Neighbor predation linked to natural competence fosters the transfer of large genomic regions in Vibrio cholerae
Figures
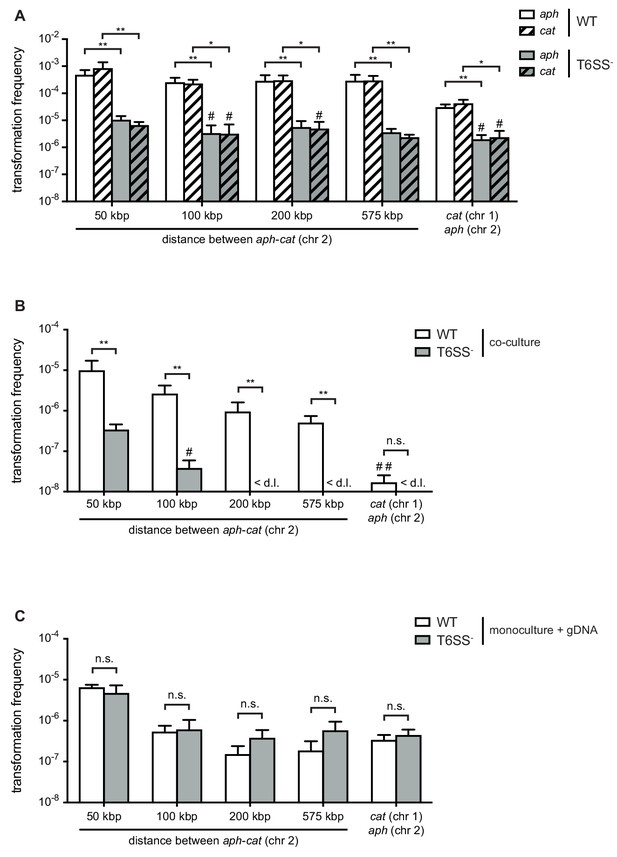
Type VI secretion system (T6SS) enhances horizontal gene transfer (HGT) of single- and double-resistance cassettes if carried in cis.
(A–B) Transformation occurs in predator/prey co-cultures. To induce natural competence, the WT or a T6SS-negative derivative (A1552ΔvasK; T6SS -) was co-cultured on chitin with different prey strains (Sa5Y-derived) that carried two antibiotic resistance cassettes: aph in vipA (chr 2) and cat at variable distances from aph on the same chromosome or on chr 1, as indicated on the X-axis. Transformation frequencies (Y-axis) indicate the number of transformants that acquired (A) a single resistance cassette or (B) both resistance cassettes divided by the total number of predator colony forming units (CFUs). (C) Natural transformation is not impaired in the T6SS- acceptor strain. Purified genomic DNA (gDNA) was added to competent WT or T6SS- strains. (A–C) Data represent the average of three independent biological experiments (± SD, as depicted by the error bars). For values in which one (#) or two (##) experiments resulted in the absence of transformants, the detection limit was used to calculate the average. <d .l., below detection limit. Statistical significance is indicated (*p<0.05; **p<0.01; n.s., not significant).
-
Figure 1—source data 1
Raw data for Figure 1.
- https://doi.org/10.7554/eLife.48212.004
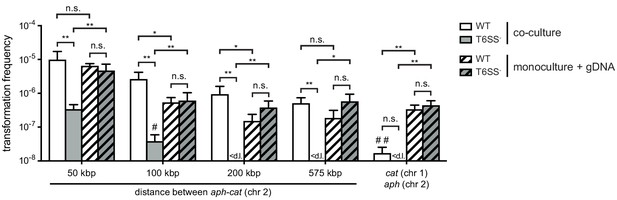
Combined data from Figure 1B and C comparing double-resistance acquisition efficiencies in co-cultures or in gDNA-supplemented monocultures.
Data represent the average of three independent biological experiments (± SD). For values in which one (#) or two (##) experiments resulted in the absence of transformants, the detection limit was used to calculate the average and for statistical analyses. < d .l., below detection limit. Statistical significance is indicated (*p<0.05; **p<0.01; n.s., not significant).
