Evolution of Yin and Yang isoforms of a chromatin remodeling subunit precedes the creation of two genes
Figures
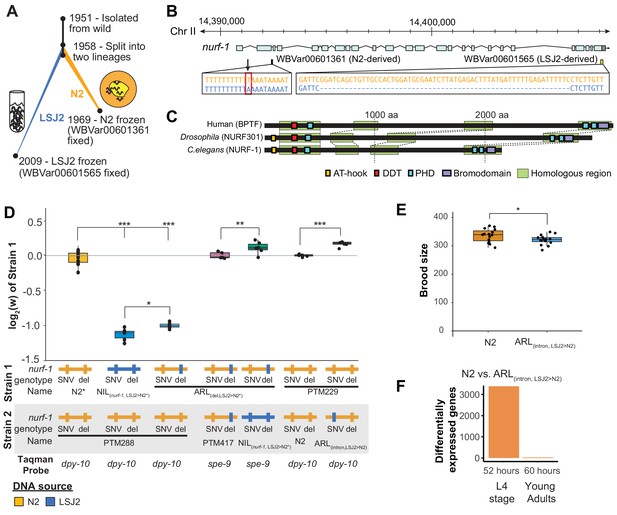
An N2-derived genetic variant in the intron of nurf-1 increases fitness in laboratory conditions.
(A) History of two laboratory adapted C. elegans strains N2 and LSJ2, which descend from the same individual hermaphrodite isolated in 1951. The N2 and LSJ2 lineage split sometime around 1958. N2 grew on agar plates with E. coli OP50 as a food source for around 11 years until they were cryopreserved. LSJ2 animals were cultured in liquid axenic media containing sheep liver extract and soy extract peptone as a food source for about around 51 years until they were cryopreserved. 302 genetic variations were fixed between these two strains, including two that fall in the nurf-1 gene – WBVar00601361 and WBVar00601565. (B) Genetic location of two nurf-1 variations. WBVar00601361 (in red box) is an N2-derived intron single nucleotide substitution T/A (N2/ancestral) in the 2nd intron of nurf-1. WBVar00601565 is an LSJ2-derived 60 bp deletion in the 3’ end of nurf-1 that removes the last 18 amino acids and part of the 3’-UTR. (C) Comparison of NURF-1 orthologs from Drosophila and humans showing position of protein domains and conserved regions as determined by Blastp and Clustal Omega. (D) Boxplot of pairwise evolutionary fitness differences between the indicated strains measured by directly competing the indicated strains against each other for five generations. PTM288 and PTM229 were created from the N2* and N2 strains, respectively, by a engineering DNA barcode in the dpy-10 gene. PTM417 is the same genotype as ARL(del_LSJ2>N2), with the exception of a background mutation in the spe-9 gene that occurred during the construction of the ARL(del_LSJ2>N2) strain (for details see Methods). This mutation was crossed out of the PTM417 strain and used as a barcode for the digital PCR reaction. The genotype of each nurf-1 allele (shown in B) is indicated by color. The NIL strain also contains LSJ2 alleles of additional linked mutations, which is indicated by the blue horizontal line. (E) Total brood size of the N2 and ARL(intron,LSJ2>N2) strains. (F) Number of differentially expressed genes between synchronized N2 and ARL(intron,LSJ2>N2) animals harvested 52 hr (L4 stage - when spermatogenesis is active) or 60 hr (young adults - when oogenesis is active) after hatching. For all figures, each dot represents an independent replicate, the box indicates the interquartile values of all data, and the line indicates the median of all data. Positive values indicate strain one is more fit than strain two. Negative values indicate strain two is more fit than strain one. For all figures, n.s. indicates p>0.05, one star indicates significant difference at p<0.05 level, two stars indicate significant difference at p<0.01 level, and three stars indicate significant difference at p<0.001 level.
-
Figure 1—source data 1
Source data for Figure 1.
- https://doi.org/10.7554/eLife.48119.006
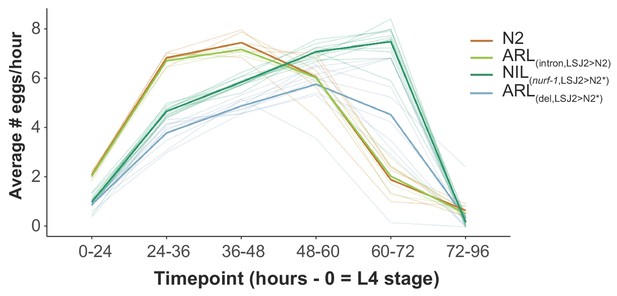
Egg-laying rate of four strains.
Egg-laying rate was calculated at the indicated time points. Six L4 animals were picked onto each assay plate (time = 0) for the indicated times. The total number of eggs was counted for each plate and used to calculate the average number of eggs laid per hour for each animal. Each individual trial is shown as a dim line. The mean for each strain is shown as the bold, colored lines. The effect of the intron SNV on this trait was subtle, especially when compared to the difference in reproductive rate caused by the nurf-1 60 bp deletion (ARL(del,LSJ2>N2*)). We speculate this is explained by epistasis between the two nurf-1 mutations, with the LSJ2 combination of both alleles having a non-linear effect on reproductive rate. We are unable to test this hypothesis as we did not construct a double ARL strain containing LSJ2 alleles of both of the nurf-1 mutations due to the difficulty in creating the intron edit (see Methods). Alternatively, additional mutations in NIL(nurf-1,LSJ2>N2*) could contribute to reproductive rate.
-
Figure 1—figure supplement 1—source data 1
Source data for Figure 1—figure supplement 1.
- https://doi.org/10.7554/eLife.48119.004
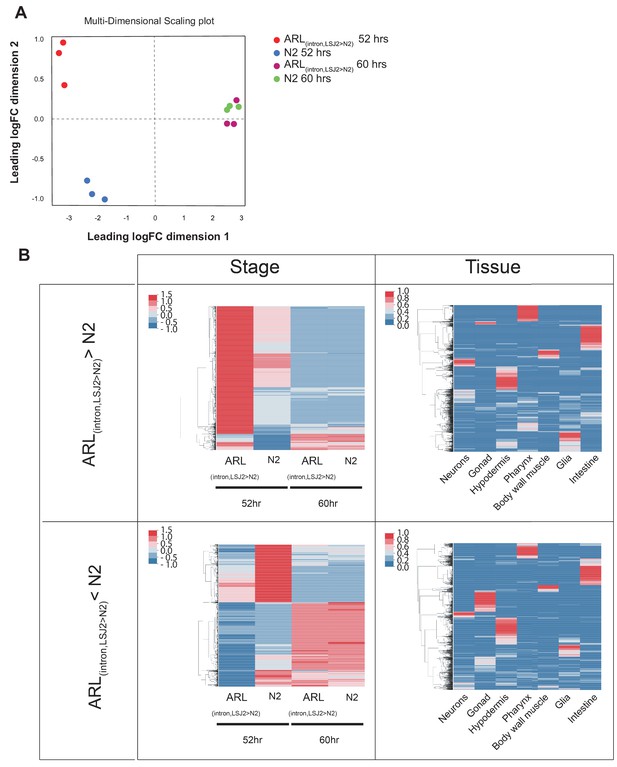
Transcriptional analysis of N2 and ARL(intron,LSJ2>N2) at 52 and 60 hr.
(A) Multi-dimensional scaling analysis of transcriptional responses indicates that the N2 and ARL(intron,LSJ2>N2) strains show different expression patterns at 52 hr but not 60 hr. (B) Analysis of genes upregulated (top) or downregulated (bottom) at all four strain/conditions (left) and also by tissue-specific expression).
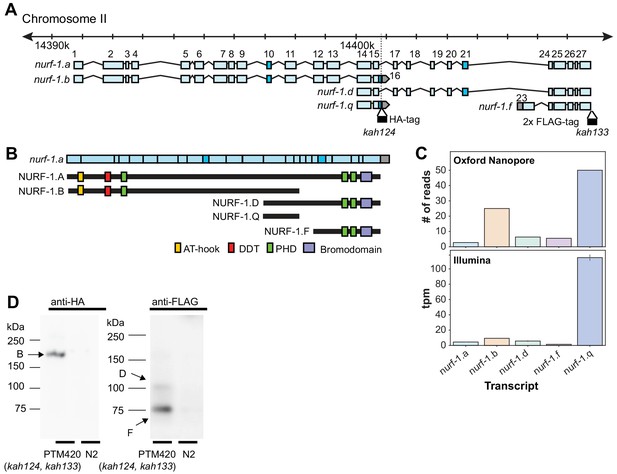
nurf-1 encodes five major transcripts.
