Mamo decodes hierarchical temporal gradients into terminal neuronal fate
Figures
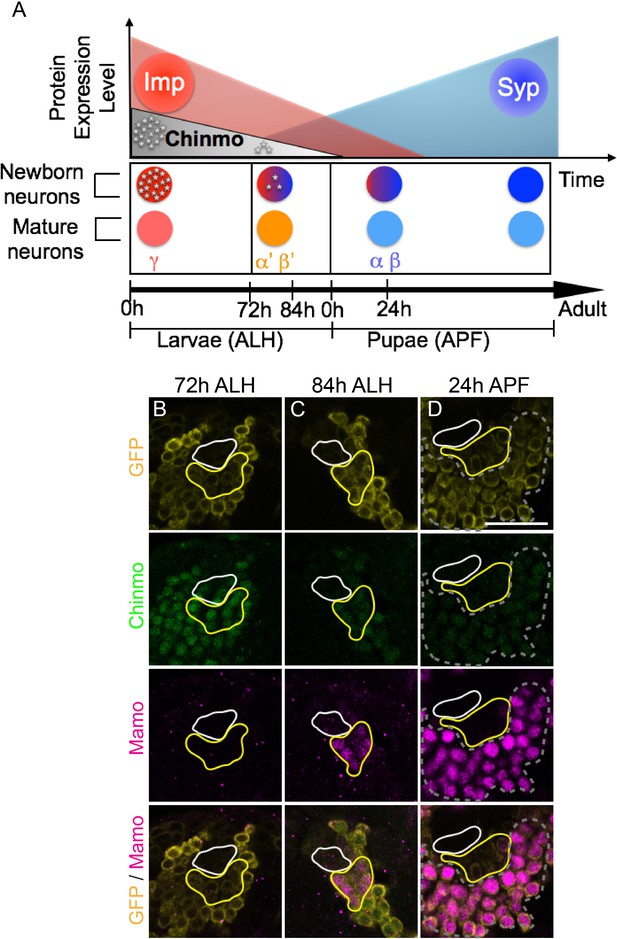
Mamo expression coincides with the generation of α’/β’ neurons in the Mushroom Body (MB) neuronal lineages.
(A) Temporal gradients specify postembryonic neurons of the MB lineages into three sequential neuronal classes (Lee et al., 1999; Liu et al., 2015). Newborn neurons are colored to illustrate expression levels of Imp (red), Syp (blue), and Chinmo (gray stars). ALH = after larval hatching, APF = after pupal formation. (B–D) MB lineages (OK107 > GFP) immunostained for GFP, Chinmo (Rat-anti-Chinmo), and Mamo at different developmental times. A single focal plane near the MB NB is shown. Newborn neurons (NN) are identified by the very dim GFP expression near the NB as described by Zhu et al. (2006) and outlined in white. Young/maturing neurons are immediately adjacent to the NNs with a slightly higher GFP intensity and outlined in yellow. Chinmo levels in NNs decline over time. Mamo staining is visible at 84 hr ALH in young/maturing neurons (C). At 24 hr APF, Mamo expression is strong in older neurons (gray dashed outline), but absent from young/maturing neurons (D). Scale bar = 20 μm. Images are representative of n > 18. The quantification of Chinmo and Mamo staining is in Figure 1—figure supplement 1.
-
Figure 1—source data 1
Intensity of Chinmo and Mamo staining at different developmental times.
- https://doi.org/10.7554/eLife.48056.004
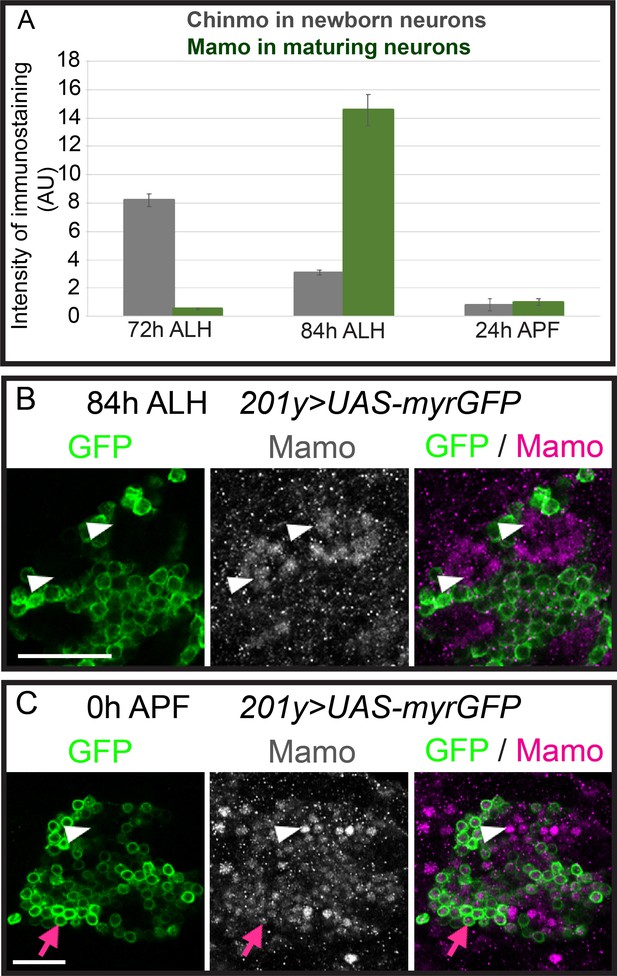
Mamo expression coincides with the generation of α’/β’ neurons in the Mushroom Body (MB) neuronal lineages.
(A) The intensity of Chinmo (gray line) and Mamo (Green line) antibody staining in MB neurons at different developmental times. The Chinmo levels were measured in the newborn neurons and Mamo levels were measured in the young/maturing neurons (mean ± SEM, n = 6 brains). AU, arbitrary fluorescent intensity units. (B–C) Mushroom body lineages (201y > GFP) immunostained for GFP (green), and Mamo (gray/magenta) at 84 hr ALH (B) and 0 hr APF (C). A single focal plane near the MB NB is shown. Mamo (white arrows) staining is visible at 84 hr ALH (B). However, Mamo staining with GFP-positive neurons is evident (MB γ neuron types, red arrow) at 0 hr APF (C). Note that white arrow indicates Mamo staining is not in the GFP-positive neurons. Scale bar = 20 μm. Images are representative of n > 6.
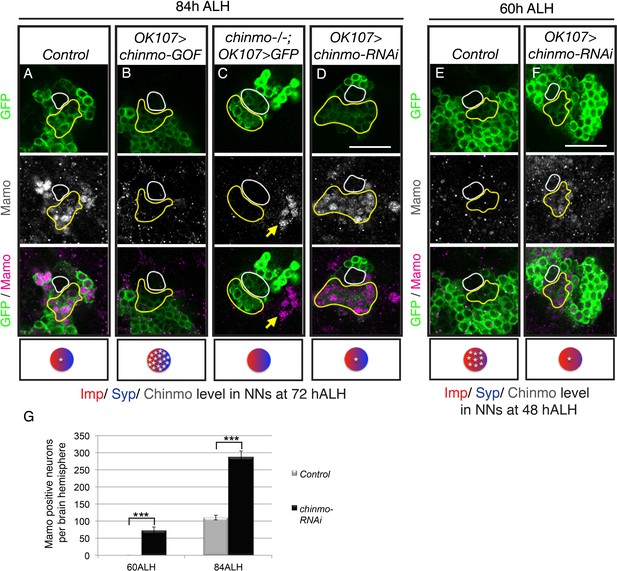
Weak Chinmo initiates Mamo protein expression.