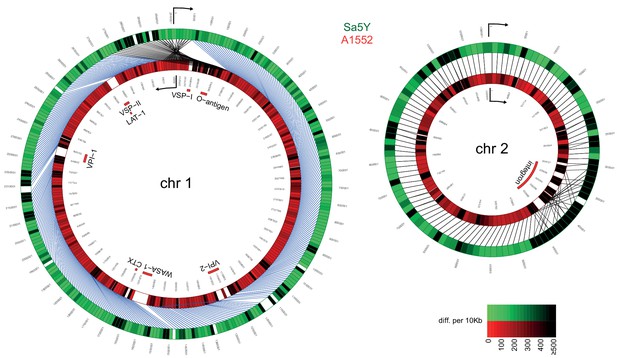
Comparative genomics of pandemic strain A1552 and the environmental isolate Sa5Y.
The genomic sequences of chr 1 and 2 of Sa5Y (green) were segmented in 10-kbp-long fragments and aligned against the respective chromosome of the reference A1552 (red). To simplify visualization, chr 1 of strain A1552 was inverted and plotted counter-clockwise relative to Sa5Y (due to the large inversion in this strain; see Materials and methods section), as indicated by the arrow. To represent the differences between the two genomes, a color intensity scale was used that corresponded to the number of differences (SNP or indel), from 0 to ≥500 as measured per 10 kbp fragment (same values are indicated for both colors). White regions show no homology. Important genomic features of pandemic V. cholerae are highlighted inside the rings.
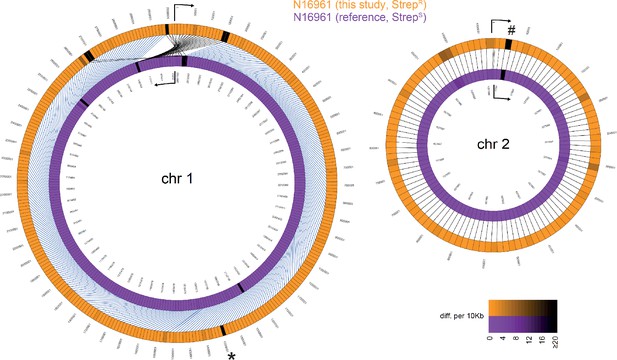
Comparative genomics of V. cholerae reference strain N16961 and a newly sequenced laboratory stock of the same strain.
The newly sequenced genome of the streptomycin-resistant laboratory stock of strain N16961 (long-read PacBio sequencing technology; Matthey et al., 2018) was compared to the genome sequence of the reference genome (Heidelberg et al., 2000). Detailed explanations on the comparison are as described for Figure 2. The shadings (for both colors) reflect differences (SNP or indel) from 0 to ≥20 per 10 kbp fragment. Marked discrepancies are based on repetitive sequences (*) or a sequencing error (#) as discussed in the Materials and methods section.
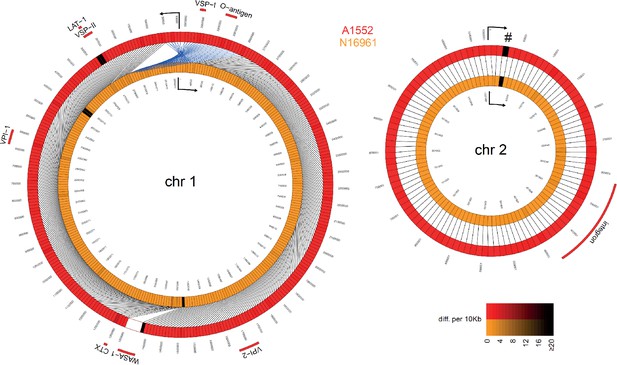
Comparative genomics of pandemic V. cholerae strains N16961 and A1552.
The genome sequences of our laboratory stock of pandemic strains N16961 and A1552 were compared. Details are as described in Figure 2. The shadings (for both colors) reflect differences (SNP or indel) from 0 to ≥20 per 10 kbp fragment. The marked discrepancy (#) resulted from a sequencing error, as discussed in the Materials and methods section.