(A) Genomic position of the five nurf-1 transcripts supported by Illumina short read and Oxford Nanopore long reads. Each blue box is an exon. Exon number is indicated on the figure. Dark blue exons (10, 16, and 21) are alternatively spliced, resulting in a 6–9 bp difference in length (see Figure 2—figure supplement 1 for details). Genomic location of the HA and FLAG epitope tag insertion site are shown in black along with their associated allele names. (B) The predicted protein isoforms produced by each of the five major transcripts and along with the domains each isoform contains. Immunoblots only supported translation of the B, D, and F isoforms (see panel D for details). For reference, the spliced nurf-1.a transcript is also shown. (C) Relative expression levels of each transcript, determined by number of Oxford Nanopore reads from a mixed population (top panel) or analysis of Illumina short reads from L2 staged animals using kallisto (bottom reads). tpm = transcripts per million. (D) Western blots of N2 and PTM420 strains. PTM420 contains the HA and FLAG epitope tags shown in panel A. Anti-HA antibody detected a band matching the expected size of the NURF-1.B isoform (arrow). Anti-FLAG antibody detected bands matching the expected size of the NURF-1.D and NURF-1.F isoforms (arrows).
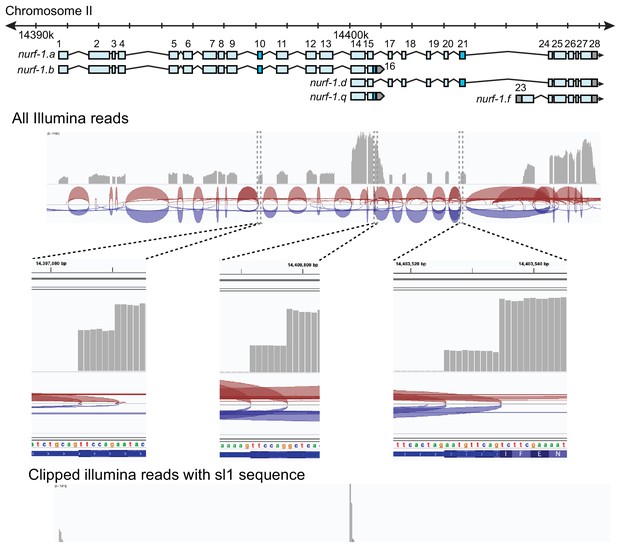
RNA-seq analysis of nurf-1.
Coverage plot of reads from RNA. Note the high expression of the 14th, 15th, and 16th exons, supporting the existence of the nurf-1.q transcript. Reads covering the 23rd exon were also observed, supporting the expression of the nurf-1.f transcript. Zoomed in view of the 10th, 16th, and 21 st exons indicates alternative splicing sites are used at these exons. Clipped reads containing sl1 sequence support transcriptional start sites at the 1 st and 14th exon.
-
Figure 2—figure supplement 1—source data 1
Source data for Figure 2—figure supplement 1.
- https://doi.org/10.7554/eLife.48119.010
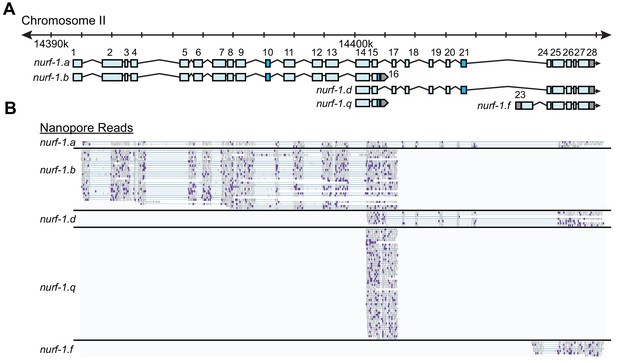
nurf-1 encodes multiple transcripts.
(A) Subset of nurf-1 transcripts analyzed in this paper. Each blue box is an exon. Exon number is indicated on the figure. Dark blue exons are alternatively spliced, resulting in a 6–9 bp difference in length. (B) Nanopore sequencing reads aligned to nurf-1. Reads were grouped by the nurf-1 transcripts they support. Dark purple marks are mismatches from the reference sequence.
-
Figure 2—figure supplement 2—source data 1
Source data for Figure 2—figure supplement 2.
- https://doi.org/10.7554/eLife.48119.012
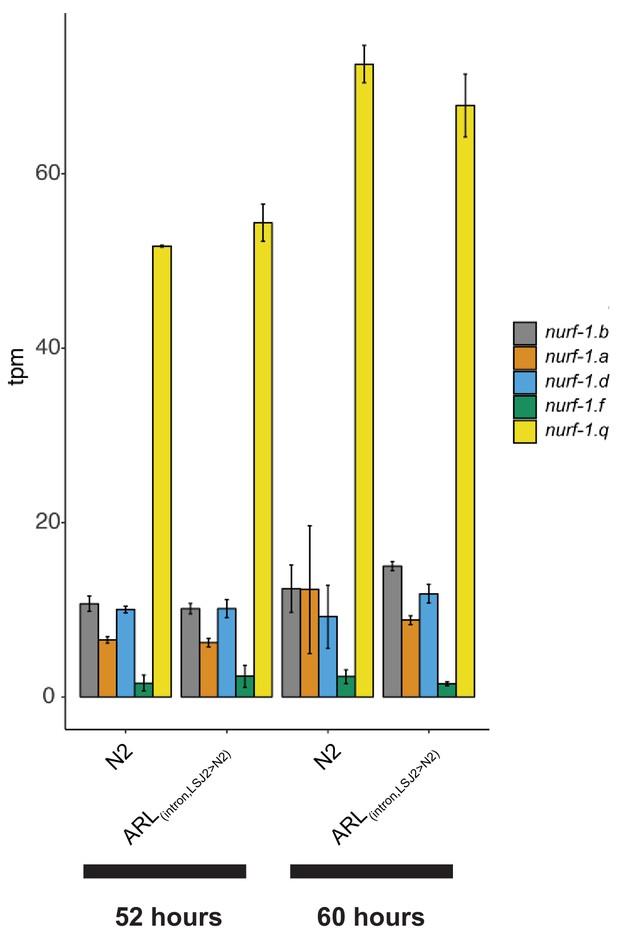
Relative expression levels of each nurf-1 transcript, determined by analysis of Illumina short reads using kallisto (bottom reads).
Strains and timepoints (relative to the L1 stage) are indicated on the x-axis. No significant difference in transcript levels was observed between the N2 and ARL(intron, LSJ2>N2) strains.
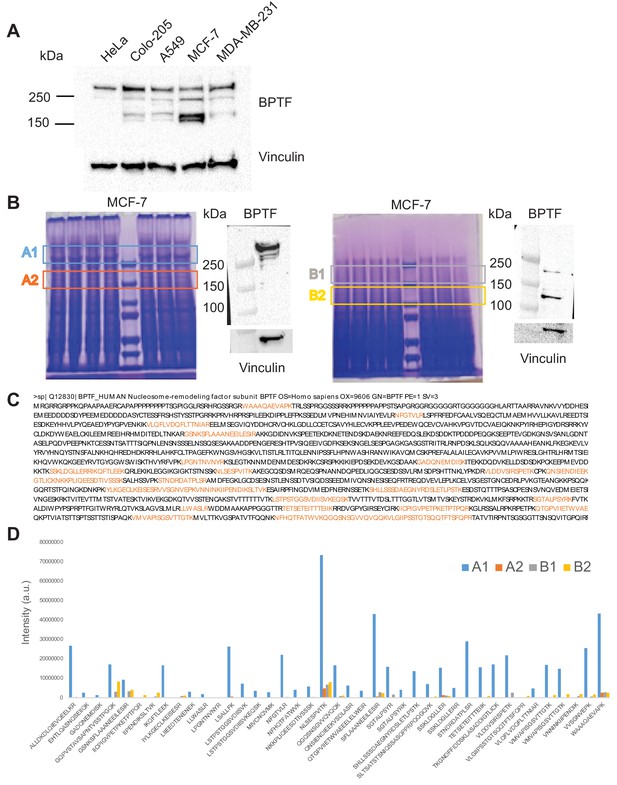
Identification of alternative BPTF species in human cancer cells.
