Evolution of neuronal anatomy and circuitry in two highly divergent nematode species
Figures
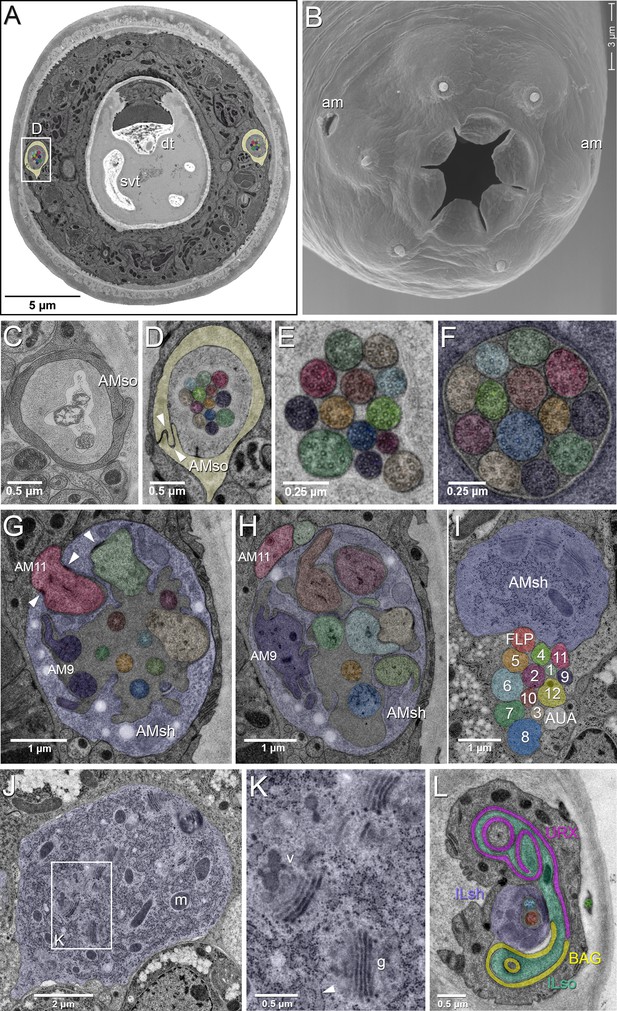
EM of the amphid sensillum and two other sensory neurons of P. pacificus hermaphrodite adults.
All images are from specimen 107 (Series 14), except B and C. As specimens were sectioned from the head, left structures appear on the right side in the images and vice versa. (A) Complete transverse section 6.8 µm from the tip of the head showing the amphid socket cells (light yellow) with false-colored amphid cilia bundles inside the channels. Note the dorsal tooth (dt) and sub-ventral tooth (svt) in the buccal cavity. (B) Scanning electron micrograph of the head of an adult animal showing the two amphid openings (am). (C–H) Transverse TEM sections through the amphid cilia at various levels from anterior to posterior (C) Left amphid channel close to the pore, containing the tips of the longest three cilia; the channel is formed by the amphid socket cell (AMso). (Specimen DB-9–1) (D) Right amphid channel slightly more posterior than in A (7.1 µm from the tip of the head) with all 13 cilia visible in the channel matrix. Arrowheads indicate the autocellular junction of the amphid socket cell (AMso). (E) Distal segments of the amphid cilia with singlet microtubules in the left channel 6.85 µm from the tip. (F) Middle segments of the right amphid cilia with discernable nine outer doublet microtubules that make up the core axoneme 11.25 µm from the tip; here the channel is formed by the amphid sheath cell (lilac). (G) Section through the region of sheath entry 13.1 µm from the tip, showing the thick periciliary membrane compartment (PCMC) of AM11 (red) entering the left amphid sheath cell (AMsh), the green and beige PCMCs have just completed their entry. Additionally, the sheath lumen harbors the basal parts of the double cilia of AM9 (dark lilac) with typical transition zone (TZ) arrangement of microtubules and seven further TZ cilia. Arrowheads mark the adherens junctions between the PCMCs and the amphid sheath. (H) A slightly more posterior section through the left sheath cell (13.85 µm from the tip), showing the distinctively large and irregular outline of the PCMC of AM9 (dark lilac) and the PCMCs of 6 other dendrites. The base of the AM8 cilium (blue) and the TZ of AM5 (orange) are found in the lumen. AM11 is still outside the sheath. (I–K) Transverse TEM sections through the posterior AMsh process and cell body (lilac). (I) Right-side amphid nerve directly anterior of the nerve ring (88.3 µm from the tip) consisting of the AMsh process (sh) with prominent Golgi stacks and the 12 amphid neuron dendrites. AM1 to AM12 are labeled by number and color. The color code is used throughout the paper. FLP (reddish, just below sheath) and AUA (white) join the amphid process bundle until they diverge from it or end. (J) AMsh cell body (100.8 µm from tip) with numerous mitochondria (m) and Golgi stacks. (K) A higher magnification of the Golgi apparatus (g) and vesicles (v) in the midst of ribosome-studded rough ER cisternae (arrowhead). (L) The bilaterally symmetrical URX and BAG neurons have extended flattened ciliary endings associated with the lateral Inner Labial socket cell (ILso) process; 2.05 µm from the tip. Comparable C. elegans EM annotations are available at SlidableWorm http://www.wormatlas.org/SW/SW.php/. See Figure 1—figure supplements 1–3, and Figure 1—videos 1–3 for details on the URX, URY, URA, and BAG neurons.
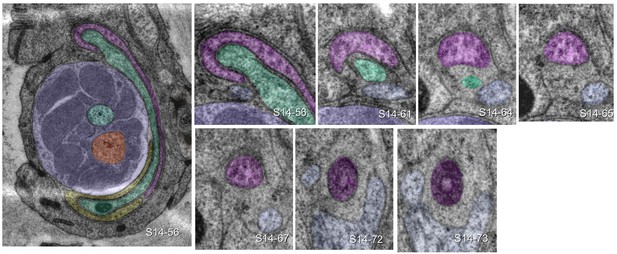
URX is ciliated.
The large image shows the left lateral IL sensillum of P. pacificus (specimen 107) with ILsh (lilac) and ILso (green), surrounded by URXL (pink) and BAGL (yellow). Small images track the URXL cilium from its extended sheet-like tip (section 56 = 2.8 µm from the tip of the nose) to transition zone (section 73 = 3.65 µm from the tip of the nose). Dendritic structures can be compared to C. elegans at SlidableWorm http://www.wormatlas.org/SW/SW.php/ (slices 8–10).