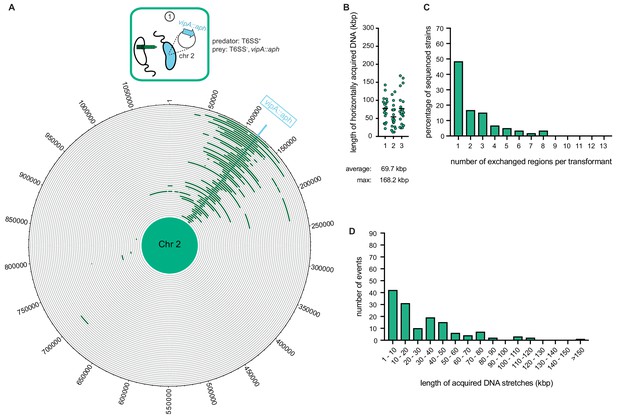
Whole-genome sequencing (WGS)-based quantification of horizontally acquired DNA.
WGS analysis of transformants after prey killing and DNA transfer. Twenty kanamycin-resistant transformants were selected per independent biological experiment (n = 3). (A) The scheme represents the experimental setup of the co-culture experiment (condition ①). Sequencing reads for each transformant were mapped onto the prey genomes to visualize the transferred DNA regions (in dark green; see Figure 4—figure supplement 2 for both chromosomes). The position of the resistance cassette (aph) is indicated by the light blue line. (B) Total DNA acquisition frequently exceeds 100 kb. The total length of horizontally acquired DNA is indicated on the Y-axis for each transformant. Data are from three biologically independent experiments as indicated on the X-axis. Average and maximum lengths are indicated below the graph. (C) Multiple transferred DNA regions were identified in the transformants. Percentage of transformants (n = 60) that exchanged one or more DNA regions, as indicated on the X-axis. (D) Large DNA stretches are transferrable by transformation. The length of individual consecutive DNA stretches was determined as indicated on the X-axis.
-
Figure 3—source data 1
Raw data for Figure 3.
- https://doi.org/10.7554/eLife.48212.009
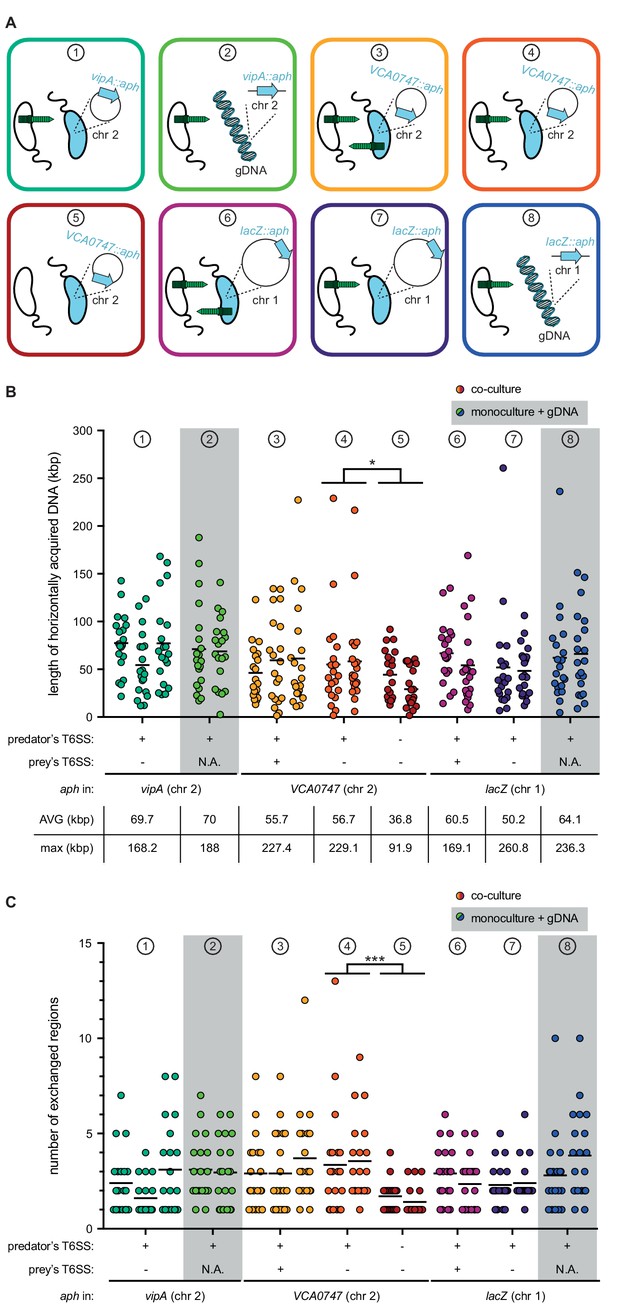
T6SS-mediated neighbor predation followed by DNA uptake enhances the frequency and length of transferred DNA stretches.