(A) Reactivity of anti-BPTF antibodies with lysates from human cancer cells. Cell line-specific patterns were observed, with bands of the same mobility being detected across lines and detected with independent antibodies. The findings suggest the occurrence of multiple BPTF-related protein species in human cancer cells. A representative western blot is shown. (B) Gel bands selected for BPTF Mass Spectrometry identification according to the localization of the BPTF signal detected by western blotting in MCF-7 cells. (Left, NP-40 lysis buffer, gel bands A1 and A2; Right, Laemmli buffer, B1 and B2). The detection of low molecular weight species upon direct cell lysis in Laemmli buffer strongly supports the notion that the findings do not result from artifactual proteolysis. (C) Sequence coverage of BPTF protein. Peptides identified by LC-MS/MS are highlighted in color. (D) BPTF peptide intensity (arbitrary units) calculated by MaxQuant in the gel bands A1, A2/B1, B2.
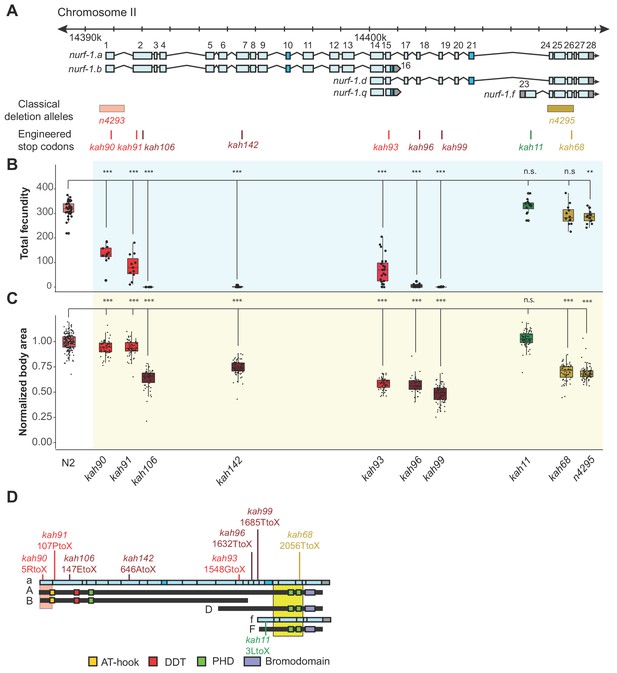
An additional isoform besides NURF-1.
B is necessary for reproduction in C. elegans. (A) Genomic positions of nurf-1 classical deletion alleles and nine engineered stop codons created using CRISPR/Cas9 based gene editing. Each allele is color-coded by the reproductive ability of homozygous strains. Green is statistically indistinguishable from wild-type, yellow indicates slightly reduced brood size and change in reproductive rate, red indicates substantially reduced brood size in the first generation and eventual sterility after multiple generations of homozygosity, and dark red indicates sterility in the first generation of homozygosity. (B) Fecundity of indicated strains (shown in x-axis of panel) (C) Normalized body area of the indicated strains. Normalized body area was calculated by thresholding video recordings of each strain to segment individual animals and registered throughout each frame of the video. Each dot represents the average area of a single worm, normalized to the N2 data. For red or dark red strains (panel A), measurements were taken on animals homozygous for a single generation. (D) Predicted amino acid change of engineered stop codons and classical alleles on the NURF-1 isoforms. The kah11 mutation only affects the F isoform.
-
Figure 3—source data 1
Source data for Figure 3.
- https://doi.org/10.7554/eLife.48119.019
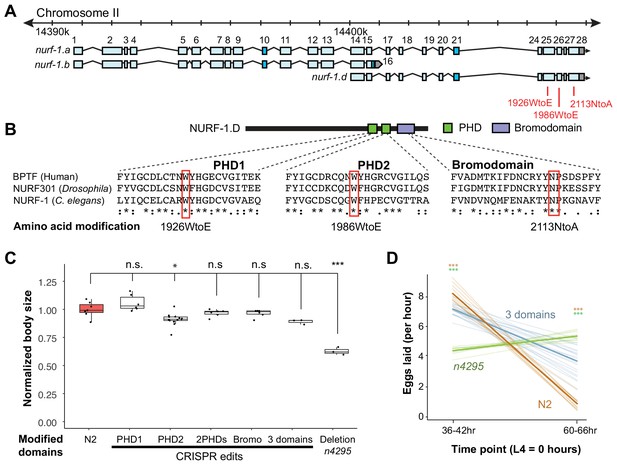
Histone recognition domains in NURF-1.D are not essential for its activity.
(A) Genomic position of CRISPR/Cas9 edits to modify conserved amino acids required for recognition of modified histone tails. (B) Partial protein alignment of the two C-terminal PHD domains and the bromodomain showing the conserved tryptophan (W) and aspagine amino acids (N). In other species, the W to E change impairs the PHD domain’s ability to bind H3K4me3 and the N to A change impairs the bromodomains ability to bind H4K16Ac. (C) Normalized body size of indicated strains. n4295 is a deletion allele predicted to delete both PHD domains and create a frame shift in the bromodomain. The 2PHDs strain contains both the 1,926 WtoE and 1,986 WtoE edits. The three domains strain contains all three edits shown in B. All animals were normalized to N2. (D) Egg laying rate of indicated strains at 36–42 hr and 60–66 hr after the L4 larval stage.
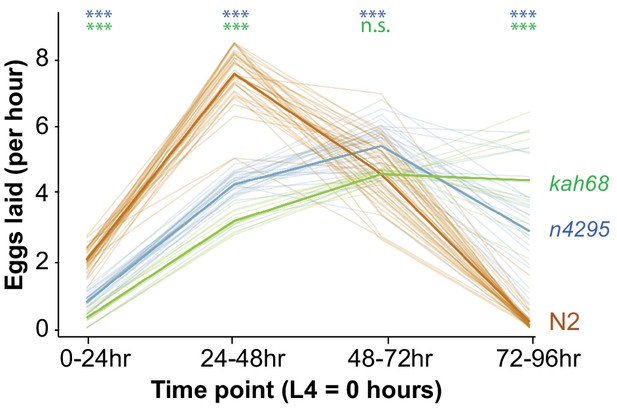
Reproductive output of indicated strains at indicated times.
Six L4 animals from each strain were synchronized and placed on assay plates for the indicated timepoints. Progeny from each plate were counted to calculate the average reproductive output. Light colored lines indicate each replicate and solid colored lines show the average egg laying rate of each strain. For statistical significance, N2 vs. kah68 comparison is in green and N2 vs. n4295 comparison is in blue.
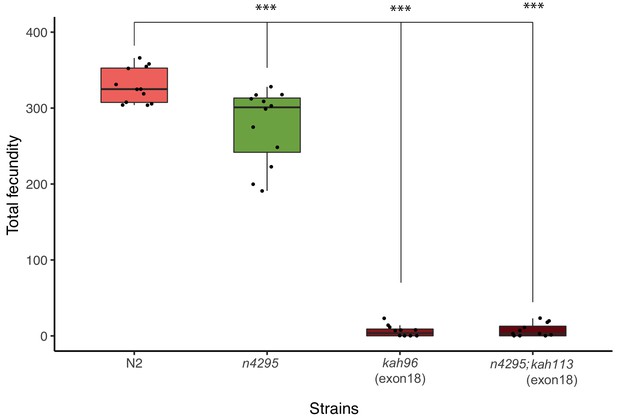
Fecundity analysis of the indicated alleles of nurf-1.
kah96 and kah113 are CRISPR-induced STOP codon replacement mutations in exon 18 of nurf-1. Despite their identical nucleotide changes, in C. elegans nomenclature, they are given unique allele names to indicate their origin from independent CRISPR-Cas9 experiments.