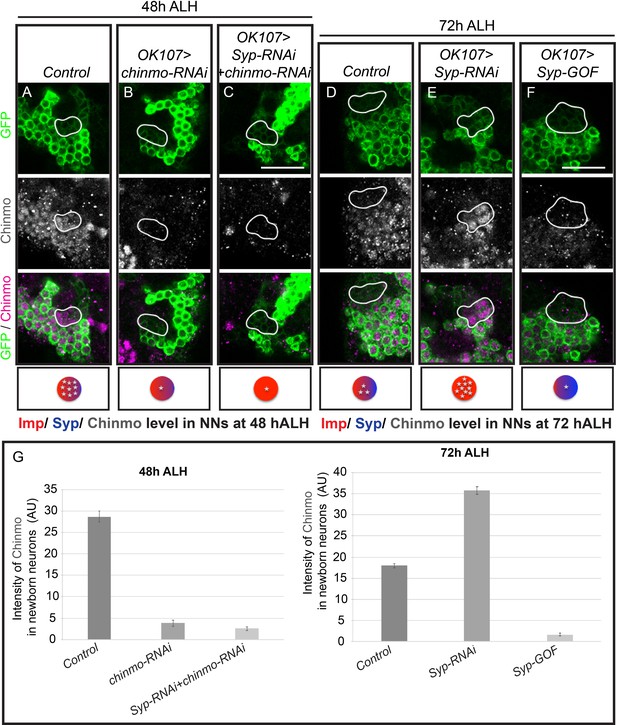
MB lineages (OK107 > GFP) with different genetic manipulations immunostained for GFP and Mamo. A single focal plane near the MB NB is shown. Newborn neurons (NN) are outlined in white. Young/maturing neurons are outlined in yellow. Images are representative of n > 18. Scale bar = 20 μm. The diagram below shows approximate levels of Imp (red), Syp (blue), and Chinmo (stars) expressed in the young/maturing neurons when they were NNs ~ 12 hr prior (as reported by Liu et al., 2015), or Figure 3—figure supplement 1). (A-F) At 84 hr ALH, Mamo staining is visible only in the young/maturing neurons in control brains (A) and brains expressing chinmo-RNAi (D). (C) chinmo-/-; OK107 > GFP is a chinmo null MARCM clone induced at newly hatched larvae (NHL). Note that OK107 drives GFP only within the clone. MB neurons outside of the chinmo-/- clone (eyeless+, data not shown) express Mamo (yellow arrow). (F) At 60 hr ALH, Mamo staining is only visible after OK107 > chinmo RNAi. (G) Mean number (± SEM) of Mamo positive neurons per brain hemisphere in control (gray) and chinmo-RNAi (black) expressing MBs (***p<0.005, n = 4–5). The quantification of Mamo staining is in Figure 2—figure supplement 1.
-
Figure 2—source data 1
Quantification of Mamo positive neurons.
- https://doi.org/10.7554/eLife.48056.007
-
Figure 2—source data 2
Mamo staining intensity in young/maturing neurons with chinmo manipulations.
- https://doi.org/10.7554/eLife.48056.008
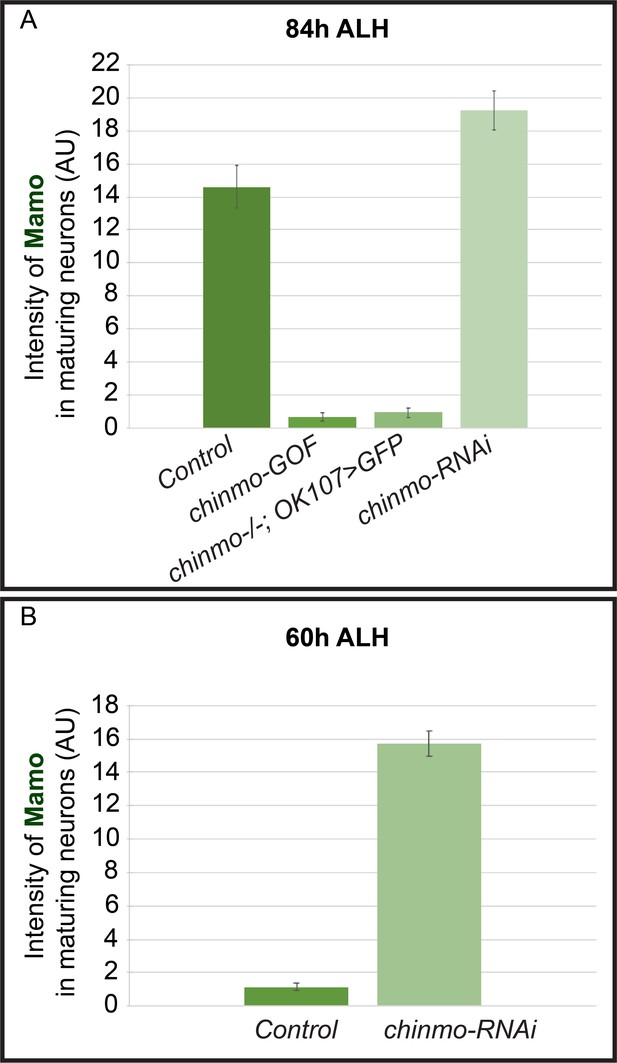
The intensity quantification of Mamo staining in different manipulations of MB neurons.
(A–B) The intensity of Mamo (Green line) staining in MB neurons at 84 hr ALH (A) and 60 hr ALH (B). The Mamo levels were measured in the young/maturing neurons (mean ± SEM, n = 5–6 brains). AU, arbitrary fluorescent intensity units.
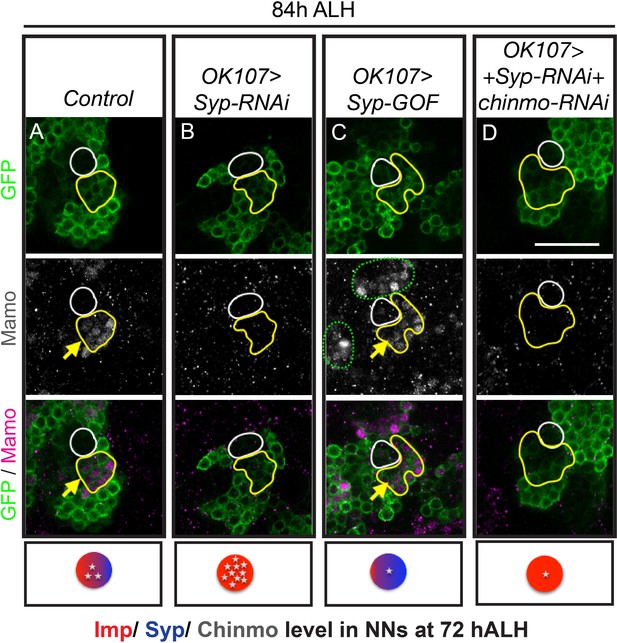
Mamo protein expression requires Syp RNA binding protein.