(A) Scheme representing the eight experimental conditions tested in this study. Each scheme indicates whether the transformants acquired the aph resistance gene from a prey bacterium (blue) (position of aph indicated on the zoomed-in circles of chr 1 or chr 2) or from purified genomic DNA (gDNA). In the former case, the killing capacity of the predator (white) and prey (blue) is shown by the presence or absence of the dark green T6SS structure. The same color code is maintained throughout all figures. (B–C) Transformants from independent biological experiments (n ≥ 2) were analyzed by WGS for each of the conditions ① to ⑧, as indicated at the top of each graph. The main features of predator and prey/gDNA are summarized below the X-axis. Panels (B) and (C) depict the total length of acquired DNA and the number of exchanged DNA stretches, respectively, for each transformant. N.A., not applicable. Statistical analysis is based on a pairwise comparison between different conditions. *p<0.05, ***p<0.001.
-
Figure 4—source data 1
Raw data for Figure 4.
- https://doi.org/10.7554/eLife.48212.021
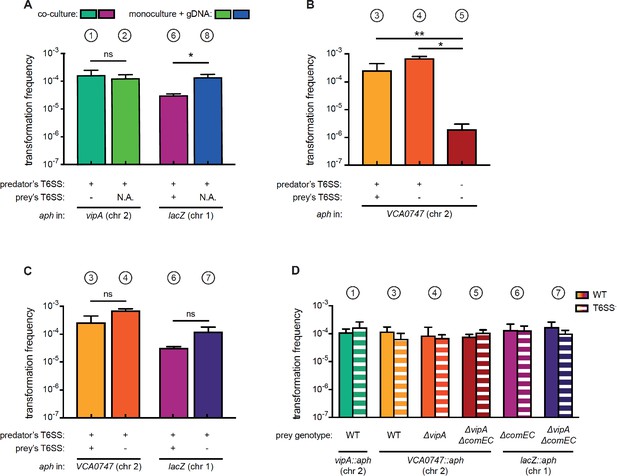
Natural transformation is enhanced by T6SS-mediated killing of prey bacteria.
(A–C) Transformation is enhanced in T6SS-positive predator cells. To induce natural competence, cultures were grown on chitin flakes. Bacteria were grown as co-cultures (predator + prey) or as monocultures (predator only). In the latter case, purified gDNA served as the transforming material. Transformants were selected based on their acquisition of the aph resistance cassette located in vipA, VCA0747, or lacZ on chr1 or chr2, as indicated below the graphs. The killing capability of each strain is indicated below the graph (e.g., T6SS + or -). Transformation frequencies are shown on the Y-axis (± SD, as indicated by the error bars) and depict averages of at least two biologically independent experiments (corresponding to the experiments described in Figure 4 and maintaining the same color code). N.A., not applicable. (D) The location of the resistance genes does not influence the transformation efficiency. Natural transformability of the WT (plain bars) or its T6SS-minus derivative (T6SS-; ΔvipA; hashed bars) was scored using gDNA as the transforming material. The gDNA samples were derived from the prey strains of conditions j and l–p and the respective genotype is shown below the graph. Transformation frequencies were scored based on the acquisition of the aph resistance cassette, which was integrated into different genes on chromosome 1 or 2 (chr 1/chr 2). Data represent the average of three independent experiments (± SD). (A–D) Statistical significance is indicated (*p<0.05; **p<0.01; ns, not significant).
-
Figure 4—figure supplement 1—source data 1
Raw data for Figure 4—figure supplement 1.