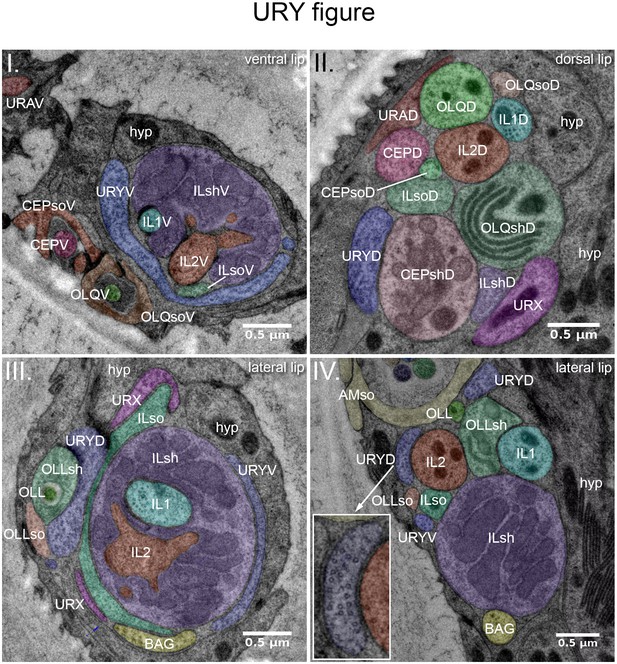
URY, URX, URA and BAG endings.
TEM cross-sections through two right labial sensilla (I., III.) and process bundles (II., IV.) of P. pacificus (specimen 107) (seen from anterior). (I) Right ventral lip, 3.8 µm from the tip of the nose. The membranous extension of a URYV branch (blue) wraps halfway round the ILV sensillum. It terminates further distally in the tip of the ventral lip. (II) Right dorsal lip, 7.8 µm from the tip of the nose. This more posterior section shows that the dendrites of all three UR neurons are part of the dorsal labial process bundle. The URYD process is found next to CEPshD, URX next to ILshD and OLQshD, and URAD next to CEPD and OLQD. (III) Right lateral lip, 3.6 µm from the tip of the nose, showing branches of URYD, URYV and the URX dendrite extending into the lateral labial process bundle. URYV is associated with ILsh, and URYD with ILsh, OLLsh and OLLso. URX and BAG endings are found apposed to ILso. For more anterior sections showing the elaborate URX and BAG extensions, see Figure 1L. (IV.) Right lateral lip, 7.05 µm from the tip of the nose. Lateral labial processes, branches of URYD and URYV, and the BAG dendrite are located directly ventral of the amphid sensillum. URYD shows many prominent singlet microtubules (see inset) but appears to be unciliated. URX will join the bundle more anteriorly.
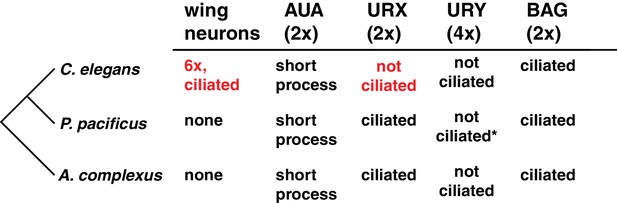
A comparison of select non-amphid ciliated neurons in three nematode species.
The derived characters in C. elegans are highlighted in red. *Many prominent singlet microtubules.
Video of URX 3D reconstruction.
https://doi.org/10.7554/eLife.47155.007Video of URY 3D reconstruction.
https://doi.org/10.7554/eLife.47155.008Video of URA 3D reconstruction.
https://doi.org/10.7554/eLife.47155.009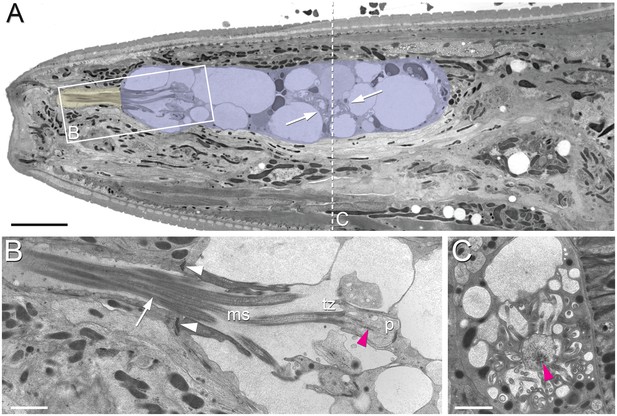
Position of amphid cilia in AMso and AMsh.
(A–B) Sagittal TEM sections of a P. pacificus young adult hermaphrodite. (A) The yellowish shading highlights the narrow amphid channel, which is formed by the socket cell process and contains the distal segments of the 13 amphid cilia. The lilac shading highlights the expanded region of the amphid sheath cell process, which harbors the proximal segments of the amphid cilia in its anterior lumen and, more posteriorly, the finger cell in its cytoplasm amidst a multitude of vesicles. Arrows point at the space taken up by the finger cell cilium with its projections. Scale bar: 5 µm. (B) Detail of A (from a neighboring section) showing the different ciliary regions: middle segment (ms), transition zone (tz), and periciliary membrane compartment (p). The arrow points at cilia in the narrow, matrix-filled amphid channel. Arrowheads mark the adherens junctions between the AMsh and AMso processes. The red arrowhead indicates the ciliary rootlet within the periciliary membrane compartment (p) of one of the amphid neurons. Scale bar: 1 µm. (C) Transverse section through the periciliary membrane compartment of the amphid finger cell with central rootlet (red arrow head) and numerous fingerlike villi in various orientations. The dotted line in A represents the approximate plane of sectioning. Scale bar: 1 µm. The order of each dendrite entry is shown in Figure 2—figure supplements 1 and 2. Comparative sagittally sectioned C. elegans nose images are present at http://www.wormatlas.org/hermaphrodite/neuronalsupport/jump.html?newLink = mainframe.htm and newAnchor = Amphidsensilla31 (Figure 35). Figure 2—figure supplements 1 and 2.
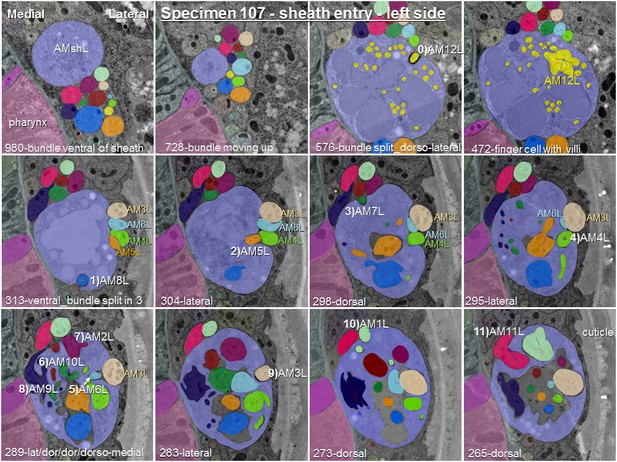
Posterior-anterior path of amphid dendrite entry into the left sheath glia of specimen 107.