MB lineages (OK107 > GFP) immunostained for GFP and Mamo. A single focal plane near the MB NB is shown. Newborn neurons (NN) are outlined in white and young/maturing neurons are outlined in yellow. Arrows indicate regions with Mamo protein expression. Images are representative of n > 18. Scale bar = 20 μm. The diagram below shows the relative protein levels of Imp (red), Syp (blue), and Chinmo (stars) expressed in the young/maturing neurons when they were newborn 12 hr prior (based on Figure 3—figure supplement 1 and Liu et al., 2015). (A-D) Mamo expression in young/maturing neurons occurs in genotypes with low Chinmo levels (A and C) with the exception of Syp-RNAi plus chinmo-RNAi (D). Note that green dashed circle is labeling other MB neurons. Chinmo immunostaining and quantifications from earlier stages can be found in Figure 3—figure supplement 1. Mamo levels are shown in Figure 3—figure supplement 2.
-
Figure 3—source data 1
Mamo staining intensity in young/maturing neurons with Syp manipulations.
- https://doi.org/10.7554/eLife.48056.012
-
Figure 3—source data 2
Chinmo staining intensity in newborn neurons with different genetic manipulations.
- https://doi.org/10.7554/eLife.48056.013
Syp gradient alter Chinmo protein expression.
(A–F) Larval MB lineages (OK107 > GFP) immunostained for Chinmo (gray/magenta, Rabbit-anti-Chinmo) and GFP (green). A single focal plane near the MB NB is shown. Newborn neurons are outlined in white. The diagram on the bottom shows the relative protein levels of Imp (red), Syp (blue) and Chinmo (stars) expressed in the young/maturing neurons when they were newborn 12 hr prior. Images are representative of n > 18 Scale bar = 20 μm. (G) The intensity of Chinmo immunostaining (gray line) in different genetic manipulations of MB neurons at different developmental times. The Chinmo levels were measured in the newborn (mean ± SEM, n = 6 brains). AU, arbitrary fluorescent intensity units.
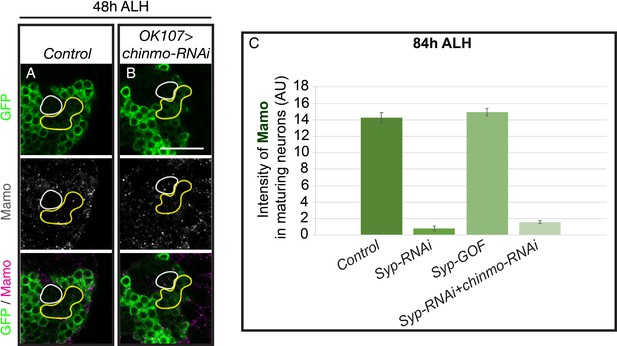
Mamo is absent with premature low Chinmo levels before 60 hr ALH.
Mushroom body lineages (OK107 >GFP) immunostained for GFP (green) and Mamo (gray/magenta). A single focal plane near the MB NB is shown. Newborn neurons (NN) are outlined in white. Yellow outlines indicate young/maturing neuron region immediately adjacent to the NNs. Images are representative of n > 18. Scale bar = 20 μm. (A-B) At 48 hr ALH, Mamo staining is absent from control (A) and OK107 >chinmo RNAi (B). (C) The intensity of Mamo (Green line) staining in different genetic manipulations of MB neurons at 84 hr ALH. The Mamo levels were measured in the young/maturing neurons (mean ± SEM, n = 5–6 brains). AU, arbitrary fluorescent intensity units.
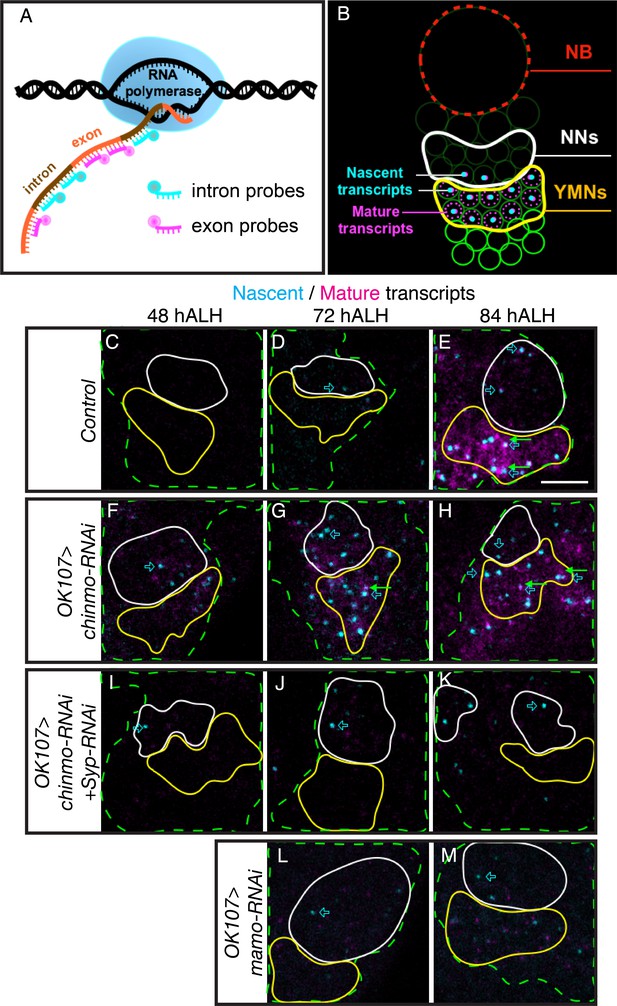
Syp promotes sustained mamo transcription.
(A) Graphic illustrating the use of intron and exon probes for single molecule florescent in situ hybridization (smFISH). Nascent transcripts are labeled by both intron and exon probes, while mature transcripts are only labeled by exon probes. (B) Diagram illustrating interpretation of smFISH data. Active transcription is seen as a single, double-labeled focus per cell. Mature transcripts (magenta only) are diffuse and cytoplasmic. (C–M) smFISH with probes targeting mamo intronic (cyan) or exonic (magenta) sequences. Images are of developing larval brains with different genetic manipulations of the MB. Maximum Intensity Z-projections (2.3–3.8 μm) near the MB NB are shown. MB cells are determined by OK107 >GFP (green dashed outline) and the newborn neuron (NN) region is outlined in white and the young/maturing neuron (YMN) region is outlined in yellow. Blue open arrows highlight examples of mamo active transcription, green arrows highlight examples of mature mamo transcripts. Images are representative of n > 6. Scale bar = 5 μm. Control brains (OK107 >GFP) show active transcription in NNs at 72 hr (D) and in NNs and young/maturing neurons at 84 hr (E). Mature transcripts are visible in young/maturing neurons at 84 hr ALH (E). OK107 >chinmo-RNAi results in a shift in the timing of mamo transcription. Active transcription is visible at 48 hr in both NNs and young/maturing neurons (F) and is abundant at 72 hr and 84 hr ALH (G and H). Note that MBs expressing chinmo-RNAi together with Syp-RNAi have active transcription in NNs at all time points, but lack mature transcripts and active transcription in young/maturing neurons (I–K). Depleting mamo (OK107 >mamo-RNAi) causes loss of mature transcripts and active transcription in young neurons (L–M). The quantification of mamo mature transcripts is in Figure 4—figure supplement 1.