- https://doi.org/10.7554/eLife.48212.012
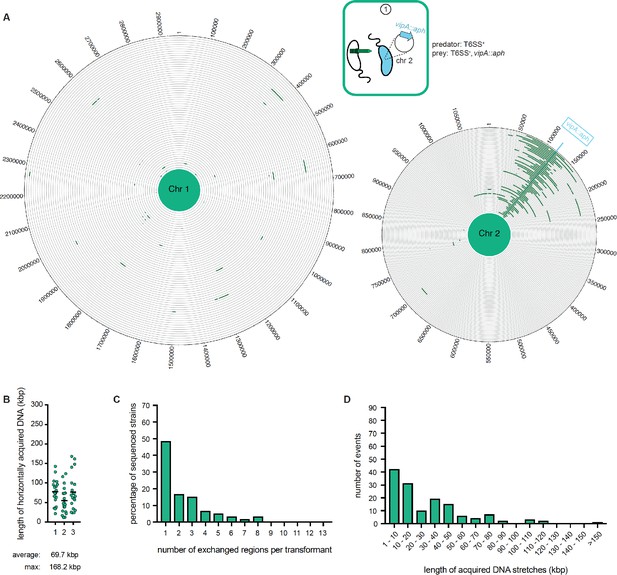
WGS-based quantification of horizontally acquired DNA under condition ①.
Data as in Figure 3 with the addition of the map of both chromosomes in panel A.
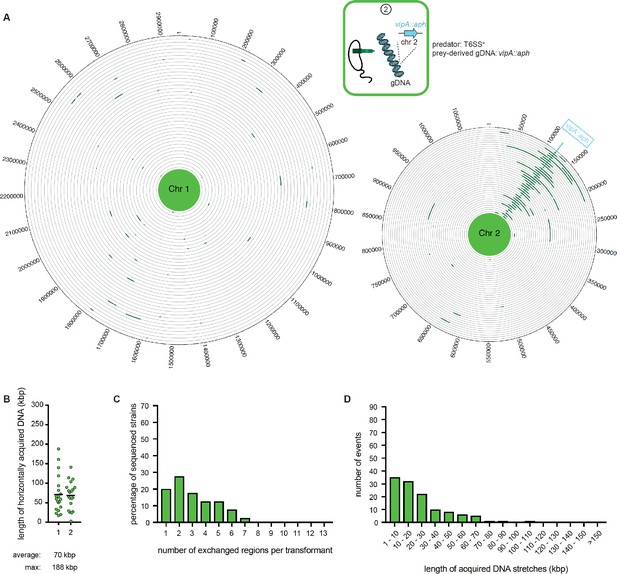
WGS-based quantification of horizontally acquired DNA under condition ②.
Details as described for Figure 3 with the addition of the map of both chromosomes in panel A.
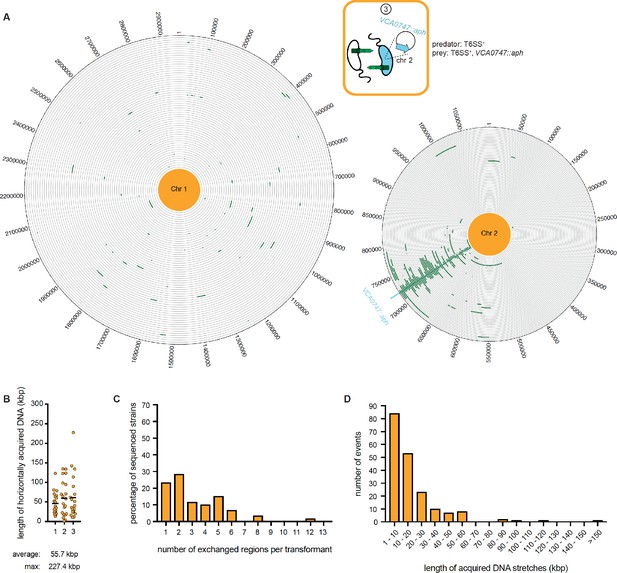
WGS-based quantification of horizontally acquired DNA under condition ③.
Details as described for Figure 3 with the addition of the map of both chromosomes in panel A.
