Cryo-EM of dynein microtubule-binding domains shows how an axonemal dynein distorts the microtubule
Figures
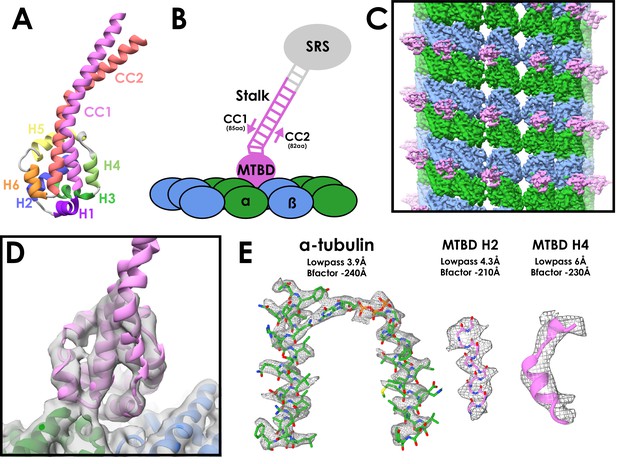
Cryo-EM Structure of the cytoplasmic dynein-1 microtubule-binding domain.
(A) Crystal structure of the cytoplasmic dynein-1 MTBD in the low-affinity β +registry (PDB 3ERR) coloured by helix. (B) Schematic of the MTBD constructs used for structure determination. A globular seryl-tRNA synthetase (SRS, grey) has a protruding coiled-coil to which 12-heptads of the dynein stalk is fused (pink). CC1 is three residues longer than CC2 to force the stalk into the high-affinity α registry, allowing the MTBD to bind to the microtubule (α-tubulin in green, β-tubulin in blue) (C) Reconstruction of the cytoplasmic dynein-1 MTBD (pink) bound to microtubule (α-tubulin in green, β-tubulin in blue), lowpass-filtered to 5 Å. (D) New models for the cytoplasmic dynein MTBD (pink) and tubulin (α-tubulin in green, β-tubulin in blue) was refined into the cryo-EM density (lowpass filtered to 5 Å) (E) Representative density of different regions of the map, filtered and sharpened according to local resolution.
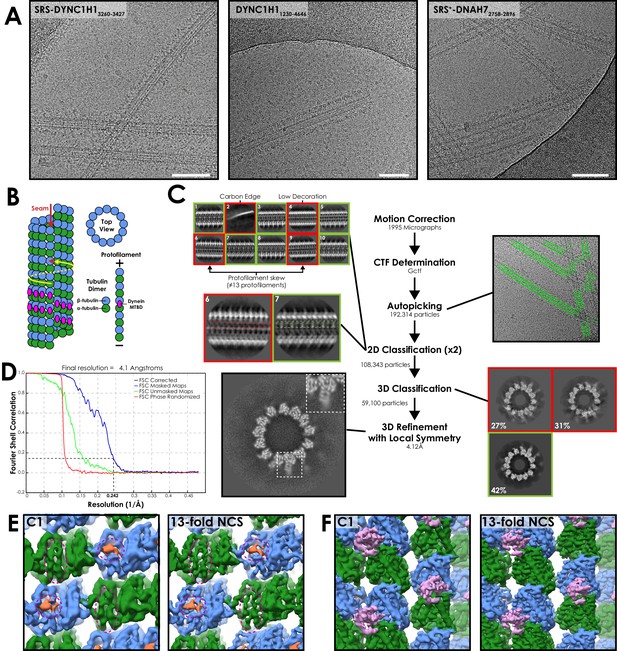
Processing workflow of the SRS-DYNC1H13260-3427 structure.