-
Figure 4—source data 1
Quantifications of mature mamo transcript.
- https://doi.org/10.7554/eLife.48056.016
-
Figure 4—source data 2
Intron and exon probe sequences.
- https://doi.org/10.7554/eLife.48056.017
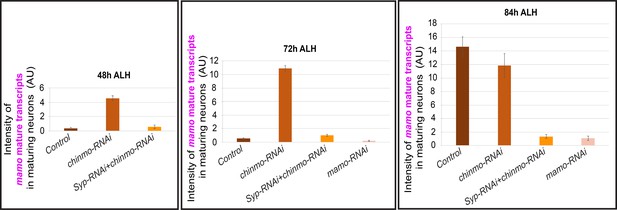
The intensity quantification of mamo mature transcripts in different manipulations of MB neurons.
The intensity of mamo (different color lines) mature transcripts in MB neurons at 48 hr, 72 hr, 84 hr ALH. The Mamo levels were measured in the young/maturing neurons (mean ± SEM, n = 4–6 brains). AU, arbitrary fluorescent intensity units.
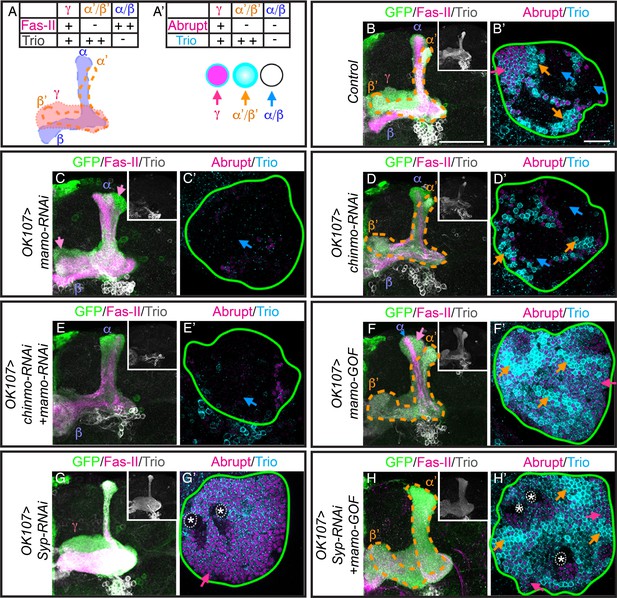
Mamo is necessary and sufficient for the α’/β’ fate.
(A) Schematic of MB lobe morphology and table of corresponding marker expression. (B–H) Adult MB lobes (OK107 > GFP) immunostained for GFP, Fas-II and Trio. Images are Z-stack projections of the axon region and are representative of n > 18. Lobes are identified based on both 3D structure and marker expression. α’/β’ lobes are outlined with orange dashed lines. Scale bar = 50 μm. Insert shows Trio staining alone. (A’) MB cell body markers that distinguish three MB neuron types. (B’–H’) Adult MB cell bodies (OK107 > GFP) immunostained for Abrupt, Trio and GFP. GFP channel is not shown, but is represented by a green outline. Colored arrows highlight MB neuron types red=γ ( TrioPM/Abrupt+ ), orange=α’/β’ (TrioPM,Cyto/Abrupt-), blue=α/β (Trio-/Abrupt-). Images are representative of n > 6. A single focal plane is shown. Scale bar = 20 μm. Note wide Fas-II++ α/β lobes and a morphologically indistinct FasII weak/negative lobe (magenta arrows) with mamo-RNAi (C). mamo-RNAi and chinmo-RNAi + mamo-RNAi both lack most cell body marker staining (C’ and E’). γ lobes and cell body markers are reduced in chinmo-RNAi alone (D and D’). Mamo overexpression (mamo-GOF) results in an expanded α’/β’ lobe (F) and increased numbers of TrioPM,Cyto/Abrupt- cell bodies (F’’). Note the Fas-II++, Trio- axons in the A/P (α), but not medial/lateral (β) portion of the axon lobe (blue arrow) which is surrounded by FasII-/weak, Trio+, morphologically indistinct axons (magenta arrow) in mamo-GOF (F). The cell body region is overwhelmingly Trio+ (F’). Syp-RNAi MB (G and G’) shows only γ neurons (note the A/P axon bundle characteristic of un-remodeled γ neurons). Syp-RNAi plus mamo-GOF produced expanded α’/β’ lobes (H) and mostly TrioPM,Cyto/Abrupt- cell bodies, with some TrioPM/Abrupt+ cells (H’). Note proliferating NBs (*) and adjoining unspecified young/maturing neurons produced with mamo-GOF (G’ and H’). The analysis of Mamo variants is in Figure 5—figure supplement 1. The analysis of hierarchical model is in Figure 5—figure supplement 2. The quantification of neuron populations is in Figure 5—figure supplement 3.
-
Figure 5—source data 1
Quantification of adult MB neuron types.
- https://doi.org/10.7554/eLife.48056.022
-
Figure 5—source data 2
Intensity of Imp/Syp/Chinmo staining.
- https://doi.org/10.7554/eLife.48056.023
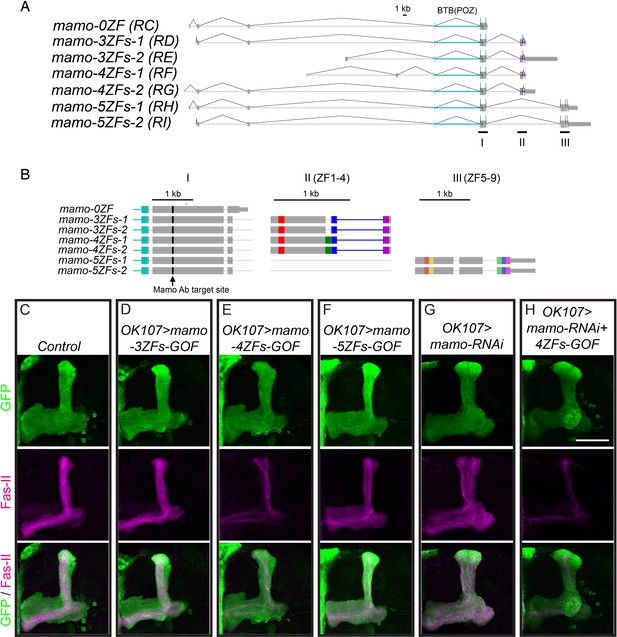
The Mamo variant containing 4ZFs is the prospective isoform acting in α’/β’ temporal fate determination.
