Diverse deep-sea anglerfishes share a genetically reduced luminous symbiont that is acquired from the environment
Figures
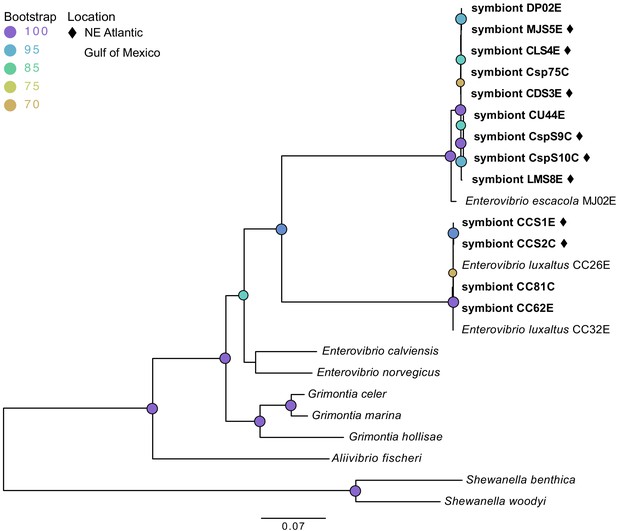
Maximum likelihood phylogenetic tree of bacterial symbionts from conserved housekeeping genes: 16S rDNA, atpA, gapA, gyrB, rpoA, and topA.
General time reversible was selected by modelfinder and a tree was constructed using IQ-TREE with 1000 bootstrap replicates. Those samples unique to this study are bolded, with samples from the Northern Atlantic denoted with ♦, and the bootstrap values over 60 are listed at tree nodes.
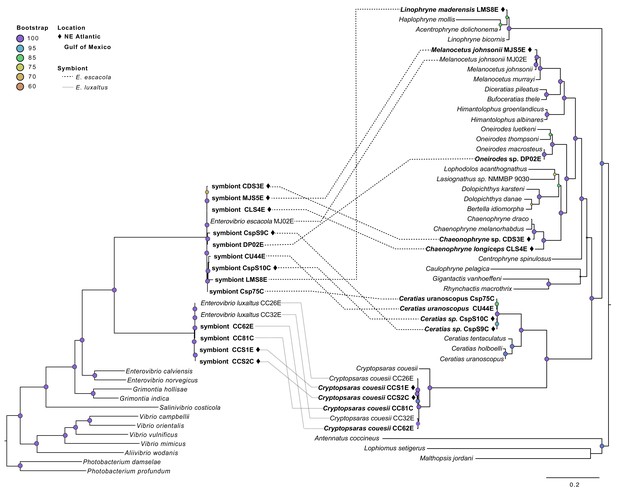
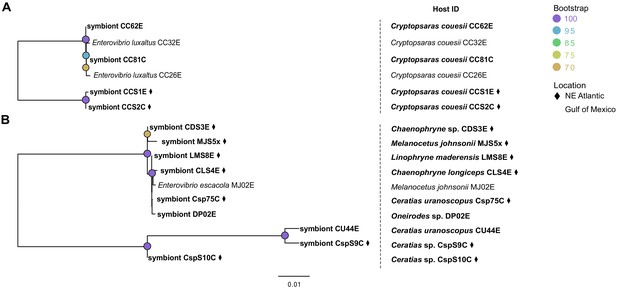
Symbiont phylogeny (left) constructed using single-copy protein-coding genes compared to the host phylogeny constructed using mitochondrial genes (right).
Bolded samples are unique to this study. Samples from the Northern Atlantic denoted with ♦, and the bootstrap values over 60 are listed at tree nodes. Linkages between symbionts and their hosts are shown with dotted lines that differentiate between symbiont species.
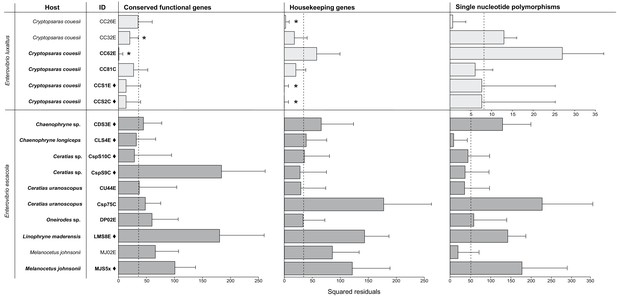
Procrustean Approach to Cophylogeny using a host matrix constructed using mitochondrial gene phylogeny compared to symbiont matrices constructed using the single-copy protein-coding gene phylogeny (p=2e-05) and housekeeping genes phylogeny (p=2e-05).