The entering dendrite is labeled in white by a number from 1) to 11) according to the order of entry followed by its P. pacificus name. Section numbers from anterior tip of head and direction of entry are given at the bottom of each image. One section is 50 nm. To demonstrate the variability of entry sites between left and right side in this specimen, AM3 to AM6 have been labeled in color.
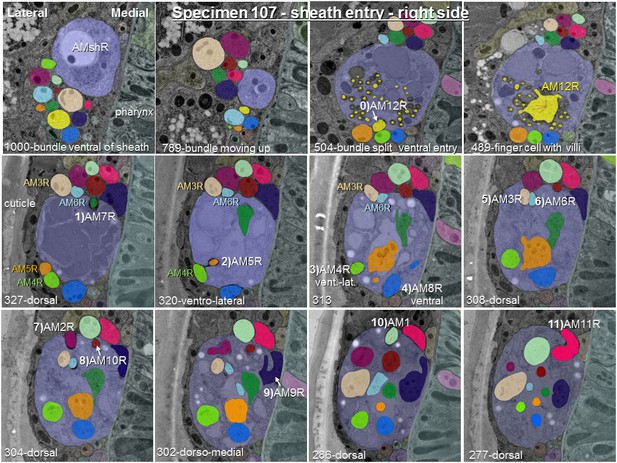
Posterior-anterior path of amphid dendrite entry into the right sheath glia of specimen 107.
The entering dendrite is labeled in white by a number from 1) to 11) according to the order of entry followed by its P. pacificus name. Section numbers from anterior tip of head and direction of entry are given at the bottom of each image. One section is 50 nm. To demonstrate the variability of entry sites between left and right side in this specimen, AM3 to AM6 have been labeled in color.
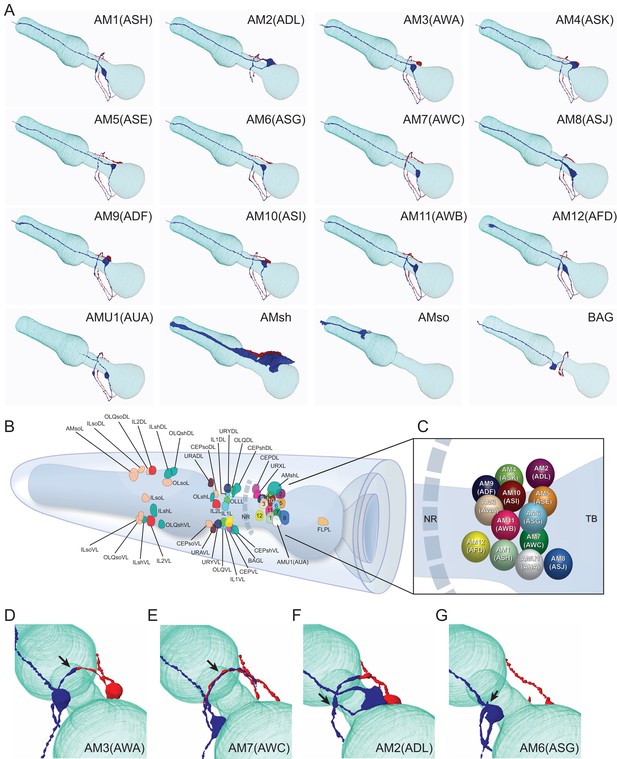
Overview of the amphid sensilla.
(A) Three-dimensional renderings of individual pairs of amphid neurons, amphid sheath, amphid socket, internal sensory receptor AUA, and BAG neuron in a P. pacificus young adult hermaphrodite (Specimen 107). Left lateral view, anterior is to the left. Blue and red renderings denote left and right counterparts, respectively. The pharynx is outlined in teal. Nomenclature is displayed with P. pacificus first and C. elegans name in parenthesis. Black arrows denote lateral nerve ring entry (not through amphid commissure) and branched axons in AM2(ASG), short axons ending at the lateral midline in AM6(ASG), dorsal overlap of axon endings in AM7(AWC), fingerlike endings of AM12(AFD) dendrite terminating within the sheath cell posterior to the nose, and short dendritic-like endings of AUA anterior to the nerve ring Figure 3—videos 1–15. (B) Schematic diagram of the positions of the nuclei of the left cuticular sensilla with sheath and socket cells and of the internal receptors. The nerve ring (NR) is indicated by a dotted line. Amphid cell bodies are labeled 1–12 by their last number, with color code the same as in Figure 1. (C) Schematic of amphid nuclei as seen from the left side with full names, nuclei slightly enlarged. (TB = terminal bulb). (D–G) The four different types of axon projections in P. pacificus amphid neurons: right and left axons: (D) meet and end with very little overlap at the dorsal midline (all except AM6/ASG and AM7/AWC, also AMU1/AUA) (AM3 is shown to represent this group); (E) cross extensively over the dorsal midline (AM7/AWC); (F) branch laterally into a dorsal and a ventral process due to lateral entry into the NR instead of the usual ventral entry of all other amphid neurons that come from the amphid commissure (AM2/ADL); (G) short axons end at the lateral midline (AM6/ASG). Cartoons of C. elegans amphid neuron anatomy are present at http://www.wormatlas.org/images/NeuronImageList.jpg. Cartoons of C. elegans ganglia are present at http://www.wormatlas.org/images/VCMNganglia.jpg. 3D renderings of individual neurons are shown in Figure 3—videos 1–15.
3D rendering of AM1(ASH) neurons.
https://doi.org/10.7554/eLife.47155.0163D rendering of AM2(ADL) neurons.
https://doi.org/10.7554/eLife.47155.0173D rendering of AM3(AWA) neurons.
https://doi.org/10.7554/eLife.47155.0183D rendering of AM4(ASK) neurons.
https://doi.org/10.7554/eLife.47155.0193D rendering of AM5(ASE) neurons.
https://doi.org/10.7554/eLife.47155.0203D rendering of AM6(ASG) neurons.
https://doi.org/10.7554/eLife.47155.0213D rendering of AM7(AWC) neurons.
https://doi.org/10.7554/eLife.47155.0223D rendering of AM8(ASJ) neurons.
https://doi.org/10.7554/eLife.47155.0233D rendering of AM9(ADF) neurons.
https://doi.org/10.7554/eLife.47155.0243D rendering of AM10(ASI) neurons.
https://doi.org/10.7554/eLife.47155.0253D rendering of AM11(AWB) neurons.
https://doi.org/10.7554/eLife.47155.0263D rendering of AM12(AFD) neurons.
https://doi.org/10.7554/eLife.47155.0273D rendering of AMU1(AUA) neurons.
https://doi.org/10.7554/eLife.47155.0283D rendering of Amphid sheath (AMsh).