(A) Representative cryo-EM images from each of the three datasets presented in this work. White bar corresponds to 100 nm (B) Cartoon depicting a 13-protofilament microtubule with a seam. α-tubulin (dark green) an β-tubulin (light green) form a constitutive dimer that polymerises end-on-end as a protofilament. 13-protofilaments are arranged in a ring to create a tube. Lateral interactions are like-for-like (α-to-α and β-to-β, yellow arrow) except at the seam (red arrows). This asymmetry means conventional helical averaging cannot be performed. The dynein MTBD (magenta) binds to the intradimer interface. (C) Processing workflow for the cytoplasmic dynein-1 MTBD structure. Motion correction and CTF determination were performed as standard. Autopicking was optimised to pick all microtubules. 2D and 3D classification was used to remove bad particles. In 2D classification, bad classes included those with incorrect features (e.g. the carbon edge, class 2), low decoration on the side of the microtubule (class 4) or identifiable non-13 protofilament architecture (classes 6 and 9). The latter can be distinguished on account of the unique lack of protofilament twist in 13-protofilament microtubules (that is, protofilaments run exactly parallel to the microtubule long axis). This means that the repeating patterns in the classes should run parallel (class 7, expanded), whereas particles with protofilament twist will have converging or diverging patterns along the microtubule (class 6, expanded). This method is then supplemented with 3D classification to remove microtubule architectures with small protofilament skews that may not show up in short 2D segments, as well as particles with poor contrast or high-resolution detail. Refinement of the good class from 3D classification followed by 3D refinement with local symmetry resulted in a 4.1 Å map (FSC0.143 cut-off, gold standard). (D) Fourier shell correlation (FSC) curve for the refined SRS-DYNC1H13260-3427 structure. (E) Luminal view of the protofilament either side of the seam in the final C1 (asymmetric) and final symmetrized SRS-DYNC1H13260-3427 maps. α- and β-tubulin (dark and light green) can be differentiated by the length of the S9-S10 loop (dotted circles). In α-tubulin the loop is extended, whereas in β-tubulin it is short, making room for the taxol binding site (taxol in blue). The asymmetry at the seam is clearly defined in both the C1 and symmetrized map. (F) Exterior view of the protofilament either side of the seam in the final C1 (asymmetric) and symmetrized SRS-DYNC1H13260-3427 maps. MTBD density (magenta) undergoes an additional rise across the seam, reconfirming the definition of the seam.
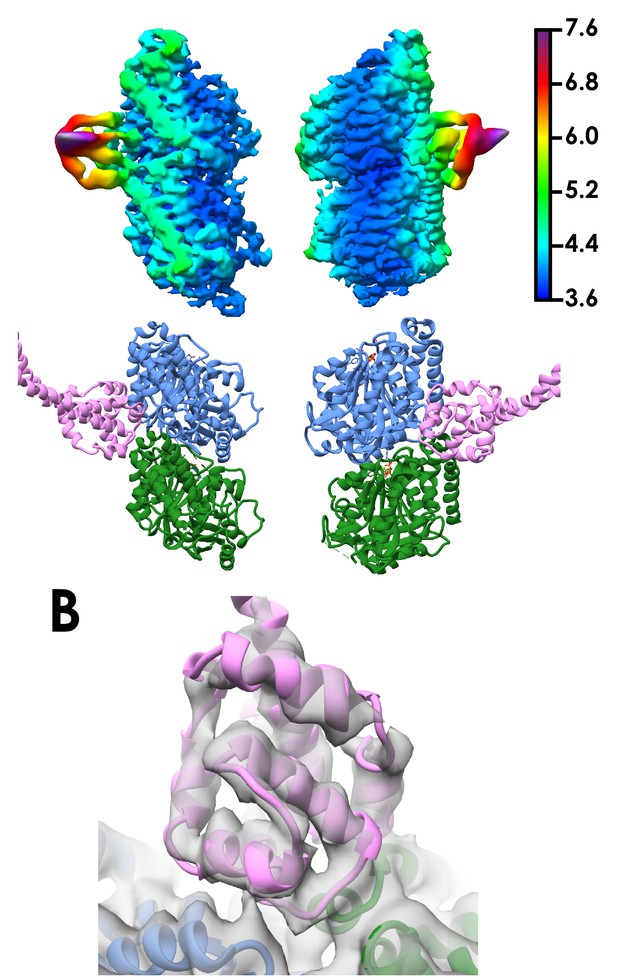
Local resolution of the cytoplasmic dynein-1 MTBD SRS structure.
(A) Density corresponding to one tubulin dimer and the cytoplasmic dynein-1 MTBD coloured and filtered according to local resolution as determined in relion_postprocess. Density (top) and the corresponding views in the model (bottom) are shown. (B) Orthogonal view of Figure 1D, showing the refined cytoplasmic dynein-1 MTBD docked into density lowpass filtered to 5 Å.