(A) mamo mRNA isoforms. Gray boxes show UTRs. Cyan labels BTB (POZ) domain at N-terminus, Different colors at C-terminus label zinc finger motifs. (B) Detailed motif descriptions of I, II, and III in (A). The Mamo antibody (Ab) target site (black arrow) is in N-terminus of the common region. There are nine ZF motifs labeled with different colors. (C–H) Adult MBs were immunostained for GFP (green) and Fas-II (magenta). Images are Z-stack projections (standard deviation) of the axon region and are representative of n > 6. Lobes are identified based on both 3D structure as well as marker expression. Scale bar = 50 μm. (D–F) Overexpression of different mamo variants with OK107. mamo-4ZFs-GOF results in altered MB lobe morphology with reduced Fas-II staining (E) compared to control (C). mamo-5ZFs-GOF expands Fas-II staining to a majority of axons. (G–H) mamo variant with 4ZFs rescues mamo-RNAi. Expression of mamo-RNAi induces expanded Fas-II staining (G). mamo-RNAi together with mamo-4ZFs-GOF results in a reduced Fas-II positive lobe, similar to mamo-4ZFs-GOF alone (E).
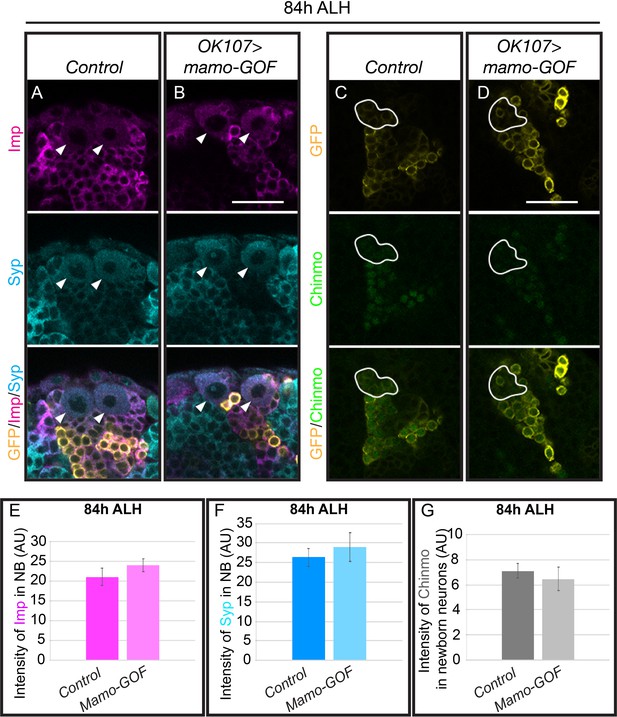
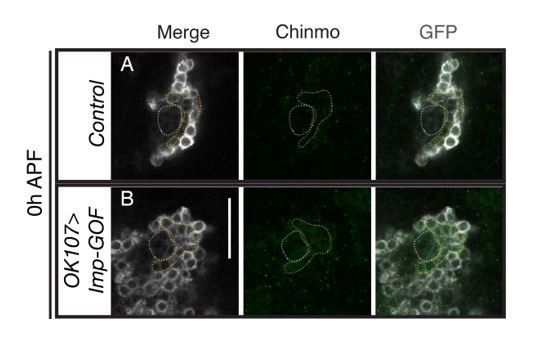
Mamo acts as a downstream factor of Imp/Syp/Chinmo gradients.
(A–B) Larval MBs (OK107 > GFP) immunostained for Imp (magenta), Syp (Cyan) and GFP (yellow). NB are indicated with arrows. (C–D) Larval MBs (OK107 > GFP) immunostained for Chinmo (green, Rat-anti-Chinmo) and GFP (yellow). Newborn neurons are outlined in white. A single focal plane near the MB NB is shown. Images are representative of n > 6 Scale bar = 20 μm. (E–G) The intensity of Imp, Syp and Chinmo immunostaining in different genetic manipulations of MB neurons at 84 hr ALH. The Imp and Syp levels were measured in the NB and the Chinmo levels were measured in the newborn (mean ± SEM, n = 5–7 brains). AU, arbitrary fluorescent intensity units.
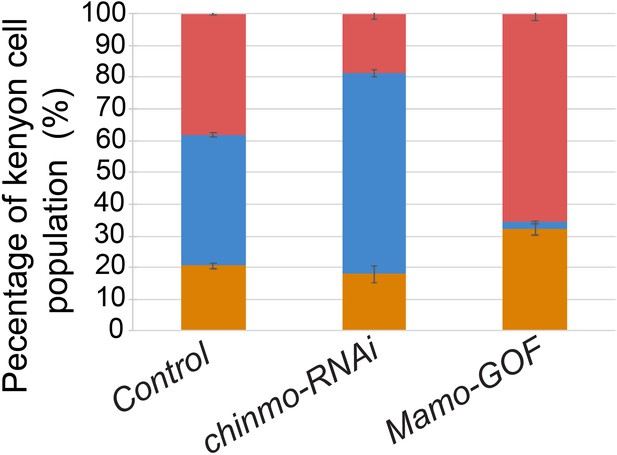
The quantification of MB neuron types.
Different populations of MB neurons were counted and showed in percentage based on marker expression (Trio/Abrupt) in control, chinmo-RNAi and Mamo-GOF. Colored lines highlight MB neuron types, red=γ, orange=α’/β’ and blue=α/β neurons (mean ± SEM, n = 3 brains).
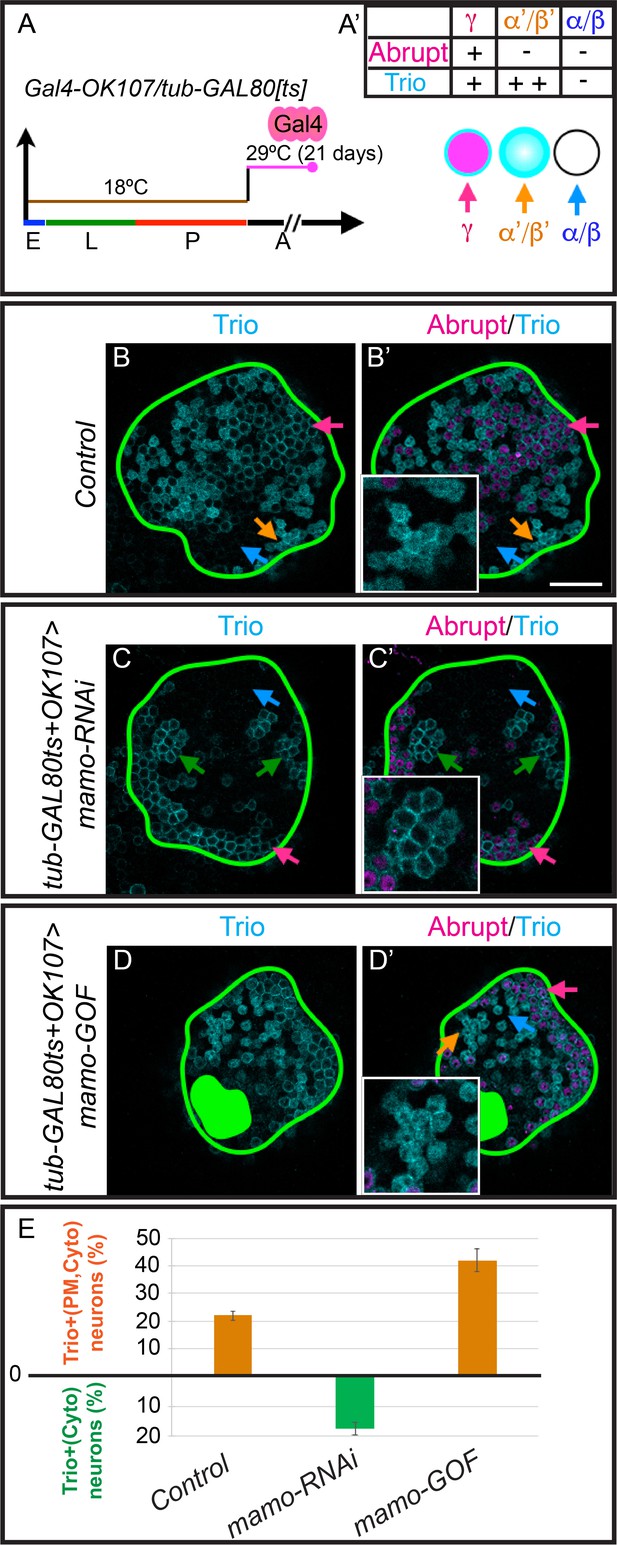
Mamo maintains α’/β’ MB neuron markers.