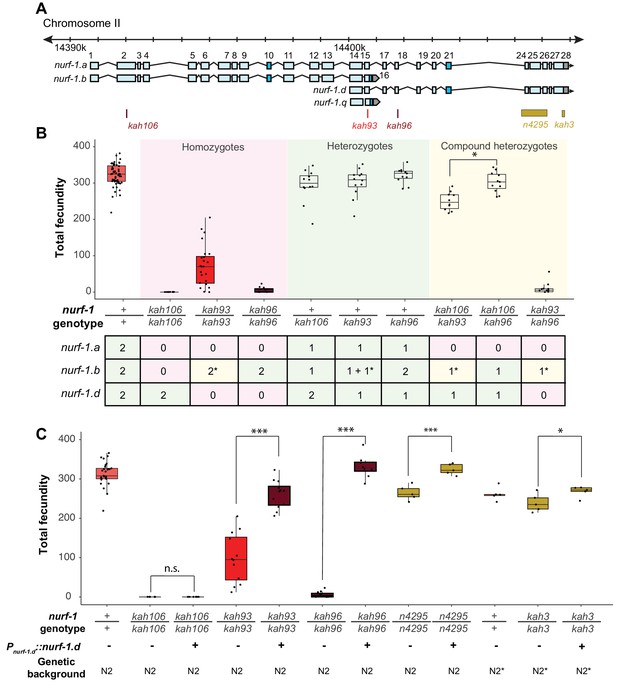
Genetic analysis suggests the NURF-1.
B and NURF-1.D isoforms are essential for reproduction in C. elegans. (A) Genomic positions of stop codon or classical deletion mutations used for compound heterozygote or transgenic rescue analysis of B and C. kah3 is a CRISPR/Cas9 genomic edit of the LSJ2-derived 60 bp deletion. (B) Fecundity of homozygote (red), heterozygote (green), and compound heterozygote mutants (yellow) as indicated in the x-axis. The table below the x-axis is the predicted effect of each mutant strain on the indicated nurf-1 isoforms. The number in the table indicates the number of functional copies. The star indicates the milder predicted effect of kah93 on NURF-1.B, as it only truncates 73 of 1621 amino acids. The y-axis shows the fecundity for each strain. (C) Fecundity of indicated strains with and without the presence of an integrated nurf-1.d transgene. The genetic background is also indicated. N2* contains ancestral introgressions of the npr-1 and glb-5 genes.
-
Figure 4—source data 1
Source data for Figure 4.
- https://doi.org/10.7554/eLife.48119.024
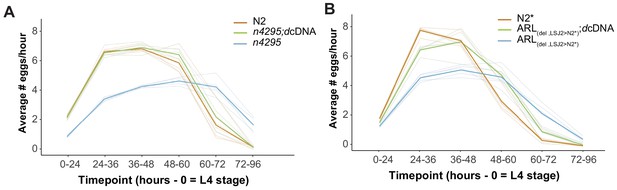
Egg-laying rate of n4295 and ARL(del, LSJ2>N2*) transgenic nurf-1.d cDNA rescue.
Egg-laying rate was calculated at the indicated time points. Six L4 animals were picked onto each assay plates (time = 0) for the indicated times. The total number of eggs was counted for each plate and was used to calculate the average number of eggs laid per hour for each animal. Each individual trial is shown as a dim line. The mean for each strain is shown as the bold, colored lines.
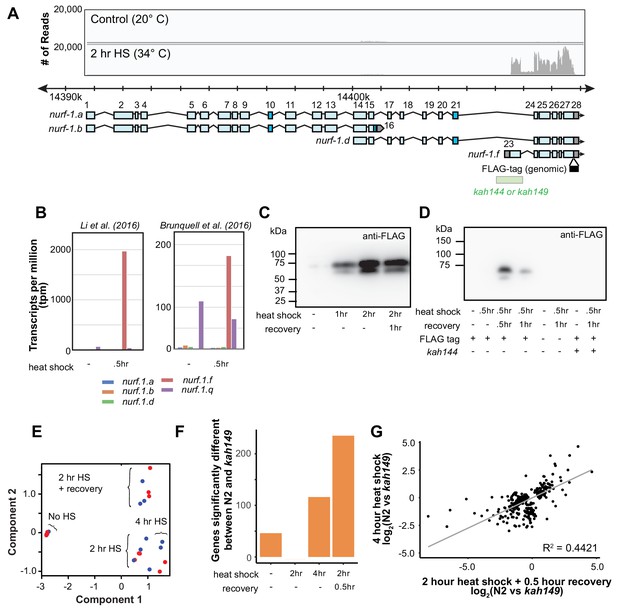
Heat shock specifically upregulates NURF-1.F.
(A) Coverage of RNA-seq reads of control and heat shocked C. elegans animals aligned to the nurf-1 genomic location. These data were taken from Brunquell et al. (2016). The x-axis shows the genomic location where the sequencing reads mapped, including the location of nurf-1 transcripts. We also show the position a precise deletion of the 23rd exon edited into two strains, created using CRISPR/Cas9. In C. elegans genetic nomenclature, each independently generated genetic mutation is given a unique allele name, even if they are genetically identical. The deletion edited into the N2 strain is named kah149. The deletion edited into a strain containing an in-frame FLAG epitope tag (shown as a black box) is named kah144. The y-axis indicates the number of reads for at each location. (B) Quantification of RNA abundance for five nurf-1 transcripts in response to heat shock. Data taken from Li et al. (2016), who heat shocked L2 animals at 34°C for 30 min and Brunquell et al. (2016), who heat shocked L4 animals at 33°C for 30 min. The y-axis is the estimated transcripts per million (tpm) for each isoform in each condition. (C) Western blotting of a strain containing the FLAG-tag fused at the position shown in panel A using an anti-FLAG antibody. We detected two bands, one matching the predicted size of the NURF-1.F isoform, that were both upregulated by heat shock (34°C). (D) Western blotting of three strains either containing a FLAG-tag and/or deletion allele predicted to ablate the nurf-1.f transcript. The x-axis shows the presence or absence of the various alleles along with the environmental condition. We detected two bands that were induced by heat shock. Observation of these bands required the FLAG epitope tag and could be ablated by the 23rd exon deletion. (E) Multi-dimensional scaling plot (MDS) of the N2 (red) and a strain carrying the kah149 deletion of the 23rd exon (blue) in response to various heat shock conditions. No HS indicates no heat shock, 2 hr or 4 hr HS indicates two or four hours of heat shock at 34°C, and 2 hr HS + recovery indicates animals experiencing two hours of heat shock at 34°C followed by 0.5 hr of recovery at 20°C. The overall transcriptional response was the same in both strains. (F) Number of genes significantly up or down regulated between N2 and kah144 animals at the indicated conditions. (G) Scatter plot of all genes differentially expressed in four hour heat shock or two hour heat shock + recovery conditions. The R2 value was 0.4421.
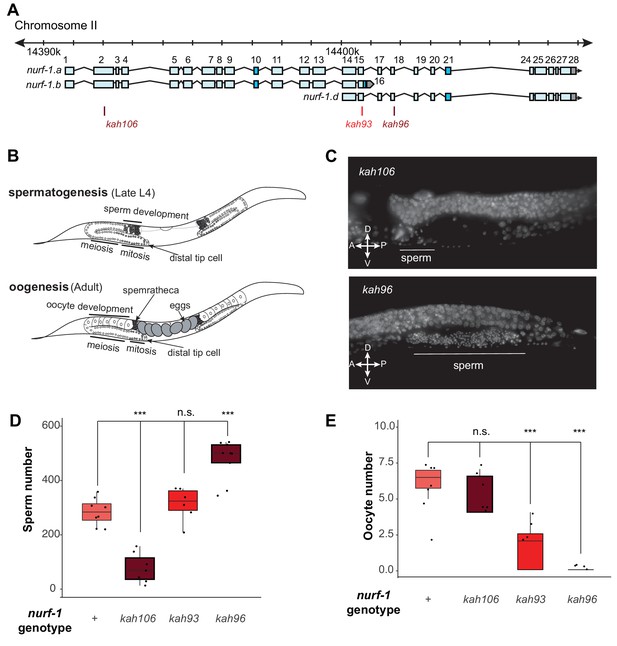
NURF-1.B and NURF-1.D have opposite effects on the sperm-to-oocyte switch in hermaphrodites.