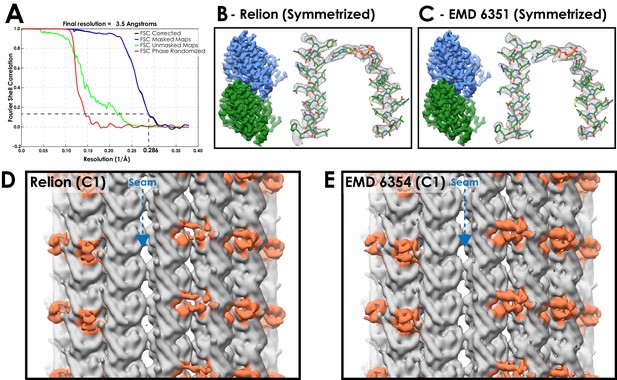
A comparison between microtubule reconstructions using Relion or previous methods.
(A) FSC curves for EMPIAR dataset 10030 (EB3 decorated microtubules) processed in the Relion pipeline following symmetrization (FSC0.143 cut-off, gold-standard). (B) Density from the Relion refined EB3-microtubule dataset, following symmetrization. Mesh representation of alpha-tubulin H4 +H7 and nucleotide. (C) Corresponding views of the published map processed from the same dataset but refined with the seam-finding protocol (EMD 6351 - Zhang and Nogales, 2015). (D) Density at the seam of the C1 (asymmetric) reconstruction from Relion (EB3 density coloured blue). The EB3 density close to the seam is weak, suggesting that the seam is poorly aligned in some particles (E) Corresponding view of a C1 reconstruction using the seam-finding protocol refinement (EMD 6354). The EB3 density is as strong around the seam as in neighbouring protofilaments.
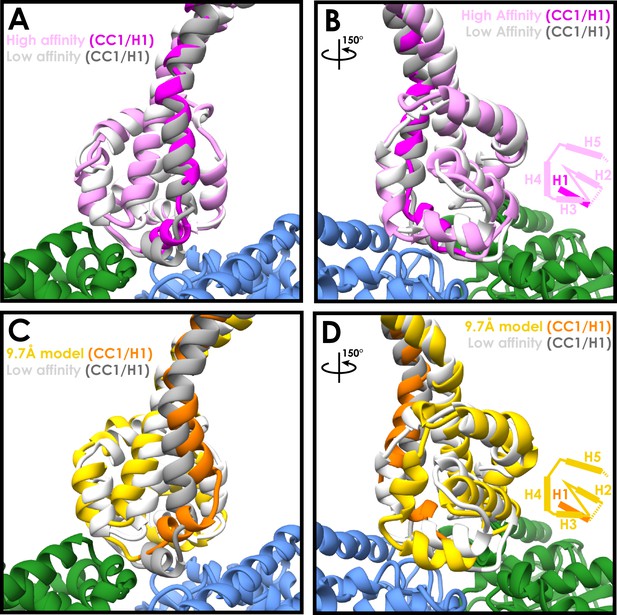
Similarities between the high- and low-affinity states of the cytoplasmic dynein-1 MTBD.
(A) A comparison between the newly refined cytoplasmic dynein-1 MTBD model (pink) and the low-affinity state crystal structure (PDB 3ERR, docked to the same density, white). CC1 and H1 (highlighted in magenta and grey for high- and low-affinity states respectively) rise in the high-affinity state to solve a steric clash with the microtubule (α-tubulin in green, β-tubulin in blue) (B) Orthogonal view of A, highlighting the similarity between the two models away from CC1/H1. Cartoon displays organisation of H2-H5 as visibile in the model. (C) A comparison the previous 9.7 Å cryo-EM microtubule-bound model (gold, PDB 3J1T) and the low-affinity state crystal structure (white, 3ERR). There is a larger movement up and to the side in CC1 and H1 (orange and grey for 3J1t and 3ERR respectively) in 3J1T compared to the new model. (D) Orthogonal view of C, showing H2, H3 and H4 all in different conformations relative to the microtubule in 9.7 Å model.