https://doi.org/10.7554/eLife.47155.0293D rendering of Amphid socket (AMso).
https://doi.org/10.7554/eLife.47155.030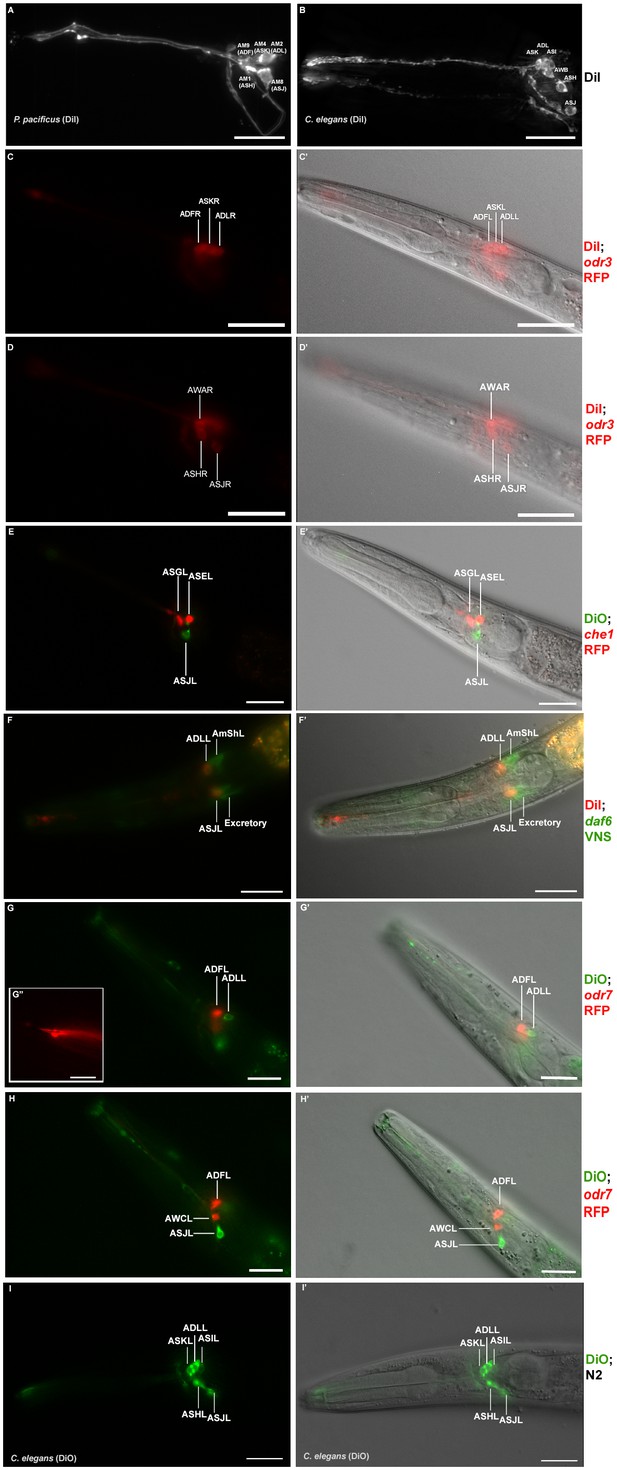
Dye filling and reporter gene expression in P. pacificus amphid neurons.
(A) Stacked and deconvoluted fluorescent images of DiI stained amphid neurons in a P. pacificus young adult hermaphrodite (left lateral view). AM9(ADF), AM4(ASK), AM2(ADL), AM1(ASH), and AM8(ASJ) amphid neurons stain robustly. (B) In C. elegans, DiI stains the ASK, ADL, ASI, AWB, ASH, and ASJ neurons. (C–H) Single plane fluorescent images in P. pacificus. (C–D) DiI stained Ppa-odr-3p::rfp transgenic J3 larvae showing the three dorsal amphid neurons and the larger ASH and ASJ neurons in two focal planes. (E) Ppa-che-1p::rfp transgenic J4 hermaphrodite showing expression in ASE and ASG. (F) A DiI stained Ppa-daf-6p::venus J4 larva showing ADL and ASJ staining, anterior to the amphid sheath and the excretory cells, respectively. (G, H) A DiO stained Ppa-odr-7p::rfp young adult in two focal planes showing dye filling in ADL and ASJ, and RFP expression in ADF and AWC. (G’ inset) The dendritic ends of another Ppa-odr-7p::rfp adult show the prominent PCMC of ADF with double cilia (bottom), and the smaller PCMC of ASK with a single cilium. (I) A DiO stained C. elegans young adult showing five dye filled neurons ASK, ADL, ASI, ASH, and ASJ. Scale bar: 5 µm. (C’–I’) DIC overlay of the same DiI fluorescence images. Anterior is left and dorsal is up. Scale bar: 20 µm. (Representative images based on: Ppa-odr-3p::rfp n = 22; Ppa-che-1p::rfp n = 31; Ppa-daf-6p::venus n = 60; PS312 n = 10; Ppa-odr-7p::venus n = 26). Additional Z-stacks are available: Figure 4—figure supplements 1–5 and Figure 1—videos 1–3.
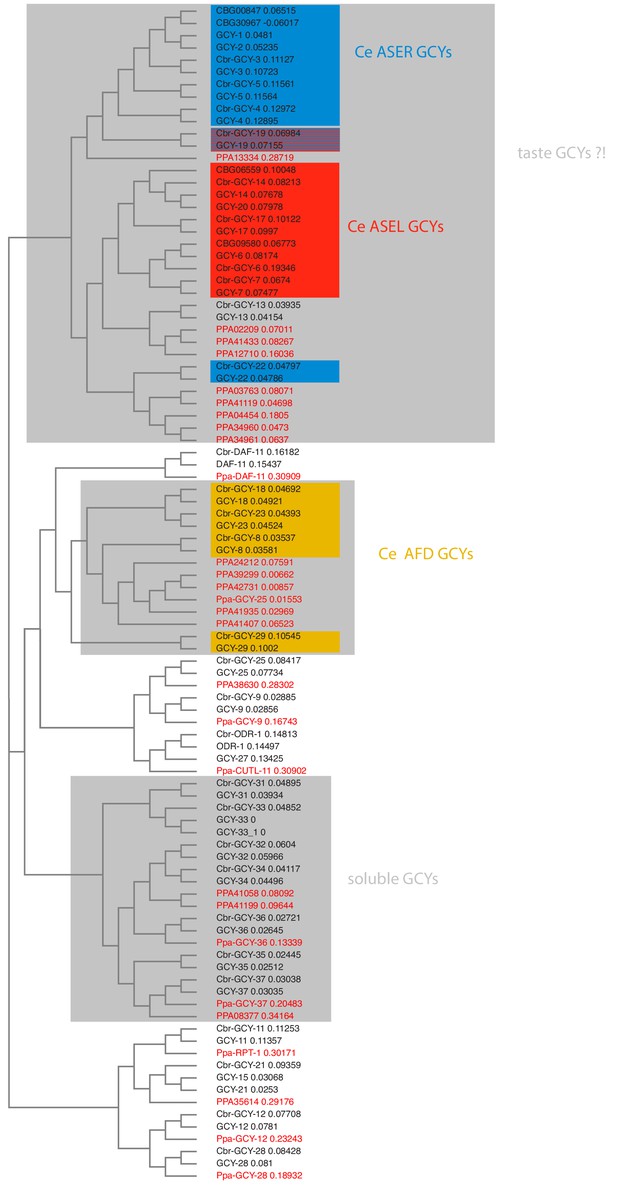
Receptor-type guanylyl cyclases in P. pacificus (red font) and Caenorhabditis species (black font).
