Proteostasis collapse, a hallmark of aging, hinders the chaperone-Start network and arrests cells in G1
Figures
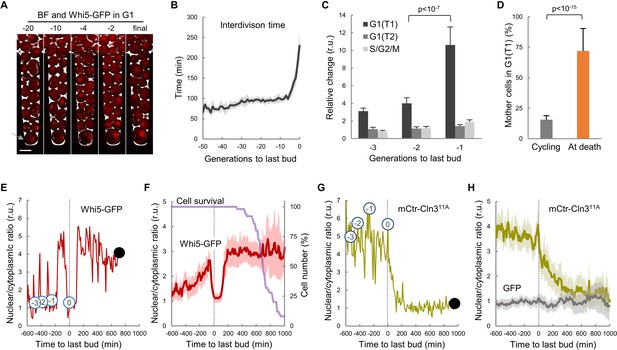
Yeast mother cells die mostly in G1 with low nuclear levels of cyclin Cln3.
(A) A yeast mother cell (arrow) expressing Whi5-GFP during aging at the G1 phase of indicated cycles before death. (B) Interdivision times (mean ±CL, n = 50) aligned to the last budding event. (C) Cell-cycle period lengths (mean ±CL, n = 50) in aging cells relative to young cells. (D) Percentage (±CL, n = 50) of cells in G1 in young cycling cultures or at death. (E) Nuclear levels of Whi5-GFP during the last generations (numbers in open circles) before death (closed circle) of an aging mother cell. (F) Nuclear levels of Whi5-GFP (mean ±CL, n = 50) in aging cells as in panel E aligned at the last budding event. (G–H) Nuclear levels of mCtr-Cln311A as in panels (E) and (F). Shown p-values were obtained using a Mann-Whitney U test. Bar = 5 µm. Results shown in this figure are representative of two replicate experiments.
-
Figure 1—source data 1
Yeast mother cells die mostly in G1 with low nuclear levels of cyclin Cln3.
- https://doi.org/10.7554/eLife.48240.004
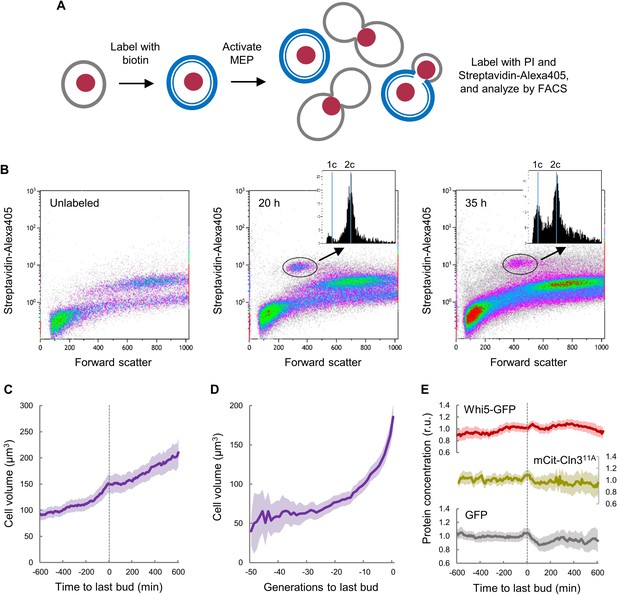
Yeast mother cells delay G1 progression with no major effects in growth during aging.
(A) Cell labeling and MEP activation steps used for FACS analysis. (B) FACS distributions of cells labeled as in A after 20 hr and 35 hr of MEP induction to identify aging mother cells (circled) and obtain the corresponding DNA content distributions (insets). A plot with unlabeled cells is also shown. (C) Cell volume (mean ±CL, n = 50) of aging cells as a function of time around the last budding event. (D) Cell volume (mean ±CL, n = 50) during the whole lifespan of aging cells aligned at the last budding event. (E) Levels (mean ±CL, n = 50) of Whi5-GFP, mCtr-Cln311A and GFP as control in aging cells aligned at the last budding event.
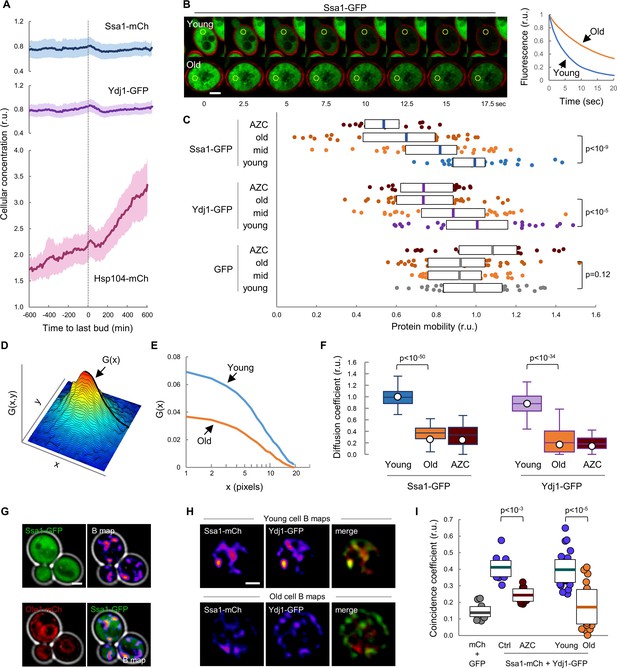
Mobility and spatio-temporal coincidence of Ssa1 and Ydj1 are reduced in aging cells.