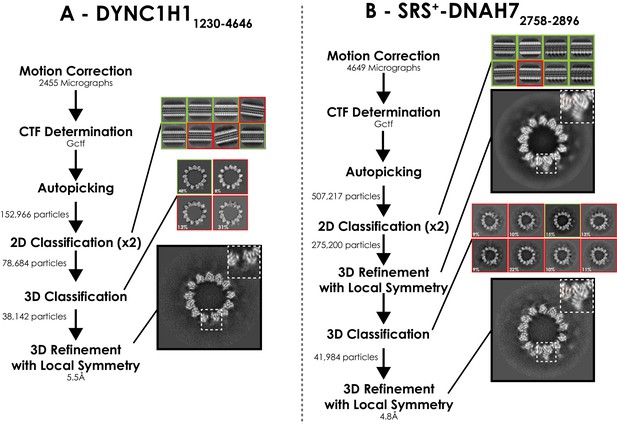
Processing pipeline for DYNC1H11230-4646 and SRS+-DNAH7 2758-2896 structures.
(A) Processing pipeline for DYNC1H11230-4646 structure. (B) Processing pipeline for SRS+-DNAH72758-2896 structure. 3D classification directly after 2D classification did not result in a good class. 3D refinement was performed, and these orientations were used for 3D classification. This good class was taken forwards for 3D refinement, creating the final DNAH7 structure.
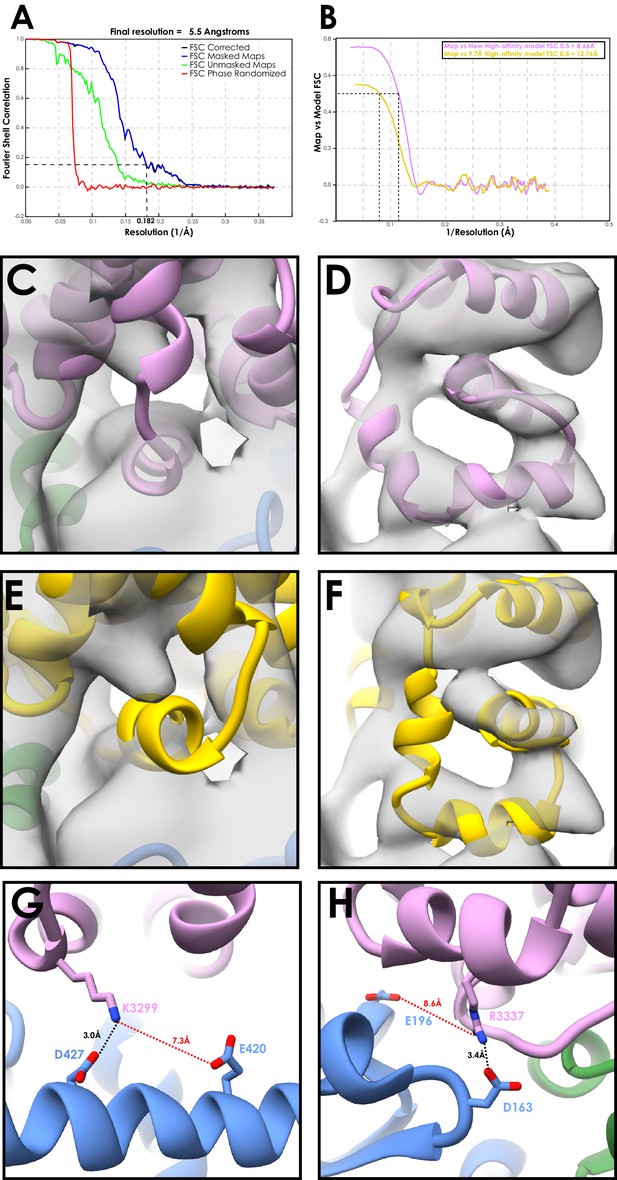
Validation of the SRS-DYNC1H13260-3427 model with the dynein motor domain.
(A) FSC curve for the DYNC1H11230-4646 decorated microtubule structure (FSC0.143 cut-off, gold-standard). (B) Model to map FSC curves for our new high-affinity cytoplasmic dynein-1 model or the previous 9.7 Å high-affinity model (3J1T) docked into the DYNC1H11230-4646 map (C) Our newly refined cytoplasmic dynein-1 high-affinity model (Figure 1D/E) docked into the DYNC1H11230-4646 map (filtered to 8 Å), viewing CC1 and H1 (D) Orthogonal view of C, viewing H2-H4 (E) Equivalent to C, but with the previous 9.7 Å cryo-EM MTBD model docked. H1 is now fully outside the density, compared to a good fit in B F) - Orthogonal view of E, showing most of H2, H3 and H4 outside the density, compared to good fits in C G) - The 4.1 Å resolution density does not show side-chain positions for most of the MTBD. However, based on our model and all possible rotamer conformations, a number of interactions between the MTBD and the microtubule identified previously (Redwine et al., 2012) are too far apart in the new model. The tubulin residue interacting K3299 was suggested to be E420, but this is now too far to interact, and we suggest that D427 is the more likely partner. (H) - Similarly, R3337 is now more likely to interact with D163 than E196.