SNPs phylogenies were analyzed for each species, and the scale for E. luxaltus was dissimilar to the E. ecacola; neither were statistically significant (p>0.5 for analysis of both species). The squared residuals below the median squared residual value (dotted line) are significantly codiverging with the host phylogenies (marked with an asterisk). Sample IDs from the Northern Atlantic are marked with a ♦ and those from the Gulf of Mexico are unmarked.
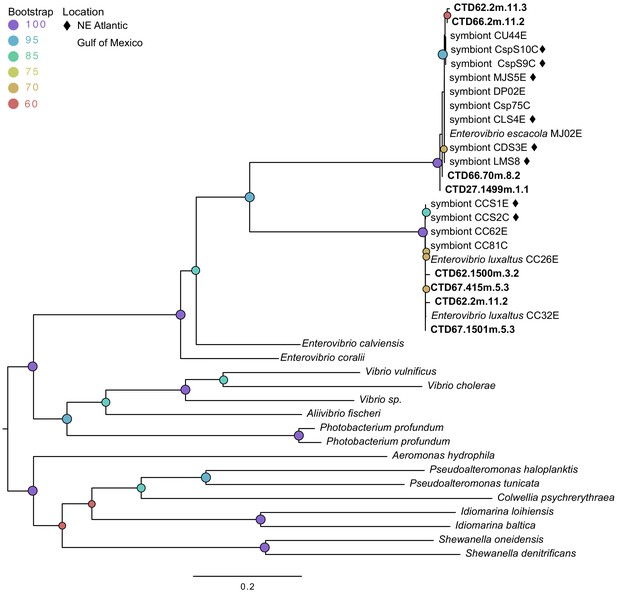
Maximum likelihood phylogenetic tree of cheAfrom environmental samples (bolded) compared to sequences from symbiont genomes isolated from fish and sequences from related species.
Modelfinder selected the general time reversible model and a tree was constructed using IQ-TREE with 1000 bootstrap replicates. Those samples from the Northern Atlantic denoted with ♦, and the bootstrap values over 60 are given at tree nodes.
Phylogenies constructed using single nucleotide polymorphisms for (A) E. luxaltus (2252 SNPs) and (B) E. escacola (15272 SNPs).
Host identifications for each sample are listed in the right-hand column. Samples unique to this study are bolded and those from the Northern Atlantic are marked with a ♦.
Tables
Statistics for symbiont genome sequences analyzed in this study.
Samples that are unique to this study are bolded. For binned genomes, the average nucleotide identity (ANI) of the genome compared to the reference sequence is shown. For E. luxaltus the reference was the CC26 symbiont and E. escacola was the MJ02 symbiont previously documented (Hendry et al., 2018). Results indicating similar species using ANI are bolded. Samples that could not be successfully binned and were not included in the ANI and completeness analysis are marked with a ‘--‘. Samples when compared to themselves are marked with ‘NA’. Statistics for total length and GC content were generated using OrthoANU, the percent completeness was generated using checkM, and the coverage was generated using BBmap. Sample location is denoted with a ♦ for those from the Northern Atlantic and without notation for those from the Gulf of Mexico.
| Sample | FishID | Light organ | Accession # | E. escacola ANI | E. luxaltus ANI | Length (Mb) | GC content (%) | Complete (%) | Ave coverage |
|---|---|---|---|---|---|---|---|---|---|
| CC26E | Cryptopsaras couesii | esca | GCA002300443.1 | 73.7 | NA | 2.14 | 37.7 | 91.3 | 25 |
| CC32E | Cryptopsaras couesii | esca | SRR8206628 | -- | -- | -- | -- | -- | 23 |
| CC81C | Cryptopsaras couesii | caruncle | SRR8206630 | -- | -- | -- | -- | -- | 19 |
| CCS1E ♦ | Cryptopsaras couesii | esca | RPOE00000000 | 73.6 | 99.9 | 2.14 | 37.7 | 90.8 | 567 |
| CCS2C♦ | Cryptopsaras couesii | caruncle | RPOF00000000 | 73.7 | 99.9 | 2.20 | 37.6 | 90.3 | 313 |
| CC62E | Cryptopsaras couesii | esca | SRR8206629 | -- | -- | -- | -- | -- | 19 |
| Csp75C | Ceratias uranoscopus | caruncle | RPGC00000000 | 99.9 | 73.8 | 2.73 | 39.8 | 91.0 | 1600 |
| CspS10C ♦ | Ceratias sp. | caruncle | RPGB00000000 | 99.2 | 73.6 | 2.72 | 39.8 | 91.1 | 99 |
| CspS9C ♦ | Ceratias sp. | caruncle | RPGE00000000 | 99.1 | 73.8 | 2.69 | 39.8 | 89.3 | 26 |
| CU44E | Ceratias uranoscopus | esca | RPGD00000000 | 99.1 | 74.0 | 3.04 | 39.8 | 88.3 | 15 |
| CLS4E ♦ | Chaenophryne longceps | esca | RPGF00000000 | 99.9 | 73.7 | 2.73 | 39.8 | 90.4 | 330 |
| CDS3E ♦ | Chaeonophryne sp. | esca | RPGG00000000 | 99.9 | 73.6 | 2.73 | 39.8 | 89.3 | 291 |
| LMS8E ♦ | Linophryne maderensis | esca | RPGH00000000 | 99.8 | 73.8 | 3.40 | 40.0 | 88.8 | 1 |
| MJ02E | Melanocetus johnsoni | esca | GCA002381345.1 | NA | 73.7 | 2.65 | 39.8 | 89.9 | 766 |
| MJS5x ♦ | Melanocetus johnsoni | esca | RPGI00000000 | 99.9 | 73.8 | 3.09 | 39.8 | 91.1 | 321 |
| DP02E | Oneirodes sp. | esca | RPGJ00000000 | 100.0 | 73.7 | 2.68 | 39.8 | 89.3 | 910 |
A summary of modes of symbiont transmission, examples of some bacterial species and the functions they perform for animal hosts, and trends in the reduction of symbiont genomes.