(A) Genomic position of the previously-described stop codon mutants used in B and C. (B) Summary of gametogenesis of C. elegans. Animals undergo spermatogenesis during the late L4 and then transition to oogenesis stage during maturation to adulthood. The number of sperm produced during spermatogenesis can be determined by counting sperm in the spermatheca when oogenesis has begun. (C) Representative fluorescence images of one spermatheca for DAPI stained young adult animals. Each tiny dot represents the condensed chromosomes of a single sperm. (D) Sperm number of indicated strains. L4 animals were synchronized and allowed to develop for an additional 12 hr. DAPI staining was used to identify and count the number of sperm in each animal. Each dot represents a single animal. (E) Oocyte number of indicated strains. L4 animals were synchronized and allowed to develop for an additional 12 hr. DAPI staining was used to identify and count the number of oocytes in each animal.
-
Figure 5—source data 1
Source data for Figure 5.
- https://doi.org/10.7554/eLife.48119.027
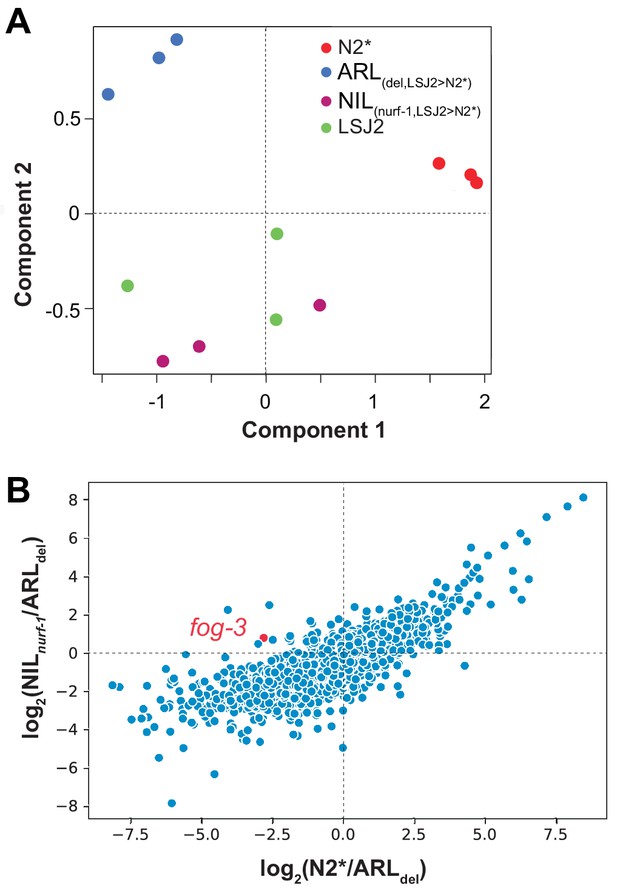
Transcriptome analysis of strains containing N2/LSJ2 genetic variation linked to nurf-1.
(A) Multi-dimensional scaling plot (MDS) of CX12311 (N2*), ARL(del,LSJ2>N2*) (PTM88), NIL(nurf-1,LSJ2>N2*) (PTM66) and LSJ2. The x-axis and y-axis are two dimensions used to separate samples from different biological conditions based upon the transcriptional change between different samples. (B) Scatter plot of all genes detected in RNA sequencing. The x-axis is the log2 of the relative expression changes of each gene in N2* vs. ARL(del,LSJ2>N2) (indicating transcriptional responses induced by the 60 bp deletion). The y-axis is the log2 of the relative expression of each gene in NIL(nurf-1,LSJ2>N2*) vs. ARL(del,LSJ2>N2*) (indicating transcriptional responses induced by other mutations linked to the 60 bp deletion including the LSJ2-derived intron SNV). The transcriptional changes are positively correlated with an R2 value of 0.63, indicating the genetic variation regulates a common subset of genes. fog-3 gene is shown in red.
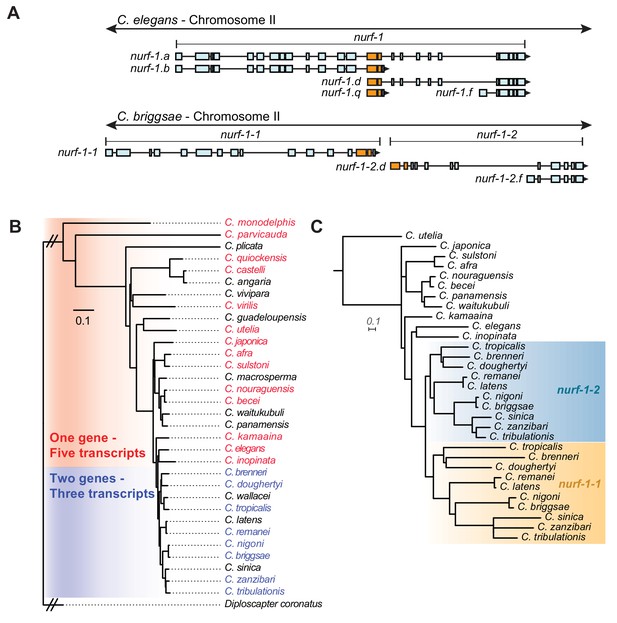
A duplication of the shared exons of the nurf-1.b and nurf-1.d transcripts resulted in the split of nurf-1 it into two separate genes in a subclade of Caenorhabditis species.
(A) Comparison of two species with different versions of nurf-1. In C. elegans, nurf-1.b and nurf-1.d overlaps in the 14th and 15th exon (shown in orange). In C.briggsae, a duplication of the orange exons resulted in separation of nurf-1.b and nurf-1.d into separate genes. C. briggsae has also lost expression of the nurf-1.a and nurf-1.q transcripts. (B) Distribution of the two versions of nurf-1 (shown in panel A) in 32 Caenorhabditis species. Red indicates the C. elegans version, blue indicates the C. briggsae version, and black indicates a nurf-1 version that could not be determined. The species phylogeny suggests that a duplication event occurred in the common ancestor of the brenneri/tribulationis clade. (C) The most well supported topology of the duplicated region is consistent with a single duplication event. Orange indicates protein sequence from the duplicated region in the nurf-1–1 gene, and turquoise indicates protein sequence from the duplicated region in the nurf-1–2 gene. Non-colored branches indicate unduplicated nurf-1 sequence. The rate of amino acid substitution in the nurf-1–1 duplicated region has also increased, as seen in the branch lengths. Scale is in substitutions per site.
-
Figure 6—source data 1
Source data for Figure 6.
- https://doi.org/10.7554/eLife.48119.035
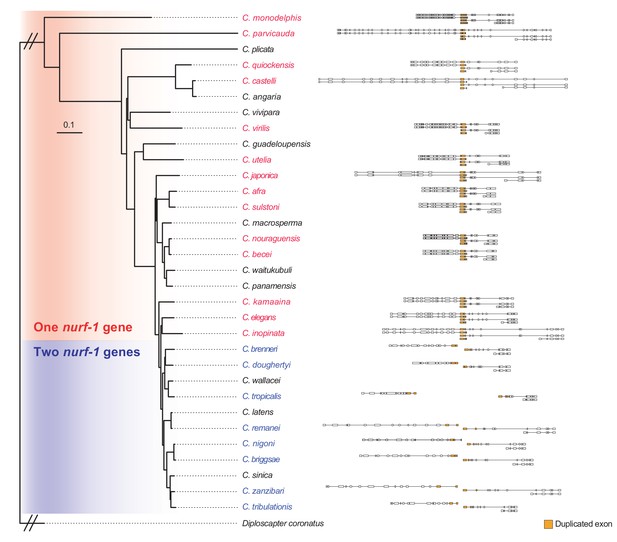
nurf-1 isoform structure for 22 Caenorhabditis species.
From the phylogenic tree of 32 Caenorhabditis species, we determined the nurf-1 gene structure of 22 species using genome and transcriptome information. Species with one nurf-1 gene (in red) are consistent with expression of five transcripts orthologous to C. elegans: nurf-1.a, nurf-1.b, nurf-1.q, nurf-1.d and nurf-1.f. Species with two nurf-1 genes (in blue) contain duplicated sequence and transcripts matching C. elegans nurf-1.b (nurf-1–1), nurf-1.d (nurf-1–2.d) and nurf-1.f (nurf-1–2 .f). Exons corresponding to the duplicated region were labeled in orange.