https://doi.org/10.7554/eLife.47155.032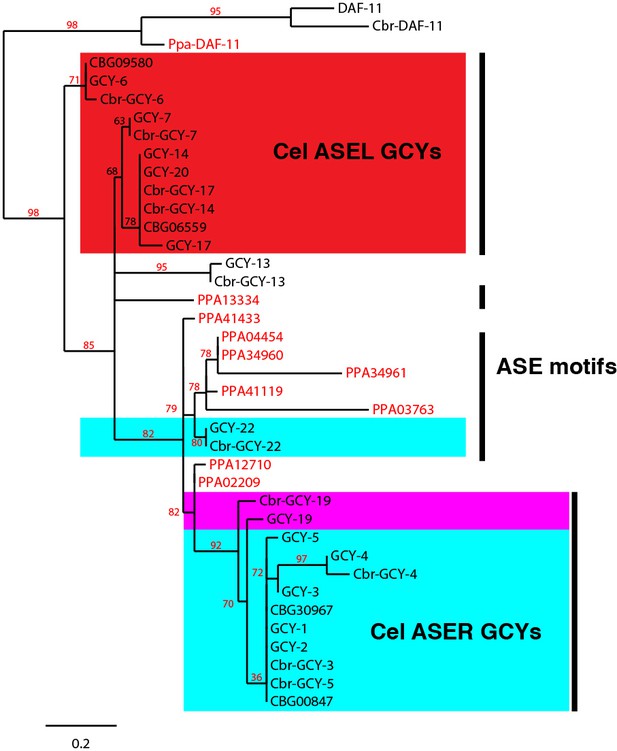
Abbreviated phylogeny of ASE taste receptor-type guanylyl cyclases in P. pacificus (red font) and Caenorhabditis species (black font).
DAF-11 orthologs form an outgroup. Only branch support ≥30% is shown.
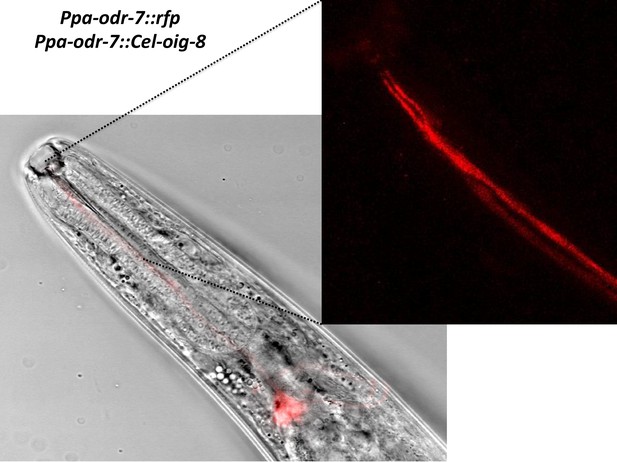
Ppa-odr-7p::Cel-oig-8 mis-expression.
An overlay of the AM7(AWC) cell body showing no change in the ciliated endings of AM7 and AM9(ADL)(blowup panel).
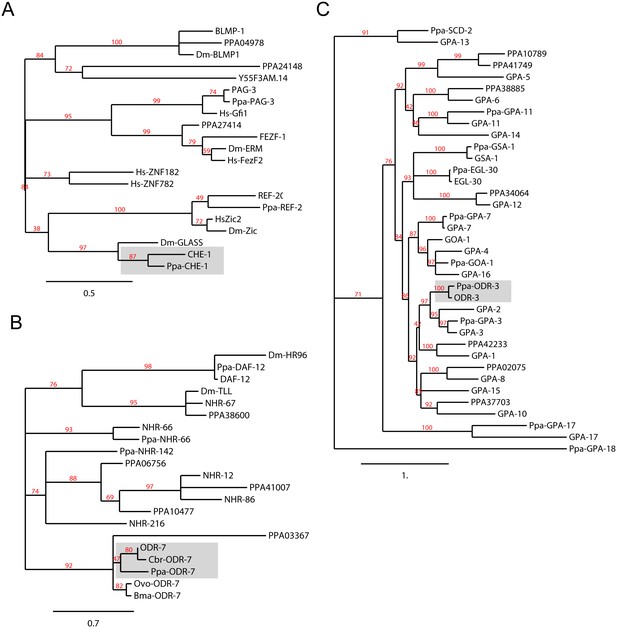
Orthology assignments for che-1, odr-7 and odr-3.
Phylogenetic trees for (A) the CHE-1 Zinc finger transcription factor, (B) the ODR-7 nuclear hormone receptor, and (C) the ODR-3 G-alpha protein subunits. The C. elegans and P. pacificus orthologs are shaded. Maximum Likelihood phylogeny trees were generated by PhyML (phylogeny.fr). For the nuclear hormone receptor tree building, only the DNA binding C4 domain was used. Midpoint rooting was used and only branch support ≥30% is shown. Protein sequences for P. pacificus are ‘Ppa-’ (orthology same as Wormbase.org), or ‘PPA’ (no orthology assignment), whereas the rest are C. elegans names. Sequences from additional species are Drosophila melanogaster (Dm), Homo sapiens (Hs), Caenorhabditis briggsae (Cbr), Brugia malayi (Bma), and Onchocerca volvulus (Ovo).
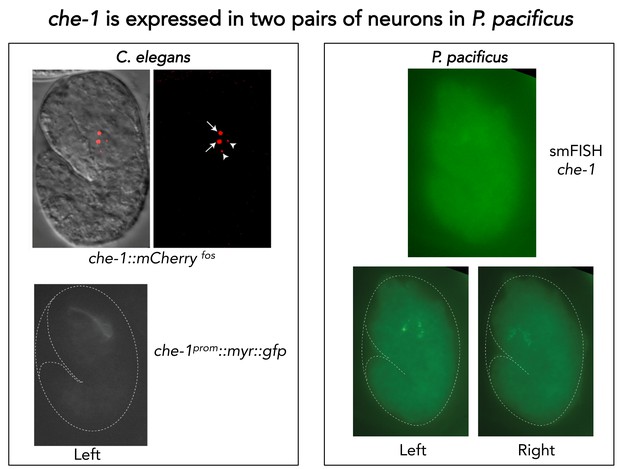
Single molecule FISH of Ppa-che-1.