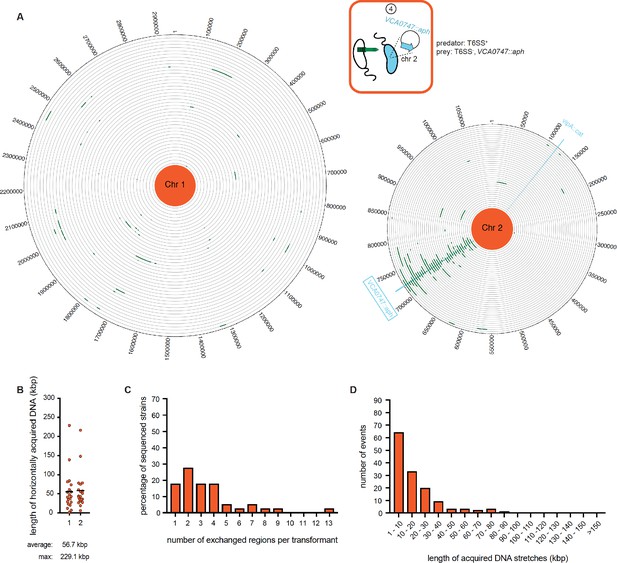
WGS-based quantification of horizontally acquired DNA under condition ④.
Details as described for Figure 3 with the addition of the map of both chromosomes in panel A.
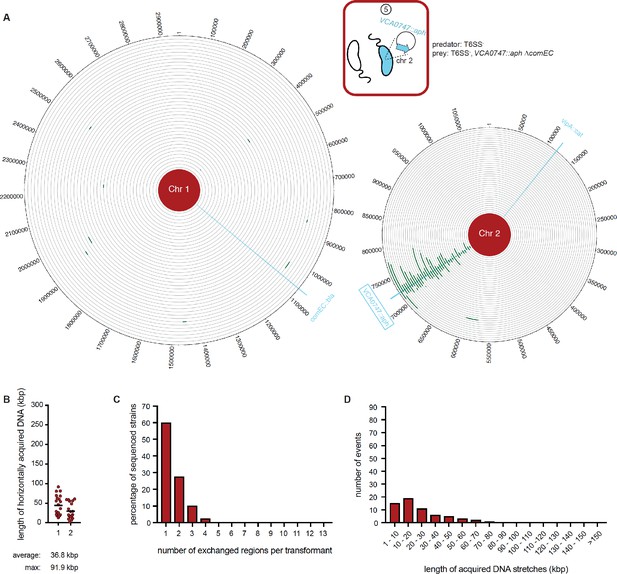
WGS-based quantification of horizontally acquired DNA under condition ⑤.
Details as described for Figure 3 with the addition of the map of both chromosomes in panel A.
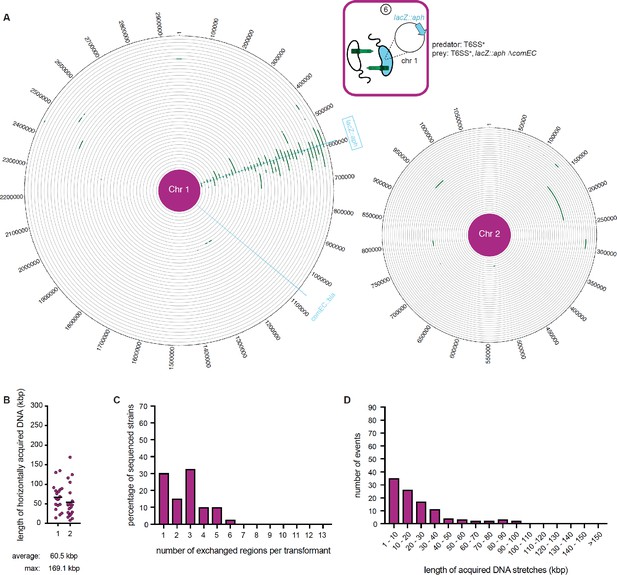
WGS-based quantification of horizontally acquired DNA under condition ⑥.
Details as described for Figure 3 with the addition of the map of both chromosomes in panel A.