https://doi.org/10.7554/eLife.47606.009| Transmission | Description | Symbiont and function | Host | Genome | References |
|---|---|---|---|---|---|
| Environmental | Acquired from free-living bacteria  | Luminescence Aliivibrio fischeri Photobacterium leiognathi Photobacterium kishitanii Nutrition "Candidatus Endoriftia persephone" Various Gammaproteobacteria Burkholderia spp. | Fish and squid Fish Fish Tubeworms Mussels Insects | Comprable to free-living relatives  | Dunlap and Urbanczyk, 2013; Gyllborg et al., 2012 Urbanczyk et al., 2011; Ast et al., 2007 Li et al., 2018; Kleiner et al., 2012 Ponnudurai et al., 2017 Kikuchi et al., 2005; Kikuchi et al., 2007 |
| Proposed Environmental | Environmentally persistant cells  | Luminescence "Candidatus Enterovibrio escacola" "Candidatus Enterovibrio luxaltus" | Anglerfish Anglerfish | Ongoing reduction | Hendry et al., 2018 Hendry et al., 2018 |
| Mixed | Pseudovertical or surface transmission  | Luminescence "Candidatus Photodesmus blepharus" "Candidatus Photodesmus katoptron" Nutrition Various Gammaproteobacteria “Candidatus Ishikawaella capsulata” | Flashlight fish Flashlight fish Clams Stink bug | Moderate toextreme reduction  | Hendry et al., 2014; Hendry and Dunlap, 2014 Hendry et al., 2014; Hendry and Dunlap, 2014 Kuwahara et al., 2007 |
| Inherited | Direct passage from parent to offspring on egg or sperm  | Nutrition Buchnera aphidicola Carsonella ruddii Portiera aleyrodidarum Varied "Candidatus Synechococcus spongiarum" | Aphids Psyllids Whiteflies Sponges | Greatly reduced  | Moran et al., 2008; Fisher et al., 2017 Moran et al., 2008; Fisher et al., 2017 Moran et al., 2008; Fisher et al., 2017 Gao et al., 2014; Burgsdorf et al., 2015 |
Additional files
-
Supplementary file 1
Collection data for anglerfish samples used in this study.
Gulf of Mexico samples were collected by the DEEPEND consortium at the given locations. Samples collected by S. Nyholm were collected by RRS Discovery expedition D243 east of the Cape Verde Islands at the given locations. A single sample (DP02E) was given a new taxonomic identification as a result of mitochondrial gene analysis.
- https://doi.org/10.7554/eLife.47606.010
-
Supplementary file 2
Lophiiform mitochondrial sequences used in this study.
Genbank accession numbers and total sequence length are shown. Samples listed in bold were generated by this study or by Hendry et al. (2018).
- https://doi.org/10.7554/eLife.47606.011
-
Supplementary file 3
Sequences from free-living Vibrionaceae species used to generate the housekeeping gene tree.
Genbank accession numbers of each gene are given.
- https://doi.org/10.7554/eLife.47606.012
-
Supplementary file 4
Genomes from free-living Vibrionaceae species used for PhyloPhlan analysis.
Genbank accession numbers of each genome assembly are given.
- https://doi.org/10.7554/eLife.47606.013
-
Supplementary file 5
The species-specific primers designed to amplify the chemotaxis protein cheA.
- https://doi.org/10.7554/eLife.47606.014
-
Supplementary file 6
Species-specific primers designed to amplify the chemotaxis protein cheA.
- https://doi.org/10.7554/eLife.47606.015
-
Transparent reporting form
- https://doi.org/10.7554/eLife.47606.016