(Left) Representative images of C. elegans embryos carrying two different Cel-che-1 reporters: a translational fusion of CHE-1 to mCherry in the context of a recombineered fosmid construct shows the expected nuclear localization in a pair of neurons identified as the ASE neurons (arrows) as well as two apoptotic bodies (arrow heads) corresponding to the dying ASE-sister cells. A 1.3 Kb fragment upstream of the Cel-che-1 coding sequence also drives expression in the ASE neurons, in this case visualized with a myristoylated GFP reporter. (Right) A representative P. pacificus embryo stained with the smFISH probe set against Ppa-che-1. Individual planes focused on the left or right sides of the embryo show two neurons on each side.
Z-stack images of DiI stained J3 larva (ventral view).
Anterior is up.
Z-stack images of Ppa-che-1p::rfp stained with DiO.
Anterior is to the left and dorsal is to the right.
Z-stack images of Ppa-odr-7p::rfp.
Anterior is to the left and dorsal is to the right.
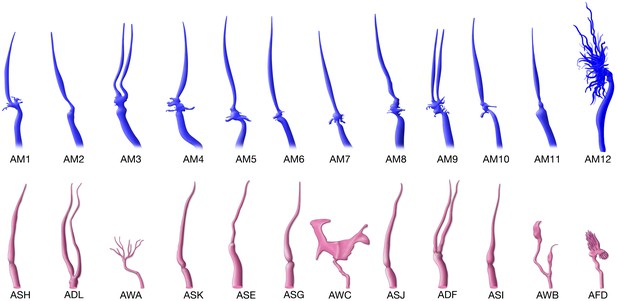
Comparison of the reconstructed cilia of P. pacificus and C. elegans amphid neurons.
P. pacificus lacks neurons with wing-like morphology.
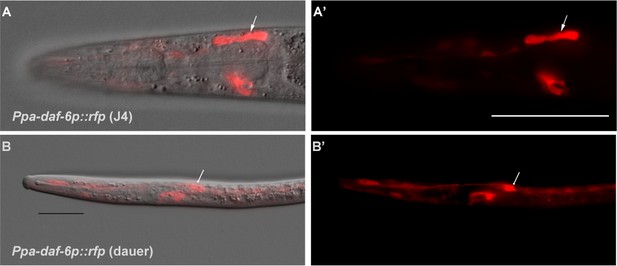
Genetic marker for the amphid glia.
(A–A’) DIC overlay and Ppa-daf-6p::rfp expression in the dorsally located amphid sheath cell as well as the excretory cell on the ventral side of a J4 hermaphrodite. Scale bar: 50 µm. (B–B’) DIC overlay and Ppa-daf-6p::rfp expression in a dauer larva with prominent amphid sheath, excretory cell, and seam cells expression. Scale bar: 25 µm. Arrows indicate the amphid sheath cell body. A Z-stack of a Ppa-daf-6p::rfp adult is available: Figure 6—video 1.
3D rendering of Ppa-daf-6::rfp J4 with confocal microscopy.
Anterior is to the left and dorsal is to the right.
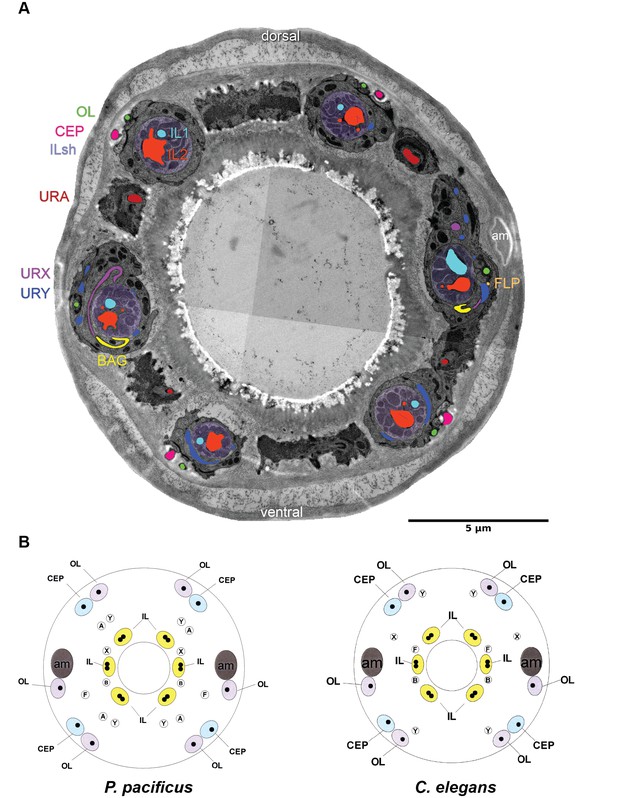
Comparison of cuticular sensilla and internal receptors.
(A) Transversal TEM section of P. pacificus cuticular sensilla and internal receptors of the mouth region. One from each sensillum type is labeled in the same false color as the neuron. Note the highly branched dendritic endings of the four URY neurons (blue), which associate with all six inner labial sensilla forming sheet-like branches around the processes of the ILsh and ILso cells. (B–C) Schematic illustrations of the cuticular sensilla in P. pacificus and C. elegans. Inner Labial 1 and 2 (IL), Inner Labial sheath (ILsh), Outer Labial (OL), Cephalic (CEP), URA (A), URY (Y), URX (X), BAG (B), FLP (F), amphid (am).
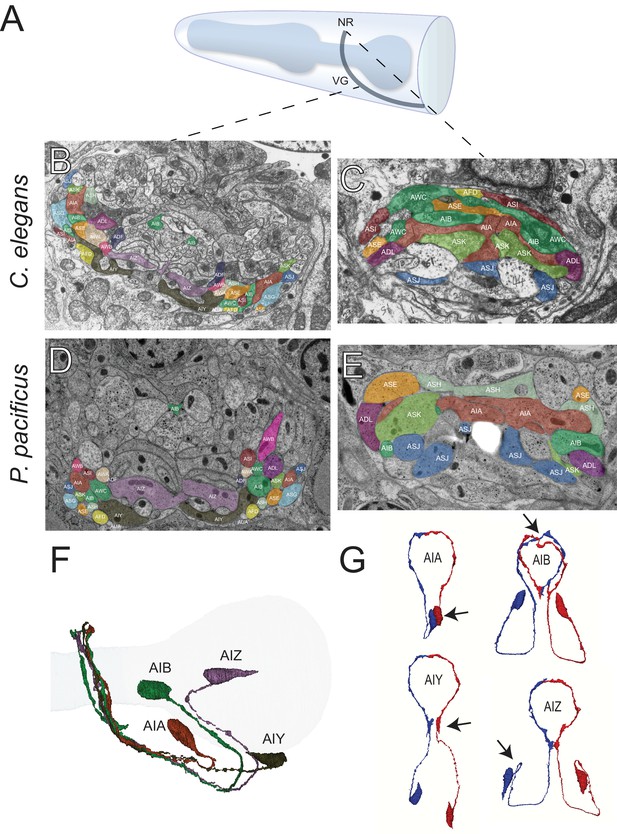
The first layer amphid interneurons.