(A) Scheme for temperature shift assay. Temperature-sensitive GAL80 was inactivated for 21 days after adult eclosion. E = embryo, L = larva, p=pupa, A = adult (A’) MB cell body marker expression of the three MB neuron types. (B–D) Adult MB cell bodies (OK107 > GFP+tub-GAL80ts) immunostained for GFP, Abrupt, and Trio. Colored arrows identify MB neuron types based on marker expression: red=γ (TrioPM/Abrupt+), orange=α’/β’ (TrioPM,Cyto /Abrupt-), blue=α/β (Trio-/Abrupt-). Images are representative of n > 6. A single focal plane is shown. Scale bar = 20 μm. Notice loss of cytoplasmic Trio staining after depleting expressing mamo-RNAi (C and C’, green arrows = TrioPM/Abrupt- neurons). Overexpressing Mamo (mamo-GOF) after adult eclosion results in increased numbers of TrioPM,Cyto/Abrupt- neurons (green filled area indicates MB calyx) (D and D’). (E) Percent of MB neurons expressing both cytoplasmic and plasma membrane Trio in orange (TrioPM,Cyto/Abrupt-) and percent of neurons expressing only plasma membrane Trio (TrioPM/Abrupt-) in green (mean ± SEM, n = 3 brains). Time course of Mamo depletion with adult induced RNAi is shown in Figure 6—figure supplement 1.
-
Figure 6—source data 1
Quantification of Trio expression (PM, Cyto).
- https://doi.org/10.7554/eLife.48056.027
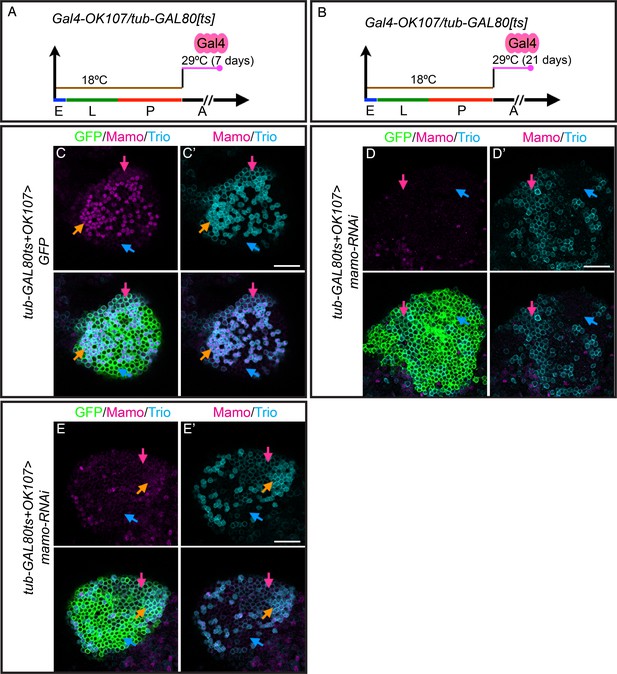
Temperature shift assay for effectively repressing Mamo.
(A and B) Scheme for temperature shift assay. Heat shock inactivated the temperature-sensitive GAL80 for 7 days after adult eclosion (A) and for 21 days after adult eclosion (B). (C–E) Adult MB cell bodies (OK107 > GFP+tub-GAL80ts) immunostained for GFP (green) and Mamo (magenta) and Trio (cyan). Colored arrows identify MB neuron types red=γ (Trio+PM/Mamo+low ), orange=α’/β’ (Trio+PM,Cyto/ Mamo+high), blue=α/β (Trio-/Mamo-) based on marker expression. Images are representative of n > 6. A single focal plane is shown. Scale bar = 20 μm. Depleting mamo (mamo-RNAi) for 7 days after adult eclosion does not eliminate Trio+PM,Cyto staining (E’). Most Mamo staining is still visible (E). Depleting mamo (mamo-RNAi) for 21 days after adult eclosion shows effectively diminish Mamo expression and shows qualitative reduction of Trio+PM,Cyto staining (D’ and Figure 6C and C’).
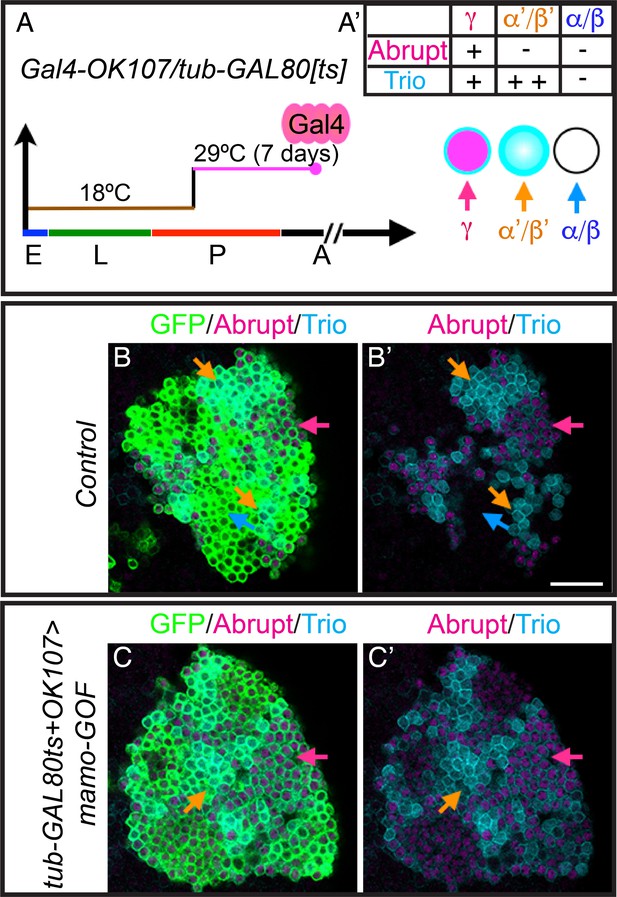
Mamo stimulates α’/β’ specific gene expression.