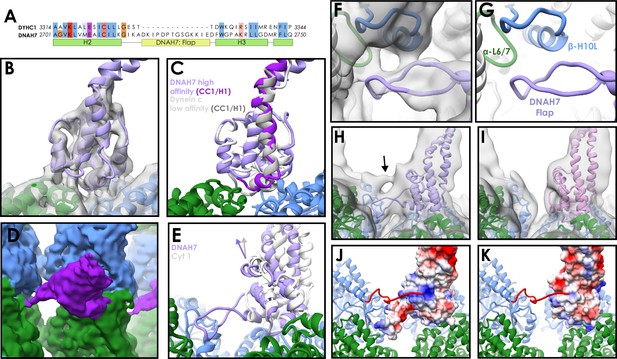
Structure of the DNAH7 MTBD.
(A) Partial sequence alignment of human cytoplasmic dynein-1 (DYHC1) and human Axonemal Dynein 7 (DNAH7). DNAH7 has a 13-residue insert between H2 and H3 compared to cytoplasmic dynein called the flap. Full MTBD sequence alignment in Figure 3—figure supplement 1 (B) A model of DNAH7 (violet) was refined into the SRS+-DNAH72758-2896 cryo-EM density (grey, lowpass filtered to 5 Å). (C) A comparison between the microtubule-bound DNAH7 model (violet) and the low-affinity Flagellar dynein C NMR structure (Kato et al., 2014, 2RR7, ensemble chain 8, docked to the same density, white). As with cytoplasmic dynein, the only major conformational changes between the two is the upward movement of CC1/H1 (purple and grey for DNAH7 and 2RR7 respectively) (D) The SRS+-DNAH72758-2896 cryo-EM density (MTBD in purple, α-tubulin in green, β-tubulin in blue) thresholded at a low level, revealing the extension of the flap to contact the adjacent protofilament (E) Rotation of the DNAH7 MTBD relative to cytoplasmic dynein-1. The tubulin was aligned in the cytoplasmic and axonemal dynein models. This reveals a rotation around the base of H6/CC2 towards the adjacent protofilament in DNAH7 (violet) compared to cytoplasmic dynein-1 (white). (F) Close-up view of the flap of the DNAH7 MTBD model with its corresponding density. (G) Corresponding view to F, without the electron density. (H) The DNAH7 map lowpass filtered to 10 Å reveals a new connecting density that cannot be explained by the flap (DNAH7 model docked, violet). (I) The extra density observed in H is not seen in the cytoplasmic dynein-1 density filtered to the same resolution (J) The C-terminal tail of β-tubulin could potentially explain the density in H, given its length and possible binding site in a positively charged pocket on the top surface of DNAH7 (surface coloured by coloumbic potential) (K) Cytoplasmic dynein-1 (surface coloured by coloumbic potential) does not have the same positive patch on DNAH7, suggesting that the C-terminal tail would not dock in the same way.
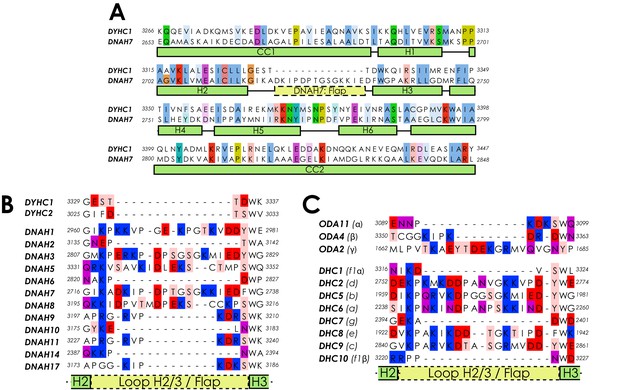
Sequence alignments of dynein microtubule-binding domains.