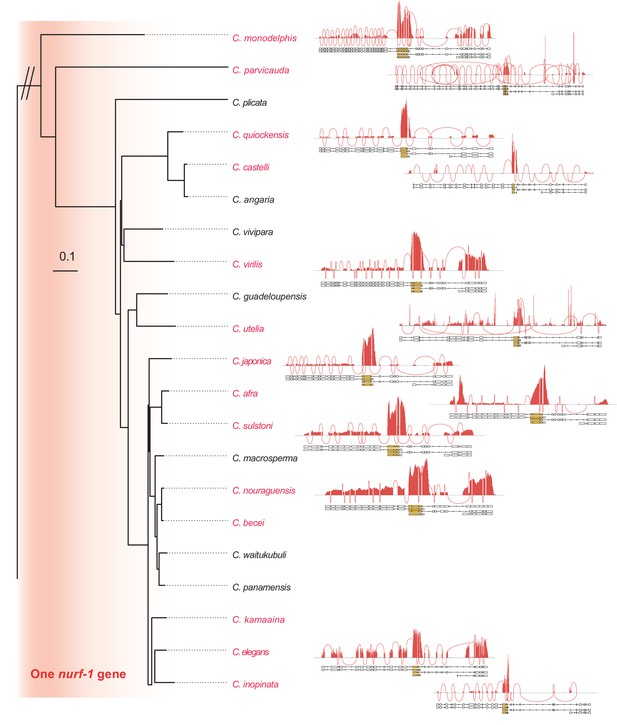
Sashimi plots for Caenorhabditis species with one nurf-1 gene.
Only species with a published genome and transcriptome were plotted. Each peak shows the coverage for each exon, each trajectory shows exon-exon junctions supported by RNA-seq reads.
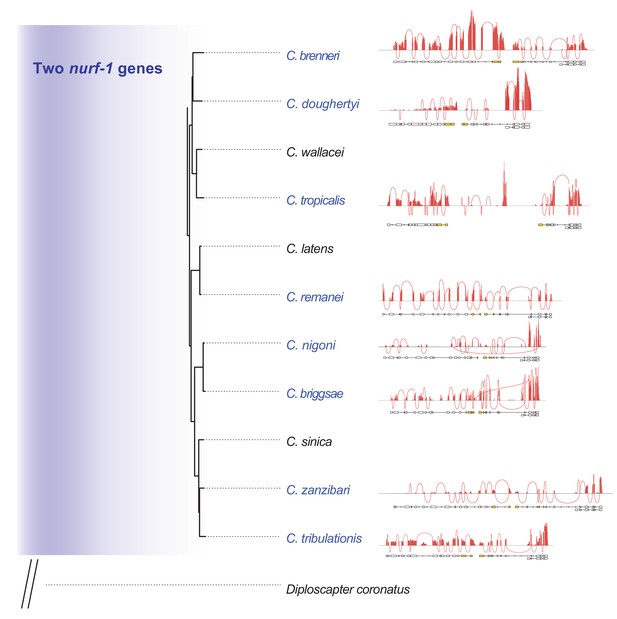
Sashimi plots for Caenorhabditis species with two nurf-1 genes.
Only species with a published genome and transcriptome were plotted. Each peak shows the coverage for each exon, each trajectory shows exon-exon junctions supported by RNA-seq reads. These plots show that no reads support the splicing from nurf-1–1 to nurf-1–2, which further suggests the split of nurf-1 in these species.
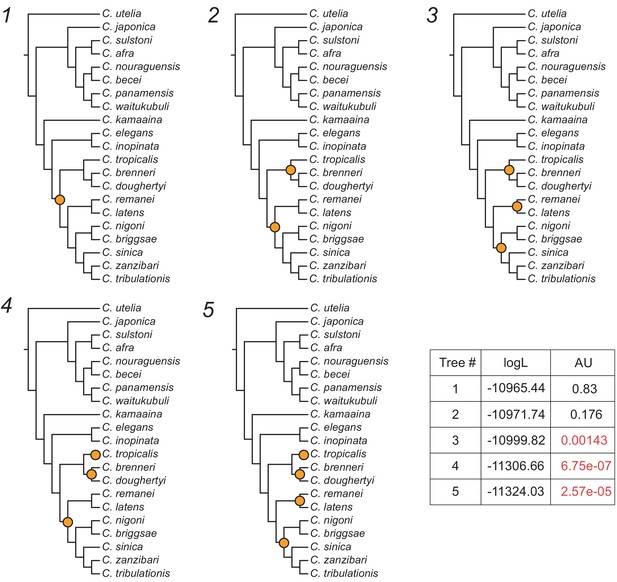
Five hypothetical topologies related to the timing and number of duplication events involved in the nurf-1 gene split.
Only those five topologies with the lowest log-likelihoods are shown. logL: log-likelihoods, AU: p-values of the approximately unbiased test. Orange circles indicate duplication events. Trees 3–5 were rejected by the AU test and are highlighted in red. Analyses were performed using IQ-TREE with the JTT substitution model with gamma-distributed rate variation among sites.
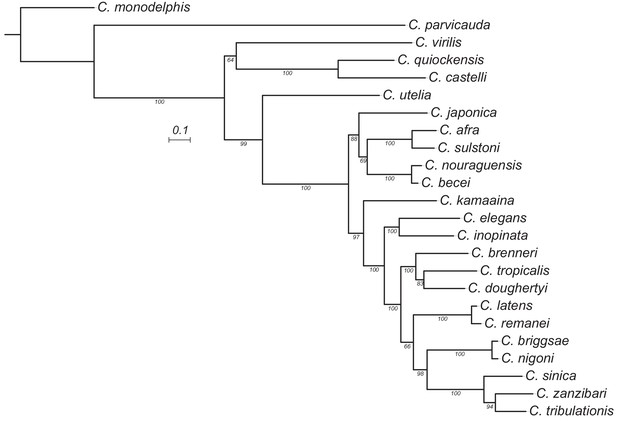
Maximum likelihood tree of the B isoform and nurf-1–1.
The duplicated region was removed from the alignment prior to inference. Estimated using IQ-TREE using the JTT substitution model with gamma-distributed rate variation among sites. Bootstrap support values are indicated as labels on branches. Scale is in amino acid substitutions per site.
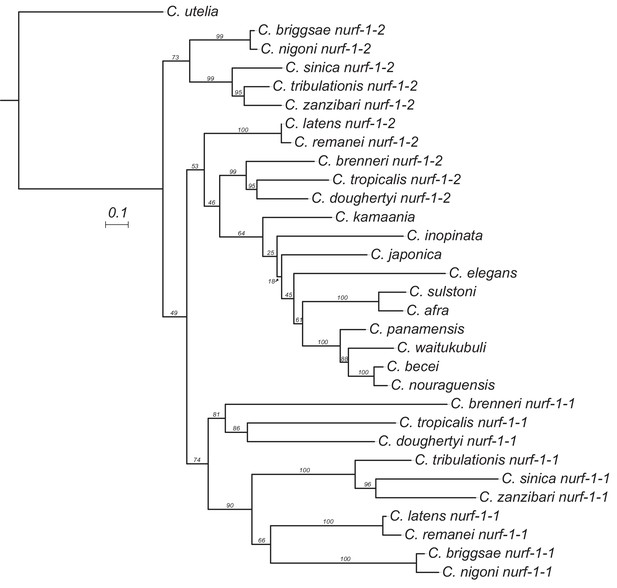
Maximum likelihood tree of the duplicated region of nurf-1 in 22 species.
Estimated using IQ-TREE using the JTT substitution model with gamma-distributed rate variation among sites. Bootstrap support values are indicated as labels on branches. Scale is in amino acid substitutions per site.