(A) Scheme for temperature shift assay. Heat shock inactivated the temperature-sensitive GAL80 for 7 days at 2 days after pupal formation. (B–C) Adult MB cell bodies (OK107 > GFP+tub-GAL80ts) immunostained for GFP (green) and Abrupt (magenta) and Trio (cyan). Colored arrows identify MB neuron types red=γ ( Trio+PM/Mamo+low ), orange=α’/β’ (Trio+PM,Cyto/ Mamo+high), blue=α/β (Trio-/Mamo-) based on marker expression. Images are representative of n > 6. A single focal plane is shown. Scale bar = 20 μm. Transgene induction at pupal development was more effective at increasing TrioPM,Cyto/Abrupt- cells (C and C’).
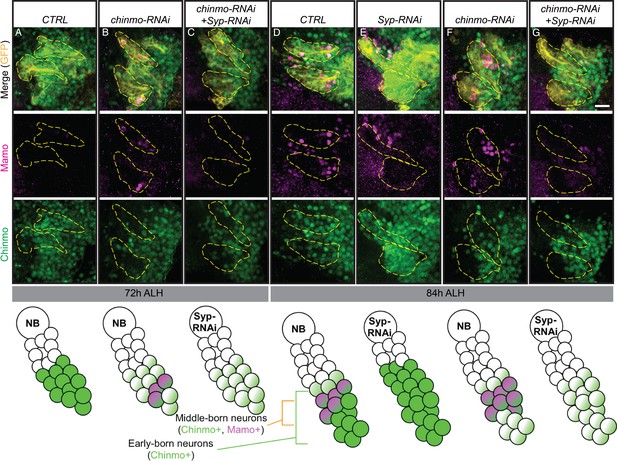
Weak Chinmo drives Mamo expression in AL lineages.
Immunostaining for GFP (yellow), Mamo (magenta) and Chinmo (green) in AL lineages expressing chinmo-RNAi, Syp-RNAi, or dual chinmo/Syp-RNAi. Images are Z-stack projections (standard deviation) of the cell body region and are representative of n > 10. Two AL lineages are outlined with yellow dashed lines based on GR44F03 lineage restricted actin >GFP expression. GR44F03 lineage restricted actin-LexA also drives RNAi transgenes. Scale bar = 10 μm. The diagrams below summarize the protein levels of Mamo and Chinmo. (A-C) 72 hr ALH larval brains. Control brains have no visible Mamo staining within the AL lineages (A). Mamo staining is visible after reducing Chinmo levels with chinmo-RNAi (B). chinmo-RNAi+Syp-RNAi results in reduced Chinmo, but no Mamo expression (C). (D-G) 84 hr ALH. Control brains have Mamo positive cells in AL lineages (D). Syp-RNAi produces expanded Chinmo and loss of Mamo expression (E). chinmo-RNAi reduces Chinmo and increases the number of Mamo positive cells (F). chinmo-RNAi + Syp-RNAi results in weak Chinmo expression and loss of Mamo staining (G).
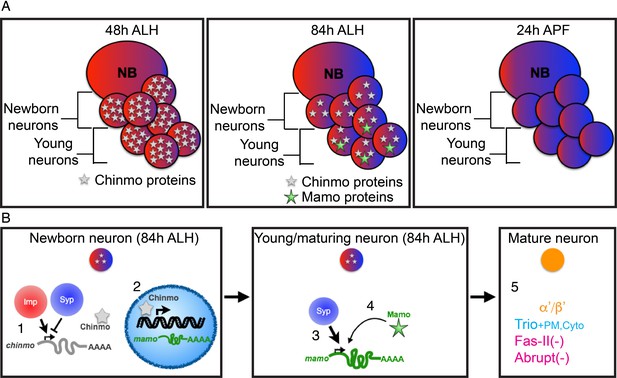
Schematic of α’/β’ neuronal fate determination.
(A) Diagram of the three temporal windows of MB development. Images are color coded to illustrate the expression level of Imp (red) and Syp (blue). Chinmo (gray stars) and Mamo (green stars) levels are also indicated. (B) Hierarchical regulation of α’/β’ neuronal fate determination. (1) Balance of Imp and Syp affects chinmo translation in the newborn neuron, producing low Chinmo levels (Liu et al., 2015). (2) Low Chinmo levels initiate mamo transcription in the newborn neuron. (3) Syp promotes mamo mRNA maturation/stabilization during neuron maturation. (4) Mamo positively autoregulates its own expression. (5) Mamo promotes α’/β’ specific gene expression in the mature neuron.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | anti-Mamo (Rabbit polyclonal) | This paper: Materials and methods | (1:1000), Lee T, Janelina Research Campus, HHMI | |
| Antibody | anti-GFP, Alexa488 (Rabbit polyclonal) | Thermo Fisher Scientific | Cat # A-21311; RRID:AB_221477 | (1:1000) |
| Antibody | anti-GFP (Chicken polyclonal) | Thermo Fisher Scientific | Cat # A10262; RRID:AB_2534023 | (1:1000) |
| Antibody | anti-Chinmo (Rabbit polyclonal) | Zhu et al., 2006 | (1:1000) | |
| Antibody | anti-Chinmo (Rat) | Wu et al., 2012 | (1:500) | |
| Antibody | anti-Trio (Rabbit) | Awasaki et al., 2000 | (1:1000) | |
| Antibody | anti-Abrupt (Rabbit) | Hu et al., 1995 | (1:200) | |
| Antibody | anti-Imp (Rabbit) | gift from Paul Macdonald | (1:600) | |
| Antibody | anti-Syp (Genia pig) | gift from Ilan Davis | (1:500) | |
| Antibody | anti-Trio (Mouse monoclonal) | Developmental Studies Hybridoma Bank | 9.4A;Registry ID:AB_528494 | (1:200) |
| Antibody | anti-Fas-II (Mouse monoclonal) | Developmental Studies Hybridoma Bank | 1D4; Registry ID:AB_528235 | (1:40) |
| Antibody | anti-nc82 (Mouse monoclonal) | Developmental Studies Hybridoma Bank | nc82; Registry ID:AB_2314866 | (1:100) |
| Antibody | anti-chicken, Alexa488 (Goat) | Thermo Fisher Scientific | Cat # A-11039; RRID:AB_2534096 | (1;500) |
| Antibody | anti-mouse, Alexa568 (Goat) | Thermo Fisher Scientific | Cat # A-11031; RRID:AB_144696 | (1;500) |
| Antibody | anti-rabbit, Alexa647 (Goat) | Thermo Fisher Scientific | Cat # A-21244; RRID:AB_2535812 | (1;500) |
| Antibody | anti-Rat, Alexa568 (Goat) | Thermo Fisher Scientific | Cat # A-11077; RRID:AB_2534121 | (1;500) |
| Antibody | anti-rabbit, Alexa568 (Goat) | Thermo Fisher Scientific | Cat # A-11036; RRID:AB_10563566 | (1;500) |
| Antibody | anti-mouse, Alexa647 (Donkey) | Jackson ImmunoResearch lab, Inc. | Cat # 715-605-151 | (1;500) |
| Antibody | anti-Rat, DyLight405 (Goat) | Jackson ImmunoResearch lab, Inc. | Cat # 112-475-167 | (1;200) |
| Chemical compound, drug | Paraformadehyde20% Solution, EM Grade | Electron Microscopy Sciences | Cat # 15713 | |
| Chemical compound, drug | Phosphate Buffered Saline 10X,Molecular Biology Grade | Thermo Fisher Scientific | Cat # 46–013 CM | |