(A) Full sequence alignment for the MTBD of human cytoplasmic dynein-1 (DYHC1) and human DNAH7. (B) A sequence alignment of the region between H2 and H3 of the MTBDs of the human dynein heavy chains. DNAH1, 3, 5, 7 and 8 all have a 18/19 residue long flap, while all other dyneins have a much shorter loop. (C) Equivalent to B for Chlamydomonas reinhardtii axonemal dynein heavy chain sequences.
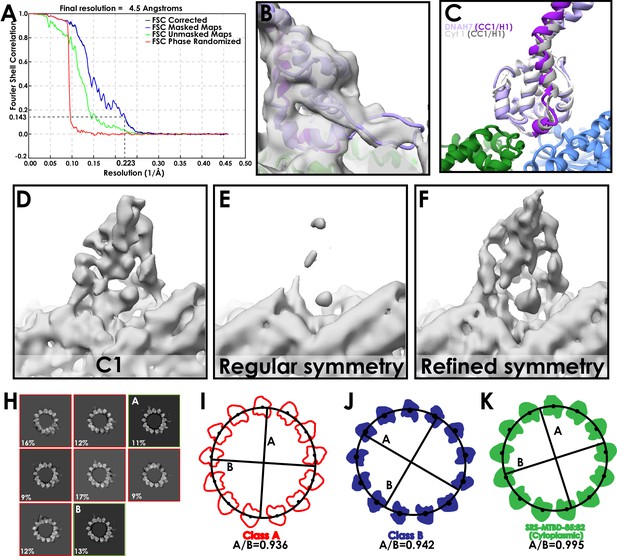
Assessment of the DNAH7 MTBD structure.
(A) FSC curve for the final, symmetrized (with refined operators) DNAH7 map (FSC0.143 cut-off, gold-standard). (B) The refined DNAH7 model docked into density, at a low threshold to visualise the flap (C) A comparison of our newly refined high-affinity cytoplasmic dynein-1 and DNAH7 MTBD models. (D) MTBD density from one protofilament of the asymmetric DNAH7 final map. (E) Corresponding view to D, following symmetrization of the DNAH7 map using the regular, circular operators used for the previous structures. The decorating density is much weaker, suggesting that the averaging was incoherent (F) Corresponding view to D, following symmetrization of the DNAH7 map using local symmetry operators refined in Relion. This finds the optimal position for each mask, and as such accounts for any deviations in curvature. The density is now much strong, and contains higher quality MTBD features than the asymmetric map. (G) Binarised representation of classes B and C, aligned at a seam-adjacent protofilament. (H) Results from 3D classification of DNAH7 when larger Tau value and local refinements were used (see Materials and methods), generating classes ‘A’ and ‘B’ (indicated) (I) Class A with protofilament coordinates. An ellipse was fit to the coordinates (black line), from which the ellipticity is shown. (J) As for H, but with Class B (K) As for H, but with the refined SRS-DYNC1H13260-3427 map.
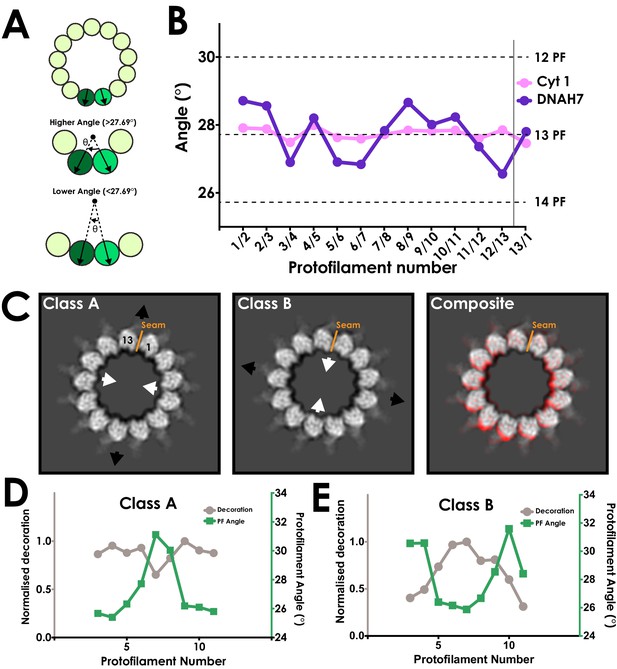
DNAH7 binding causes cross-sectional deformations in the microtubule.