(A) Model of the anterior P. pacificus highlighting the nerve ring (NR) and ventral ganglion (VG). (B) Electron micrograph of the anterior C. elegans (N2U sample) ventral ganglion and dorsal nerve ring (C) colored with the same color code as in Figure 1. Also shown are AIA (maroon), AIB (dark green), AIY (green/gray), and AIZ (purple). C. elegans neuronal nomenclature is used. Electron micrograph of the anterior P. pacificus (specimen 107) ventral ganglion (D) and dorsal nerve ring (E) colored with the same color code as in Figure 1. (F) Three dimensional rendering of the AIA, AIB, AIY, and AIZ interneurons from P. pacificus (specimen 107) with pharyngeal outline shown in light teal. (G) Individual three dimensional renderings of P. pacificus (specimen 107) amphid interneurons shown with a dorsoposterior view with the left and right neurons in blue and red, respectively. Arrows show anatomical features present in both species: AIA cell bodies are adjacent, AIB axons undergo a dorsal neighborhood change within in the nerve ring, AIY neurons have varicosities relating to synaptic output posterior to the nerve ring, and AIZ axons show anterior projections before commissural entry. Amphid neuroanatomy can be compared to C. elegans at http://www.wormatlas.org/images/NeuronImageList.jpg.
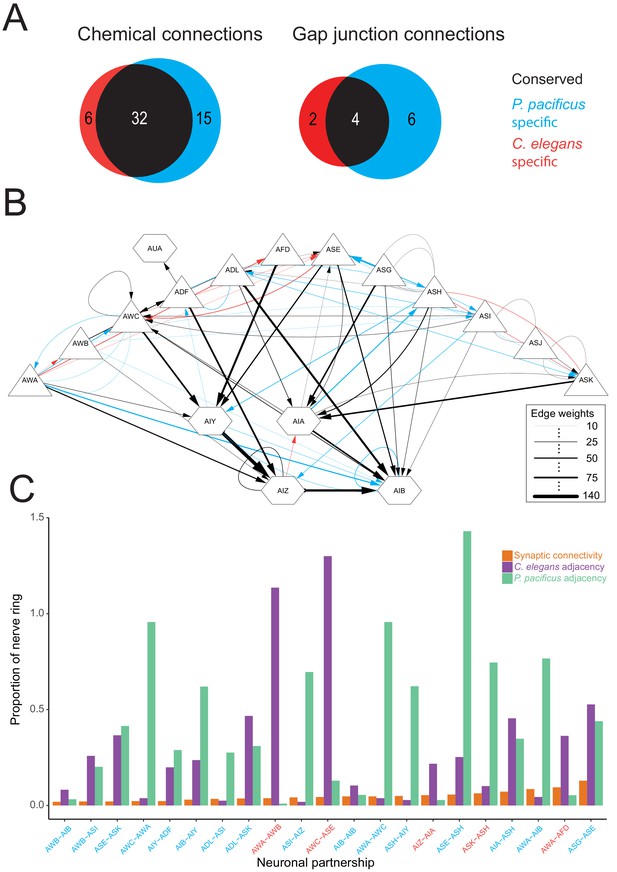
Comparisons of synaptic connectivity.
(A) Venn diagrams showing number of conserved (black), C. elegans-specific (red), and P. pacificus-specific (blue) chemical connections (directed edges) and gap junction connections (undirected edges). Threshold for edge weight was set to ≥10 serial EM sections, connectivity was determined between amphid sensory neurons, AUA, and amphid interneurons AIA, AIB, AIY, and AIZ. (B) Circuit diagram showing sensory neurons (triangles) connecting to interneurons (hexagons) by chemical synaptic connectivity (arrows) and gap junctions (lines). The thickness of the line is proportional to the weight of connection (aggregate number of serial section electron micrographs where a synaptic specialization is observed, with only connections ≥ 10 sections shown in this diagram). (C) Comparison of species-specific connections and neuronal adjacency. Bar graph showing chemical synaptic connectivity (orange), neuronal adjacency for C. elegans (purple), and neuronal adjacency for P. pacificus (green). The x axis is ordered by increasing strength of species-specific connection, and colored coded as in 8A. Adjacency was determined computationally by comparing adjacent pixels of volumetrically traced neuron profiles. Proportion was determined by dividing the number of serial EM sections showing a synaptic connection or adjacency by the total number of sections comprising the nerve ring and ventral ganglion. A proportion of 1.0 indicates the entire ipsilateral region of interest was adjacent, with proportions over 1.0 showing adjacency on both sides of the nerve ring. Each partnership evaluated is a combination of two neurons that formed a species-specific connection shown in (B). Figure 9—source datas 1–3.
-
Figure 9—source data 1
Species-specific connectivity of P. pacificus amphid neurons.
- https://doi.org/10.7554/eLife.47155.047
-
Figure 9—source data 2
Connectivity of P. pacificus amphid neurons.
- https://doi.org/10.7554/eLife.47155.048
-
Figure 9—source data 3
Connectivity of amphid neurons P. pacificus and C. elegans.
- https://doi.org/10.7554/eLife.47155.049
Tables
Comparison of amphid neurons in various nematode species.
The total number of neurons with dendritic processes encased in the sheath cell in a single amphid compartment is indicated. These neurons are further categorized as neurons with dendrites having single or double ciliated endings in the amphid channel, or as specialized ‘wing’ neurons with endings outside the channel but within the sheath cell other than the finger cell. The ASC neurons in L1 larvae of P. trichosuri described by Zhu et al. (2011) match this criterion but lack the ciliary elaborations known from C. elegans wing neurons.
| Cilia in channel | Wing | |||||
|---|---|---|---|---|---|---|
| Species | Total neurons | Dendritic ends in channel | Total count | Single | Double | Neurons |
| Pristionchus pacificus | 12 | 11 | 13 | 9 | 2 | 0 |
| Caenorhabditis elegansa | 12 | 8 | 10 | 6 | 2 | 3 |
| Haemonchus contortusb | 12 | 10 | 13 | 7 | 3 | 0 |
| Strongyloides stercoralisc | 13 | 12 | 12 | 12 | 0 | 0 |
| Parastrongyloides trichosurid | 13 | 11 | 11 | 11 | 0 | 1? |
| Acrobeles complexuse | 13 | 12 | 12 | 10 | 1 | 0 |
Provisional nominations of putative amphid neuronal homologs between C. elegans and P. pacificus.