(A) Levels of Ssa1-mCh, Ydj1-GFP and Hsp104-mCh (mean ±CL, n = 50) in aging cells aligned at the last budding event. (B) FLIP analysis of Ssa1-GFP in representative young and old (MEP-aged) cells. (C) Mobility of Ssa1-GFP, Ydj1-GFP and GFP in MEP- cells aged for 24 hr (mid) or 48 hr (old), and young control or AZC-treated cells. Median ±Q (n = 40) values are also plotted. (D) Spatial autocorrelation function (ACF) by RICS for Ssa1-GFP. G(x), the ACF in the scanning direction, is indicated. (E) Average ACFs (n = 25) obtained by RICS for Ssa1-GFP in old (MEP-aged) and young cells. (F) Diffusion coefficients (open circles) obtained by RICS for Ssa1-GFP and Ydj1-GFP in old (MEP-aged) and young control or AZC-treated cells (n = 25). The results of Monte Carlo simulations (median ±Q, n = 66) are plotted. (G) Representative fluorescence intensity (top left) and brightness (B map, top right) images obtained by RICS for Ssa1-GFP. Ole1-mCh as ER reporter (bottom left) and merged (bottom right) images are also shown. (H) Representative brightness (B) maps obtained by RICS for Ssa1-mCh and Ydj1-GFP in old (MEP-aged) and young cells. Merged B maps with Ssa1-mCh (red) and Ydj1-GFP (green) are also shown. (I) Coincidence coefficients of Ssa1-mCh and Ydj1-GFP from B maps of old (MEP-aged) and young cells (n = 20), control and AZC-treated young cells (n = 10), and cells expressing GFP and mCh (n = 10). Median ±Q values are also plotted. Shown p-values were obtained using a Mann-Whitney U test. Bar = 2 µm. Results shown in this figure are representative of at least two replicate experiments.
-
Figure 2—source data 1
Mobility and spatio-temporal coincidence of Ssa1 and Ydj1 are reduced in aging cells.
- https://doi.org/10.7554/eLife.48240.010
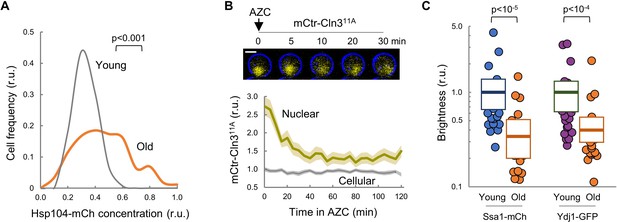
Chaperone availability is compromised in aging cells.
(A) Distribution by Hsp104 levels of young and old (MEP-aged) cells. (B) Nuclear and cellular mCtr-Cln311A levels (mean ±CL) in young cells after AZC addition. A representative cell expressing mCtr-Cln311A is show at the top. (C) Brightness values obtained by RICS for Ssa1-mCh and Ydj1-GFP in young and old (MEP-aged) cells. Median ±Q values (n = 25) are also plotted. Shown p-values were obtained using a Mann-Whitney U test. Bar = 2 µm.
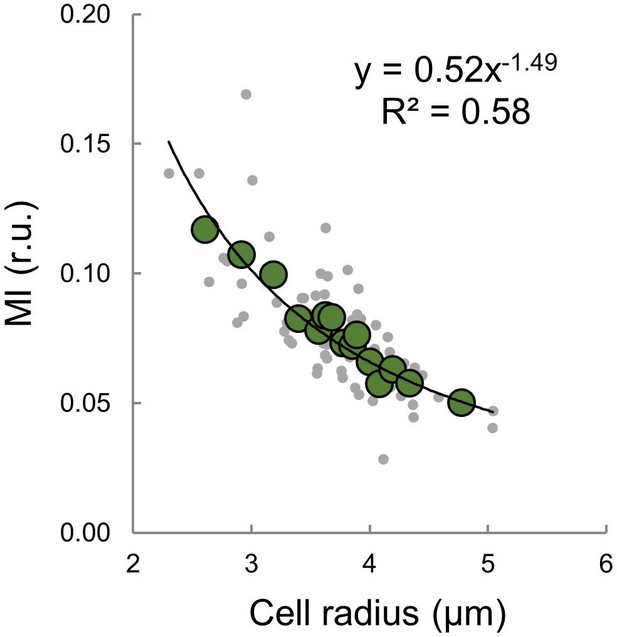
Correction of the mobility index obtained by FLIP as a function of cell size.