(A) Schematic representing measurement of protofilament angles. A PDB model for tubulin (PDB 5SYF) was docked into adjacent protofilaments, and the rotation to superimpose the two models was measured. Local curvature between each neighbouring protofilament can thus be measured to match a canonical 13-protofilament microtubule (27.69° rotation), or be more or less curved. (B) A plot of the angle between each neighbouring pair of protofilaments in the cytoplasmic dynein-1 (Cyt 1, pink) and DNAH7 (purple) maps prior to symmetrization. In DNAH7, the microtubule has been distorted from a perfect circle. Protofilament angles from canonical 12-PF (30°), 13-PF (27.69°) and 14-PF (25.71°) are shown, and the position of the seam is indicated by the grey line (C) A projection of one helical rise of Class A and Class B (left and center) with arrows indicating distortion in the curvature of the microtubule. Note that Class B and Class C are squashed and extended in reciprocal directions. (Right) A composite image showing the distortion between Class A (Red) and Class BC (white) (D) A plot of the protofilament angles (green) and relative decoration (grey) on each protofilament for class A. (E) A plot of the protofilament angles (green) and relative decoration (grey) on each protofilament for class B.

Decoration levels in classes A and B.
Density for each protofilament used for decoration measurements in Figure 4D and E.
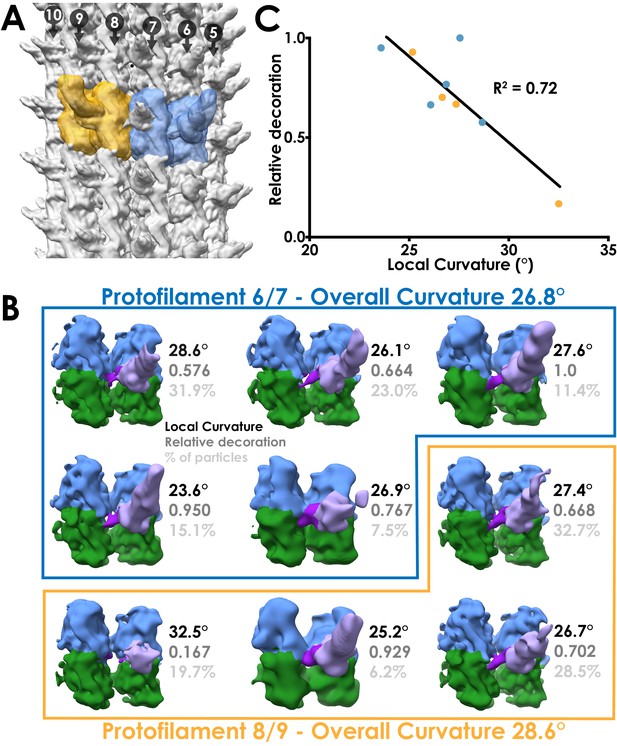
Local sub-classification of DNAH7 decorated microtubules.
(A) The two masks used for focused sub-classification of the DNAH7 dataset, encompassing one MTDD and two tubulin dimers (blue/orange). One mask (blue, protofilaments 6/7) corresponded to the region with the lowest overall curvature (26.8°, Figure 4B) and the other (orange, protofilaments 8/9) corresponded to the region with the highest overall curvature (28.6°, Figure 4B). (B) The well populated sub-classes from 3D sub-classification of the DNAH7 dataset (MTBD/flap violet/purple, α-tubulin green, β-tubulin blue). Sub-classes are in two independent groups from the low or high curvature protofilaments (blue/orange boxes). Local curvature (black), decoration relative to the most highly decorated sub-class (mid-grey) and percentage of particles contributing to the sub-class from the same job (light-grey) indicated. (C) A plot of the relationship between local curvature between tubulin dimers within a sub-class and decoration level. Line from linear regression analysis (R2 = 0.72, N = 9, Y = −0.0863*X + 3.058).