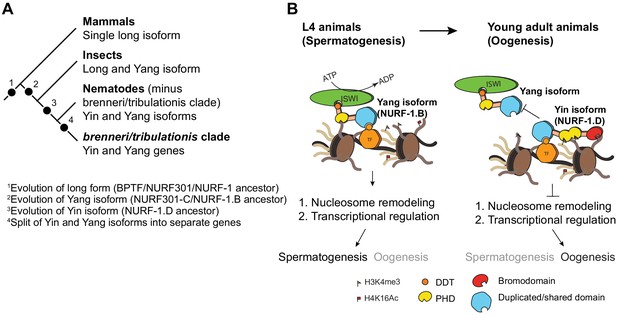
Proposed antagonistic (Yin-Yang) working model of two nurf-1 isoforms in C. elegans.
(A) Descriptive phylogeny with proposed major transitions in nurf-1 isoform evolution. Each dot indicates the timepoint of a major nurf-1 isoform evolution event. (B) Proposed molecular mechanism for NURF-1 isoforms. The NURF-1.B isoform interacts with ISWI through its DDT domain to form a NURF complex capable of remodeling chromatin at specific regions of the chromosome. NURF is recruited to these regions through interactions with specific transcription factors using protein domains encoded by the overlapping exons. This remodeling is necessary for transcriptional responses for spermatogenesis. Due to some unknown signal, after spermatogenesis has resulted in the production of ~ 300 sperm, the NURF-1 D isoform outcompetes the NURF complex away from its target loci, causing the loss of transcription of key spermatogenesis genes, resulting in gametogenesis transitioning from spermatogenesis to oogenesis. The binding affinity of PHD domains and bromodomainto histone strengthens this repression, but they are not completely necessary for the ability of the D isoform to outcompete the B isoform.
Tables
Summary of major nurf-1 transcripts identified in C. elegans.
https://doi.org/10.7554/eLife.48119.007| Name | Evidence | Size | Conservedc | Predicted biological role in C. elegansd | Other names | ||
|---|---|---|---|---|---|---|---|
| Transcripta | Proteinb | aa | kD | ||||
| nurf-1.a | N | - | 2197 | 252 | M,D | None | Full-length |
| nurf-1.b | C,N,I | W | 1621 | 186 | D | Reproduction, vulval development | N-terminal or NURF-1.A |
| nurf-1.d | C,N,I | W | 816 | 92 | - | Size, dauer, reproduction, axon guidance | C-terminal or NURF-1.C |
| nurf-1.f | C,N,I | W | 581 | 58 | - | None | NURF-1.E |
| nurf-1.q | N,I | - | 243 | 36 | - | None | - |
-
a C indicates full-length cDNA have been isolated for this transcript, N indicates evidence from direct sequencing of RNA or cDNA using Oxford Nanopore reads support this transcript, and I indicates evidence from Illumina short read RNA-seq supports this transcript
b W indicates evidence for the protein isoform was obtained using western blot
-
c M or D indicates an analogous isoform is described in mammals (mice or humans) or Drosophila, respectively
dPredictions from Andersen et al. (2006), Large et al. (2016), or Mariani et al. (2016)
Predicted effect of stop codon mutations on NURF-1 isoforms.
https://doi.org/10.7554/eLife.48119.020| Isoform | kah90 | kah91 | kah106 | kah142 | kah93 | kah96 | kah99 | kah11 | kah68 | Length |
|---|---|---|---|---|---|---|---|---|---|---|
| NURF-1.A | 5R | 107P | 147E | 646A | 1548G | 1632T | 1685Q, 1689P, 1693N | 2056T | 2197 | |
| NURF-1.B | 5R | 107P | 147E | 646A | 1548G | - | - | - | 1621 | |
| NURF-1.D | - | - | - | 170G | 254T | 307Q, 311P, 315N | 675T | 816 | ||
| NURF-1.F | - | - | - | - | - | - | 3L | 440T | 581 | |
| NURF-1.Q | - | - | - | 170G | - | - | - | 243 |
dN/dS ratio in three Caenorhabditis species pairs.
https://doi.org/10.7554/eLife.48119.036| nurf-1–1 or nurf-1.b | nurf-1–2 or nurf-1.d | ||||||
|---|---|---|---|---|---|---|---|
| Sp. pair | Duplicationa | Dup. Reg.b | Otherc | Ratiod | Dup. Reg.b | Otherc | Ratiod |
| C. afra/ C. sulstoni | N | 0.136e | 0.121 | 1.1 | 0.116e | 0.072 | 1.6 |
| C. nigoni/ C. briggsae | Y | 0.249 | 0.085 | 2.9 | 0.111 | 0.019 | 5.8 |
| C. remanei/ C. latens | Y | 0.295 | 0.121 | 2.4 | 0.177 | 0.048 | 3.7 |
-
a Duplication indicates whether the species pair contain the duplicated exons that create two nurf-1 genes
b Dup. Reg. indicates dN/dS was calculated using the region of the alignment that contains the duplication
-
c Other indicates dN/dS was calculated using the region of the alignment that does not contain the duplication
d Ratio was calculated by dividing the dN/dS value of the Dup. Reg. by the Other
-
e The dN/dS values for the nurf-1.b and nurf-1.d in the duplicated region were different due to the b transcript encoding two additional amino acids in the 14th exon (before the M initiation codon in the d isoform) and the amino acids encoded by the 16th alternatively spliced exon.
| Reagent type or resource | Designation | Source of reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (C. elegans) | nurf-1 | WormBase | Wormbase ID: WBGene00009180 | Sequence: F26H11.2 |
| Gene (human) | BPTF | National Center for Biotechnology Information | Gene ID: 2186 | |
| Strain, strain background (E. coli) | OP50 | Caenorhabditis Genetics Center (CGC) | RRID: WB-STRAIN:OP50 | |
| Strain (C. elegans) | CX12311 | PMID: 21849976 | RRID: WB-STRAIN:CX12311 | Strain Background: N2, Request a strain: please email the corresponding author |
| Strain (C. elegans) | PTM66 | PMID: 21849976 | Strain Background: N2, Request a strain: please email the corresponding author | |
| Strain (C. elegans) | PTM88 | PMID: 21849976 | Strain Background: N2, Request a strain: please email the corresponding author | |
| Strain (C. elegans) | PTM288 | PMID: 30328811 | RRID: WB-STRAIN:PTM288 | Strain Background: N2, Request a strain: please email the corresponding author |
| Strain (C. elegans) | PTM229 | PMID: 30328811 | RRID:WB-STRAIN:PTM229 | Strain Background: N2, Request a strain: please email the corresponding author |
| Strain (C. elegans) | PTM98 | This paper | RRID:WB-STRAIN:PTM98 | Strain Background: N2, Request a strain: please email the corresponding author |
| Strain (C. elegans) | PTM113 | This paper | RRID:WB-STRAIN:PTM113 | Strain Background: N2, Request a strain: please email the corresponding author |
| Strain (C. elegans) | PTM116 | This paper | RRID:WB-STRAIN:PTM116 | Strain Background: N2, Request a strain: please email the corresponding author |
| Strain (C. elegans) | PTM117 | This paper | RRID:WB-STRAIN:PTM117 | Strain Background: N2, Request a strain: please email the corresponding author |
| Strain (C. elegans) | PTM118 | This paper | RRID:WB-STRAIN:PTM118 | Strain Background: N2, Request a strain: please email the corresponding author |
| Strain (C. elegans) | PTM167 | This paper | RRID:WB-STRAIN:PTM167 | Strain Background: N2, Request a strain: please email the corresponding author |
| Strain (C. elegans) | PTM170 | This paper | RRID:WB-STRAIN:PTM170 | Strain Background: N2, Request a strain: please email the corresponding author |
| Strain (C. elegans) | PTM189 | This paper | RRID:WB-STRAIN:PTM189 | Strain Background: N2, Request a strain: please email the corresponding author |
| Strain (C. elegans) | PTM203 | This paper | RRID:WB-STRAIN:PTM203 | Strain Background: N2, Request a strain: please email the corresponding author |
| Strain (C. elegans) | PTM211 | This paper | RRID:WB-STRAIN:PTM211 | Strain Background: N2, Request a strain: please email the corresponding author |