| Chemical compound, drug | Triton X-100 | Sigma-Aldrich | Cat # 329830772 | |
| Chemical compound, drug | SlowFadeTM Gold antifade Mountant | Thermo Fisher Scientific | Cat # S36936 | |
| Chemical compound, drug | RNase-free 1x PBS | Thermo Fisher Scientific | Cat # BP2438-4 | |
| Chemical compound, drug | Acetic Acid, Glacial | Thermo Fisher Scientific | Cat # A38S-500 | |
| Chemical compound, drug | Sodium borohydride | Acros Organics/Thermo Fisher Scientific | Cat # AC448481000 | |
| Chemical compound, drug | Invitrogen SSC (20X) | Thermo Fisher Scientific | Cat # AM9763 | |
| Chemical compound, drug | Hi-Di formamide | Applied Biosystems/Thermo Fisher Scientific | Cat # 4311320 | |
| Chemical compound, drug | Alfa Aesar Denhardt's solution (50X) | Alfa Aesar/Thermo Fisher Scientific | Cat # AAJ63135AD | |
| Chemical compound, drug | tRNA from Baker's yeast | Roche | Cat # 10109495001 | |
| Chemical compound, drug | UltraPure Salmon Sperm DNA Solution | Thermo Fisher Scientific | Cat # 15632011 | |
| Chemical compound, drug | Corning 10% SDS | Corning/Thermo Fisher Scientific | Cat # 46–040 CI | |
| Chemical compound, drug | Deionized formamide | Ambion/Thermo Fisher Scientific | Cat # AM9342 | |
| Chemical compound, drug | RNaseZap RNase Decontamination Solution | Thermo Fisher Scientific | Cat # AM9780 | |
| Chemical compound, drug | Poly-L-lysine hydrobromide | Sigma-Aldrich | Cat # P1524-25MG | |
| Chemical compound, drug | Cy3 Mono-Reactive Dye Pack | GE Healthcare Life Sciences | Cat # PA23001 | |
| Chemical compound, drug | Cy5 Mono-Reactive Dye Pack | GE Healthcare Life Sciences | Cat # PA25001 | |
| Chemical compound, drug | Ethyl alcohol, pure | Sigma-Aldrich | Cat # 459844 | |
| Chemical compound, drug | Xylenes | Thermo Fisher Scientific | Cat # X5-500 | |
| Chemical compound, drug | DPX mountant | Electron Microscopy Sciences | Cat # 13512 | |
| Genetic reagent (D. melanogaster) | tub-Gal80ts | Bloomington Drosophila stock center | BDSC:7018; FLYB:FBst0007018; RRID:BDSC_7018 | FlyBase symbol:P{w[+mC]=tubP-GAL80[ts]}ncd[GAL80ts-7] |
| Genetic reagent (D. melanogaster) | UAS-Syp-RNAi | Vienna Drosophila RNAi Center | VDRC:v33012; FLYB:FBst0459886 | FlyBase symbol: P{GD9477}v33012 |
| Genetic reagent (D. melanogaster) | UAS-mamo-RNAi | Bloomington Drosophila stock center | BDSC:51770; FBti0157732; RRID:BDSC_51770 | FlyBase symbol:P{TRiP.HMC03325}attP40 |
| Genetic reagent (D. melanogaster) | UAS-mamo-RNAi | Bloomington Drosophila stock center | BDSC: 44103; FBti0158705; RRID:BDSC_44103 | FlyBase symbol:P{TRiP.HMS02823}attP40 |
| Genetic reagent (D. melanogaster) | UAS-mCD8-GFP; +; GAL4-OK107 | Connolly et al., 1996 | ||
| Genetic reagent (D. melanogaster) | UAS-chinmo-RNAi | Liu et al., 2015 | ||
| Genetic reagent (D. melanogaster) | UAS-chinmo-GOF (UAS-chinmo-3UTR) | Zhu et al., 2006 | ||
| Genetic reagent (D. melanogaster) | UAS-Syp-GOF | Liu et al., 2015 | ||
| Genetic reagent (D. melanogaster) | UAS-mamo-3ZFs-GOF | Mukai et al., 2007 | ||
| Genetic reagent (D. melanogaster) | UAS-mamo-4ZFs-GOF | This paper: Materials and methods | Lee T, Janelina Research Campus, HHMI | |
| Genetic reagent (D. melanogaster) | UAS-mamo-5ZFs-GOF | This paper: Materials and methods | Lee T, Janelina Research Campus, HHMI | |
| Genetic reagent (D. melanogaster) | Dpn > KDRT-stop-KDRT>Cre PEST; act > loxP-stop-loxP>LexA::P65, lexAop2-myr::GFP; GR44F03-KD | Awasaki et al., 2014 | ||
| Genetic reagent (D. melanogaster) | LexAop2-chinmo-RNAi | This paper: Materials and methods | Lee T, Janelina Research Campus, HHMI | |
| Genetic reagent (D. melanogaster) | LexAop2-Syp-RNAi | Ren et al., 2017 | ||
| Genetic reagent (D. melanogaster) | UAS-mCD8-GFP-insu-UAS-rCD2-RNAi, chinmo1, FRT40A | Kao et al., 2012 | ||
| Genetic reagent (D. melanogaster) | hs-FLPop; tub-GAL80, FRT40A; +; GAL4-OK107 | This paper: Materials and methods | Lee T, Janelina Research Campus, HHMI | |
| Software, algorithm | Fiji | NIH; Schindelin et al., 2012 | https://fiji.sc/ | |
| Software, algorithm | Adobe Photoshop | Adobe Systems, San Jose, CA | https://www.adobe.com/products/photoshop.html | |
| Software, algorithm | Adobe Illustrator | Adobe Systems, San Jose, CA | https://www.adobe.com/products/illustrator.html | |
| Software, algorithm | Python | Python Software Foundation | https://www.python.org/ | |
| Software, algorithm | Flybase 2.0 | Thurmond et al., 2019 | http://flybase.org | |
| Software, algorithm | Matplotlib | Hunter, 2007 | https://matplotlib.org |
Additional files
-
Source code 1
The code for mamo gene structure diagram.
- https://doi.org/10.7554/eLife.48056.030
-
Transparent reporting form
- https://doi.org/10.7554/eLife.48056.031