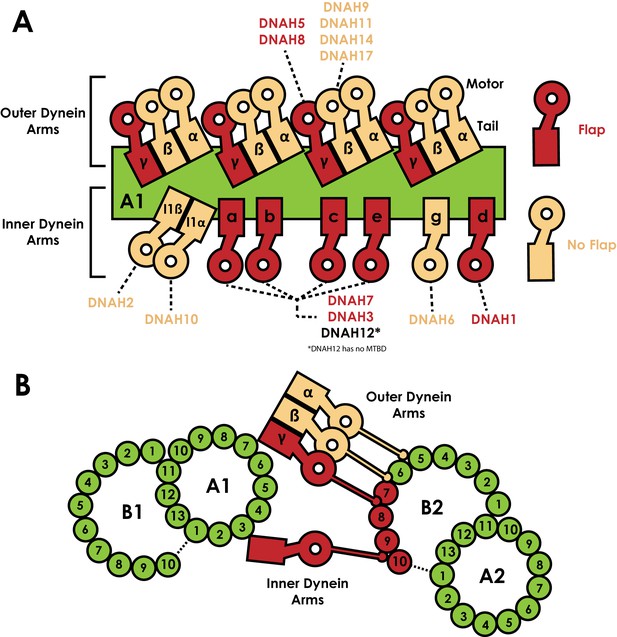
Distribution of dyneins in the axoneme.
(A) Schematic view of the positions the dynein heavy chain tails dock on the A-tubule of the microtubule doublet (green) in the 96 nm axonemal repeat, based on previous structural data on C. reinhardtii and Sea urchin sperm flagella (Bui et al., 2008; Lin and Nicastro, 2018; Nicastro et al., 2006). Heavy chains are named according to the C. reinhardtii nomenclature, and the human orthologues are linked. Heavy chains are colour coded based on whether they possess a MTBD flap (Red) or not (beige). (B) Orthogonal view of A, now looking down the flagella. Links between the inner and outer dynein arms and the exact protofilament of the adjacent doublet has also been established by structural work (Song et al., 2018). The position of the tails on tubule A1 positions the MTBDs on tubule B2. We note each of protofilaments 7 to 10 on tubule B2 are contacted by flap-containing dynein MTBDs.
Videos
A morph between Classes A and B, depicting their cross-sectional distortion.
https://doi.org/10.7554/eLife.47145.018Tables
Data collection statistics for the presented structures.
* N-fold non-crystallographic symmetry applied.
| SRS-DYNC1H13260-3427 | DYNC1H11230-4646 | SRS+-DNAH72758-2896 | |
|---|---|---|---|
| Microscope | Krios | Polar | Krios |
| Detector | Falcon III (Linear) | Falcon III (Linear) | Falcon III (Linear) |
| Voltage (kV) | 300 | 300 | 300 |
| Exposure time (s) | 1.5 | 1.5 | 1.5 |
| Total dose (e-/Å2) | 60 | 67.5 | 55.5 |
| Pixel Size (Å2) | 1.04 | 1.34 | 1.085 |
| Defocus range (μm) | -1.5 to -4.5 | -1.5 to -4.5 | -1.5 to -4.5 |
| Sessions | 1 | 1 | 1 |
| Micrographs | 1995 | 2455 | 5527 |
| Symmetry | 13* | 13* | 9* |
| Final particles | 59100 | 38142 | 41984 |
| Resolution (Å) | 4.1 | 5.5 | 4.5 |
-
Table 1—source data 1
Data collection statistics for the presented structures.
- https://doi.org/10.7554/eLife.47145.007
Model refinement statistics for the presented structures.
https://doi.org/10.7554/eLife.47145.008| SRS-DYNC1H13260-3427 | SRS+-DNAH72758-2896 | |
|---|---|---|
| Map | ||
| Map resolution (Å) | 5 | 5.4 |
| Map sharpening B factor (Å2) | -150 | -200 |
| Map CC | 0.87 | 0.84 |
| Map:Model FSC0.5 (Å) | 5.06 | 5.48 |
| Model Composition | ||
| Non-hydrogen atoms | 8001 | 14908 |
| Protein residues | 1001 | 1867 |
| Ligands (GTP/GDP) | 1/1 | 2/2 |
| R.m.s deviations | ||
| Bond lengths | 0.009 | 0.01 |
| Bond angles | 1.91 | 1.91 |
| Validation | ||
| MolProbity score | 2.35 | 2.49 |
| Clashscore | 9.05 | 9.43 |
| Rotamer Outliers (%) | 5.37 | 6.9 |
| Ramachandran plot | ||
| Favored (%) | 95.5 | 94.9 |
| Outliers (%) | 0.3 | 0.65 |
| CB Outliers (%) | 0.53 | 0.98 |
-
Table 2—source data 1
Model refinement statistics for the presented structures.
- https://doi.org/10.7554/eLife.47145.009
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.47145.021