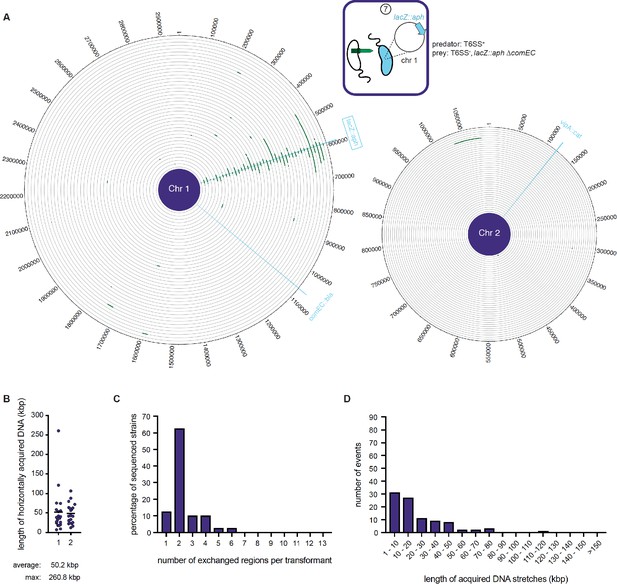
WGS-based quantification of horizontally acquired DNA under condition ⑦.
Details as described for Figure 3 with the addition of the map of both chromosomes in panel A.
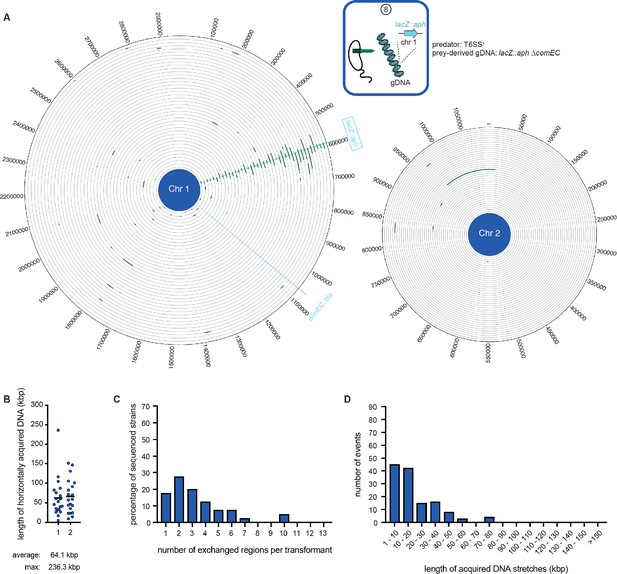
WGS-based quantification of horizontally acquired DNA under condition ⑧.
Details as described for Figure 3 with the addition of the map of both chromosomes in panel A.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Vibrio cholerae) | V. cholerae O1 El Tor, strain A1552 (Primary strain) | PMID: 9473029 PMID: 30574591 (genome sequence) | See Supplementary file 1 for strains and plasmids used in this study | |
| Strain, strain background (Vibrio cholerae) | V. cholerae, strain Sa5Y (Secondary strain) | PMID: 17449702 PMID: 30574591 (genome sequence) | See Supplementary file 1 for strains and plasmids used in this study | |
| Other | TCBS agar, selective medium | Sigma-Aldrich | 86348–500G | Additional, standard growth media are described under growth conditions |
| Peptide, recombinant protein | Pwo SuperYield DNA Polymerase | Roche / Sigma-Aldrich | 4340850001 | |
| Peptide, recombinant protein | GoTaq G2 DNA Polymerase | Promega | M7848 | |
| Commercial assay or kit | Genomic-tip 100/G (DNA purification) | Qiagen | 10243 | |
| Commercial assay or kit | Genomic DNA Buffer Set | Qiagen | 19060 |
Additional files
-
Supplementary file 1
Strains and plasmids used in this study.
- https://doi.org/10.7554/eLife.48212.022
-
Supplementary file 2
Details of eight experimental conditions and corresponding strain numbers.
- https://doi.org/10.7554/eLife.48212.023
-
Supplementary file 3
Sequence Read Archive (SRA) submission details.
- https://doi.org/10.7554/eLife.48212.024
-
Transparent reporting form
- https://doi.org/10.7554/eLife.48212.025