| Strain (C. elegans) | PTM316 | This paper | RRID:WB-STRAIN:PTM316 | Strain Background: N2, Request a strain: please email the corresponding author |
| Strain (C. elegans) | PTM317 | This paper | RRID:WB-STRAIN:PTM317 | Strain Background: N2, Request a strain: please email the corresponding author |
| Strain (C. elegans) | PTM319 | This paper | RRID:WB-STRAIN:PTM319 | Strain Background: N2, Request a strain: please email the corresponding author |
| Strain (C. elegans) | PTM322 | This paper | RRID:WB-STRAIN:PTM322 | Strain Background: N2, Request a strain: please email the corresponding author |
| Strain (C. elegans) | PTM325 | This paper | RRID:WB-STRAIN:PTM325 | Strain Background: N2, Request a strain: please email the corresponding author |
| Strain (C. elegans) | PTM332 | This paper | RRID:WB-STRAIN:PTM332 | Strain Background: N2, Request a strain: please email the corresponding author |
| Strain (C. elegans) | PTM354 | This paper | RRID: WB-STRAIN:PTM354 | Strain Background: N2, Request a strain: please email the corresponding author |
| Strain (C. elegans) | PTM371 | This paper | RRID:WB-STRAIN:PTM371 | Strain Background: N2, Request a strain: please email the corresponding author |
| Strain (C. elegans) | PTM372 | This paper | RRID:WB-STRAIN:PTM372 | Strain Background: N2, Request a strain: please email the corresponding author |
| Strain (C. elegans) | PTM373 | This paper | RRID:WB-STRAIN:PTM373 | Strain Background: N2, Request a strain: please email the corresponding author |
| Strain (C. elegans) | PTM376 | This paper | RRID: WB-STRAIN:PTM376 | Strain Background: N2, Request a strain: please email the corresponding author |
| Strain (C. elegans) | PTM416 | This paper | RRID:WB-STRAIN:PTM416 | Strain Background: N2, Request a strain: please email the corresponding author |
| Strain (C. elegans) | PTM417 | This paper | RRID:WB-STRAIN:PTM417 | Strain Background: N2, Request a strain: please email the corresponding author |
| Strain (C. elegans) | PTM420 | This paper | RRID:WB-STRAIN:PTM420 | Strain Background: N2, Request a strain: please email the corresponding author |
| Strain (C. elegans) | PTM487 | This paper | RRID:WB-STRAIN:PTM487 | Strain Background: N2, Request a strain: please email the corresponding author |
| Strain (C. elegans) | PTM489 | This paper | RRID:WB-STRAIN:PTM489 | Strain Background: N2, Request a strain: please email the corresponding author |
| Strain (C. elegans) | PTM512 | This paper | RRID:WB-STRAIN:PTM512 | Strain Background: N2, Request a strain: please email the corresponding author |
| Strain (C. elegans) | PTM517 | This paper | RRID:WB-STRAIN:PTM517 | Strain Background: N2, Request a strain: please email the corresponding author |
| Cell line (Human) | Colo-205 | American Type Culture Collection (Rockville, MD) | ||
| Cell line (Human) | MCF-7 | American Type Culture Collection (Rockville, MD) | ||
| Cell line (Human) | MDA-MB-231 | American Type Culture Collection (Rockville, MD) | ||
| Cell line (Human) | Hela | American Type Culture Collection (Rockville, MD) | ||
| Cell line (Human) | A549 | G. Roncador, CNIO | ||
| Sequence-based reagents (Plasmid) | Plasmid: pSM | Cori Bargmann Lab (Rockefeller University) | ||
| Sequence-based reagents (Plasmid) | Plasmid: pDD162 PrU6::dpy-10_sgRNA | PMID: 27467070 | CRISPR/Cas9 gene editing | |
| Sequence-based reagents (Plasmid) | Plasmid: pDD162 Preft3::Cas9 | PMID: 27467070 | CRISPR/Cas9 gene editing | |
| Sequence-based reagents (Plasmid) | Plasmid: pCFJ90 | Addgene | http://www.wormbuilder.org/test-page/about-mossci/ | |
| Sequence-based reagents (Plasmid) | Plasmid: pCFJ104 | Addgene | http://www.wormbuilder.org/test-page/about-mossci/ | |
| Sequence-based reagents (Plasmid) | Plasmid: pCFJ151 | Addgene | http://www.wormbuilder.org/test-page/about-mossci/ | |
| Sequence-based reagents (Plasmid) | Plasmid: pCFJ601 | Addgene | http://www.wormbuilder.org/test-page/about-mossci/ | |
| Antibody | (mouse monoclonal) anti-HA | Life Technologies | Cat. No.: 326700 | (1:500) |
| Antibody | (mouse monoclonal) anti-DYKDDDDK | Life Technologies | Cat. No.: MA191878 | (1:1000) |
| Antibody | (mouse monoclonal) anti-FLAG | Millipore Sigma | Cat. No.: F3165 | (1:1000) |
| Antibody | Horseradish peroxidase-conjugated secondary antibodies | Dako Glostrup | (1:10000) | |
| Peptide, recombinant protein | BPTF | Novus Biologicals | Cat. No.: NB100-41418 | |
| Peptide, recombinant protein | Vinculin | Sigma | Cat. No.: V9131 | |
| Sequence-based reagents (Oligonucleotide) | dpy-10 (cn64) | PMID: 25161212 | CRISPR/Cas9 gene editing | |
| Commercial assay, kit | Taqman probe:dpy-10 (kah82/kah83) | Thermal: Custom TaqMan SNP Genotyping Assays | PTM09 | |
| Commercial assay, kit | NEB Q5 site directed mutagenesis kit | NEB | Cat. No.: E0554 | |
| Commercial assay, kit | Next Ultra II Directional RNA Library Prep Kit | NEB | Cat. No.: E7760S | |
| Commercial assay, kit | Zymo DNA isolation kit | Zymo | Cat. No.: D4071 | |
| Commercial assay, kit | Zymo DNA cleanup kit | Zymo | Cat. No.: D4064 | |
| Commercial assay, kit | ddPCR Supermix for Probes | BIORAD | Cat. No.: 1863010 | |
| Commercial assay, kit | Droplet Generation Oils | BIORAD | Cat. No.: 1863005 | |
| Commercial assay, kit | ddPCR Droplet Reader Oil | BIORAD | Cat. No.: 1863004 | |
| Commercial assay, kit | VECTASHIELD antifade Mounting Medium with DAPI | VECTOR | Cat. No.: H-1200 | |
| Software, Algorithm | edgeR | PMID: 19910308 | RRID:SCR_012802 | Opensource: https://bioconductor.org/packages/release/bioc/html/edgeR.html |
| Software, Algorithm | SARtools | PMID: 27280887 | RRID:SCR_016533 | Opensource: https://github.com/PF2-pasteur-fr/SARTools |
| Software, Algorithm | IGV | PMID: 21221095 | RRID:SCR_011793 | https://software.broadinstitute.org/software/igv/ |
| Software, Algorithm | Kallisto | PMID: 27043002 | RRID:SCR_016582 | https://pachterlab.github.io/kallisto/ |
| Software, Algorithm | HISAT2 | PMID: 25751142 | RRID:SCR_015530 | https://ccb.jhu.edu/software/hisat2/index.shtml |
| Software, Algorithm | Samtools | PMID: 19505943 | RRID:SCR_002105 | http://samtools.sourceforge.net/ |
| Software, Algorithm | Jalview | PMID: 19151095 | RRID:SCR_006459 | http://www.jalview.org/ |
| Software, Algorithm | MAFFT | PMID: 23329690 | RRID:SCR_011811 | https://mafft.cbrc.jp/alignment/software/ |
| Software, Algorithm | IQ-Tree | PMID: 25371430 | RRID:SCR_017254 | http://www.iqtree.org |
| Software, Algorithm | ITOL | PMID: 27095192 | https://itol.embl.de/ | |
| Software, Algorithm | PAL2NAL | PMID: 16845082 |
Additional files
-
Supplementary file 1
RNA-seq counts for each gene.
- https://doi.org/10.7554/eLife.48119.038
-
Supplementary file 2
GO Category analysis for intron SNV regulon.
- https://doi.org/10.7554/eLife.48119.039
-
Supplementary file 3
Guide RNAs for CRISPR-Cas9 genome edits.
- https://doi.org/10.7554/eLife.48119.040
-
Transparent reporting form
- https://doi.org/10.7554/eLife.48119.041





