoprotein’ |
| Recombinant DNA reagent | pLenti--Nxph4-4Q-HA-mCherry | This paper | N/A | Results subsection ‘Nxph4 is a secreted glycoprotein’ |
| Recombinant DNA reagent | pLenti--Nxph4-6A-HA-mCherry | This paper | N/A | Results subsection ‘Nxph4 is a secreted glycoprotein’ |
| Recombinant DNA reagent | pCMV-Nxph4-HA | This paper | N/A | Results subsection‘Nxph4 is a secreted glycoprotein’ |
| Recombinant DNA reagent | pCMV-Nxph4-4Q-HA | This paper | N/A | Results subsection‘Nxph4 is a secreted glycoprotein’ |
| Recombinant DNA reagent | pCMV-Nxph4-6A-HA | This paper | N/A | Results subsection‘Nxph4 interacts with α-neurexin in vivo’ |
| Recombinant DNA reagent | pCAG-HA-Nrxn1β S4(+) | Addgene | Cat# 59410, RRID:Addgene_59410 | |
| Recombinant DNA reagent | pCAG-HA-Nrxn1β S4(-) | Addgene | Cat# 59409, RRID:Addgene_59409 | |
| Recombinant DNA reagent | pCAG-HA-Nrxn1α | Addgene | Cat# 58266, RRID:Addgene_58266 | |
| Recombinant DNA reagent | pCAG-HA-Nrxn1α-LNS1 | This paper | N/A | Results subsection‘Nxph4 interacts with α-neurexin in vivo’ |
| Recombinant DNA reagent | pCAG-HA-Nrxn1α-LNS2 | This paper | N/A | Results subsection‘Nxph4 interacts with α-neurexin in vivo’ |
| Recombinant DNA reagent | pCAG-HA-Nrxn1α-LNS3 | This paper | N/A | Results subsection‘Nxph4 interacts with α-neurexin in vivo’ |
| Recombinant DNA reagent | pCAG-HA-Nrxn1α-LNS4 | This paper | N/A | Results subsection‘Nxph4 interacts with α-neurexin in vivo’ |
| Recombinant DNA reagent | pCAG-HA-Nrxn1α-LNS5 | This paper | N/A | Results subsection ‘Nxph4 interacts with α-neurexin in vivo’ |
| Recombinant DNA reagent | pCMV-GABARα1N-HA | This paper | N/A | Results subsection ‘Nxph4 interacts with α-neurexin in vivo’ |
| Recombinant DNA reagent | pCMV-GABARα6N-HA | This paper | N/A | Results subsection ‘Nxph4 interacts with α-neurexin in vivo’ |
| Chemical compound, drug | X-Gal | Thermo Fisher | Cat# 15520034 | |
| Chemical compound, drug | Lipofectamine 2000 | Thermo Fisher | Cat# 11668500 | |
| Commercial assay, kit | Antigen Unmasking Solution (citrate based) | Vector Laboratories | Cat# H-3300 | |
| Commercial assay, kit | Avidin/Biotin Blocking Kit | Vector Laboratories | Cat# SP-2001 | |
| Commercial assay, kit | TSA Kit | Invitrogen | Cat# T-20932 | |
| Commercial assay, kit | Papain Dissociation System Kit | Worthington Biochemical Corp. | Cat# LK003150 | |
| Commercial assay, kit | PNGase F | NEB | Cat# P0704S | |
| Commercial assay, kit | miRNeasy Mini Kit | Qiagen | Cat#: 217004 | |
| Commercial assay, kit | M-MLV Reverse Transcriptase | Thermo Fisher | Cat#: 28025013 | |
| Commercial assay, kit | iTaq Universal SYBR Green SuperMix | BIO-RAD | Cat#: 1725124 | |
| Cell line (Homo sapiens) | HEK293T cells | ATCC | RRID:CVCL_0063 | |
| Strain, strain background (Mus musculus) | Mouse: FVB | The Jackson Laboratory | Stock No: 001800 | |
| Strain, strain background (Mus musculus) | Mouse: Nxph4 βgeo+/- | This paper | N/A | The Jackson Laboratory: 033791 |
| Strain, strain background (Mus musculus) | Mouse: Nxph4 FLAG/FLAG | This paper | N/A | The Jackson Laboratory: 033792 |
| Sequence-based reagent | Nxph4βgeo+/- genotyping forward primer: 5’-AAAGACTAGCAGACGCAGCA | This paper | N/A | Materials and methods subsection ‘Nxph4-βgeo mouse’ |
| Sequence-based reagent | Nxph4βgeo+/- genotyping reverse primer: 3’-CCCTAACTCCCCCAAACAGA | This paper | N/A | Materials andmethods subsection ‘Nxph4-βgeo mouse’ |
| Sequence-based reagent | Nxph4 FLAG/FLAGgenotyping forward primer: 5’-TCAAGTTCTCGCTGTTGGTG | This paper | N/A | Materials and methods subsection‘Nxph4FLAG/FLAG knock-in mouse’ |
| Sequence-based reagent | Nxph4 FLAG/FLAG genotypingreverse primer: 3’-TTCCACGTGGCAATTAAAAG | This paper | N/A | Materials and methods subsection‘Nxph4FLAG/FLAG knock-in mouse’ |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.46773.019