Morphological data based on TEM series of specimens 107 and 148. Features supporting cellular homology include axon projections, lipophilic dye uptake, cell body positions, and chemical synapses to the four first layer amphid interneurons (AIA, AIB, AIY, AIZ) or the AUA. DiI filling neurons in C. elegans are ASI, ASJ, ASK, ADL, ASH, AWB (weak), *ADF (shows only weak FITC uptake, not DiI). DiO filling neurons in C. elegans are ASI, ASJ, ASK, ADL, ASH, AWB. PCMC: periciliary membrane compartment. Structures of C. elegans amphid neurons are available at http://www.wormatlas.org/images/NeuronImageList.jpg.
| P. pacificus neuron | Axon termination site in nerve ring | Dye filling | Feature(s) supporting homology | Likely C. elegans homolog | Main difference compared to C. elegans |
|---|---|---|---|---|---|
| AM1 | dorsal midline | DiI only | DiI uptake, cell position, cell body morphology | ASH | lack DiO uptake |
| AM2 | dorsal midline | DiI + DiO | DiI + DiO uptake; branched axon not through commissure; AIA/AIB | ADL | single ciliated vs. dual ciliated |
| AM3 | dorsal midline | none | none | AWA | dual ciliated vs. wing; Ppa-odr-3 expression |
| AM4 | dorsal midline | DiI only | DiI uptake; AIA/AIB | ASK | lack of DiO uptake; Ppa-odr-3 expression; |
| AM5 | dorsal midline | none | Ppa-che-1 expression | ASE | axons cross the midline in C. el. |
| AM6 | lateral midline | none | short axons;cell position | ASG | Ppa-che-1 expression |
| AM7 | cross dorsal midline | none | axons overlap dorsally; dorsal sheath entry;cell position; AIA/AIB/AIY/AIZ | AWC | single ciliated vs. wing; Ppa-odr-7 expression |
| AM8 | dorsal midline | DiI + DiO | DiI and DiO uptake; ventral sheath entry;cell position | ASJ | none |
| AM9 | dorsal midline | DiI only | dual ciliated; DiI uptake*; prominent PCMC; connect to AUA | ADF | Ppa-odr-7 expression |
| AM10 | dorsal midline | none | AIA/AIB/AIY/AIZ | ASI | lack DiI and DiO uptake |
| AM11 | dorsal midline | DiI only | weak DiI uptake; cell position; AIA/AIB/AIY/AIZ | AWB | single ciliated vs. wing; lack DiO uptake |
| AM12 | dorsal midline | none | finger dendrite morphology; cell position; first to enter the AMsh | AFD | more posterior position in AMsh due to lack of winged neurons |
| AMU1 | dorsal midline | none | short ‘dendritic’ process; cell position; exclusive output of ADF | AUA | none |
Dye filling properties of individual amphid neurons in young adult hermaphrodites.
orange = clearly conserved; blue = clearly divergent; in brackets()=weak staining, *weak FITC uptake in C. elegans.
| Amphid Homologs | P. pacificus | C. elegans | |||
|---|---|---|---|---|---|
| Ppa | Cel | DiI | DiO | DiI | DiO |
| AM1 | ASH | + | - | + | + |
| AM2 | ADL | + | + | + | + |
| AM4 | ASK | + | - | + | + |
| AM8 | ASJ | + | + | + | + |
| AM9 | ADF | + | - | -* | - |
| AM10 | ASI | - | - | + | + |
| AM11 | AWB | (+) | - | (+) | + |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Pristionchus pacificus) | Ppa-daf-6p::rfp | this study | tuEx231 | extrachromosomal array transgenic strain |
| Genetic reagent (Pristionchus pacificus) | Ppa-daf-6p::venus | this study | tuEx250 | extrachromosomal array transgenic strain |
| Genetic reagent (Pristionchus pacificus) | Ppa-odr-3p::rfp | this study | tuEx265 | extrachromosomal array transgenic strain |
| Genetic reagent (Pristionchus pacificus) | Ppa-odr-7p::rfp | this study | tuEx296 and tuEx297 | extrachromosomal array transgenic strain |
| Genetic reagent (Pristionchus pacificus) | Ppa-che-1p::che-1:rfp | this study | lucEx367 | extrachromosomal array transgenic strain |
| Recombinant DNA reagent | Ppa-che-1 mRNA probe | Stellaris, Biosearch Technologies; this study | PPA01143 | single-molecule in situ fluorescence probe |
| Sequence-based reagent | Ppa-daf-6 promoter forward primer | this study | PPA15978 | CTCGCCCGTGGATCATGTG |
| Sequence-based reagent | Ppa-daf-6 promoter reverse primer | this study | PPA15978 | TGCAAATCATTGATTGAATCATGG |
| Sequence-based reagent | Ppa-odr-3 promoter forward primer | this study | PPA14189 | GAGCGAGTGAAATGAGCTCAGTCC |
| Sequence-based reagent | Ppa-odr-3 promoter reverse primer | this study | PPA14189 | GGGTGATCGATACGAGGAGTGTTC |
| Sequence-based reagent | Ppa-odr-7 promoter forward primer | this study | Contig1-aug1055.t1 | AACCAATGCATTGGCTTAGTTGGTTTCACTAATCACTACTG |
| Sequence-based reagent | Ppa-odr-7 promoter reverse primer | this study | Contig1-aug1055.t1 | CCCTTGTCATTCAGATGAGCGAGCTGATCAAGGAG |
| Sequence-based reagent | Ppa-che-1 promoter reverse primer | this study | PPA01143 | CAGGAAACAGCTATGACCATG |
| Sequence-based reagent | Ppa-che-1 intron reverse primer | this study | PPA01143 | CTGTGATAAGATCATTATTGGTAC |
| Chemical compound, drug | DiI | Molecular Probe | V22889 | 1:150 dilution |
| Chemical compound, drug | DiO | Molecular Probe | V22886 | 1:150 dilution |
| Software, algorithm | TrakEM2 | PMID: 22723842 | EM section alignment | |
| Software, algorithm | 3D reconstruction | bioRxiv 485771 | volumetric reconstruction | |
| Software, algorithm | Photoshop | Adobe | image processing | |
| Software | Image J | Imagej.net | image processing | |
| Algorithm | www.phylogeny.fr | PMID: 18424797 | maximum liklihood phylogeny |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.47155.050