BF305 (cln1,2 GALp-CLN3) cells expressing GFP from endogenous sequences were grown in glucose for different periods of time to obtain a large range of cell volumes. The plot shows the obtained raw mobility index (see Materials and methods) by FLIP analysis as a function of the cell radius. Exponential regression of these data were used to obtain an expression to correct the mobility index as a function of cell size MIc = MI .r1.49 where MI is the raw mobility index, r is the cell radius and MIc is the corrected mobility index.
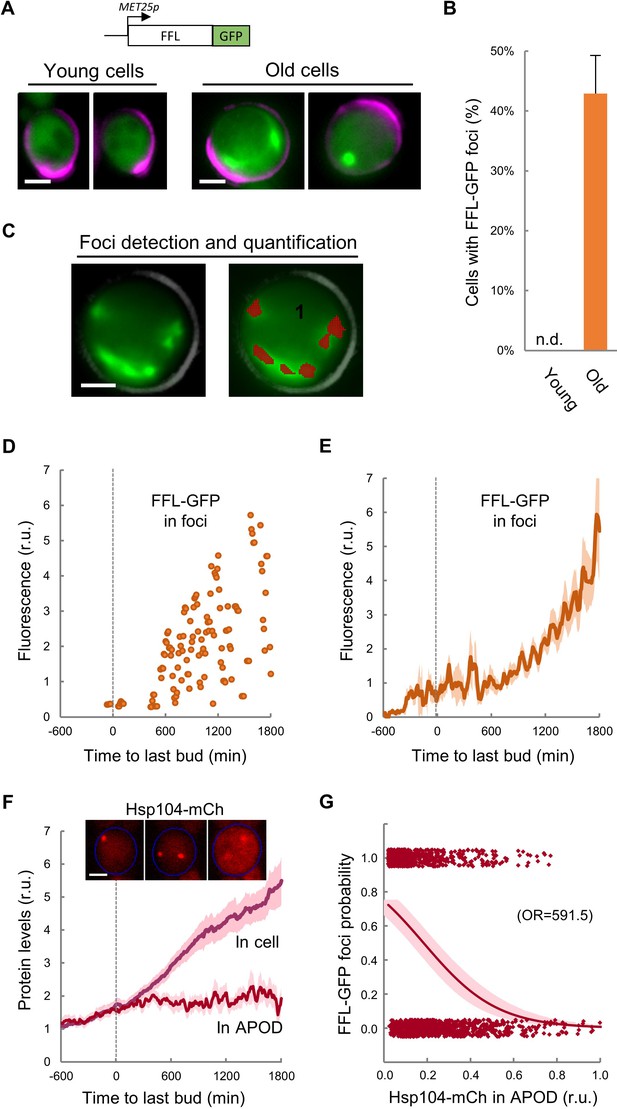
Firefly luciferase aggregates become visible during the last cycle before cell death.
(A) Representative images of FFL-GFP expressed from a regulatable promoter in young and old (MEP-aged) cells. (B) Percentage (±CL, n = 230) of young and old (MEP-aged) cells with FFL-GFP foci. (C) Representative image of an old (MEP-aged) cell expressing FFL-GFP (left) with the foci overlay (red) obtained from BudJ. (D–E) FFL-GFP levels in foci (n = 50) aligned at the last budding event. Individual (D) and mean ±CL (E) data are plotted. (F) Cellular and APOD Hsp104-mCh levels (mean ±CL, n = 50) in aging cells aligned at the last budding event. A representative aging cell (inset) at 0, 600 and 1200 min after the last budding event is shown. (G) Probability of FFL-GFP foci within the following 180 min (twice the generation time of young mother cells) after reading the relative Hsp104-mCh levels in the APOD. Sampled single-cell data (closed circles) and the logistic regression line (mean ±CL) are plotted, and the obtained odds ratio is indicated. Bar = 2 µm. Results shown in this figure are representative of two replicate experiments.
-
Figure 3—source data 1
Firefly luciferase aggregates become visible during the last cycle before cell death.
- https://doi.org/10.7554/eLife.48240.013
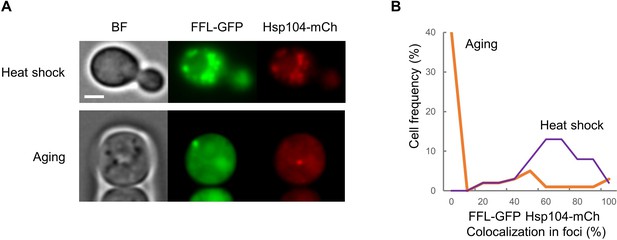
FFL-GFP and Hsp104 co-localization in heat-shocked and aging cells.
(A) Representative images of heat-shocked young or aging cells expressing FFL-GFP and Hsp104-mCh. (B) Distribution of FFL-GFP/Hsp104-mCh co-localization percentages in heat-shocked and aging cells. Bar = 2 µm.
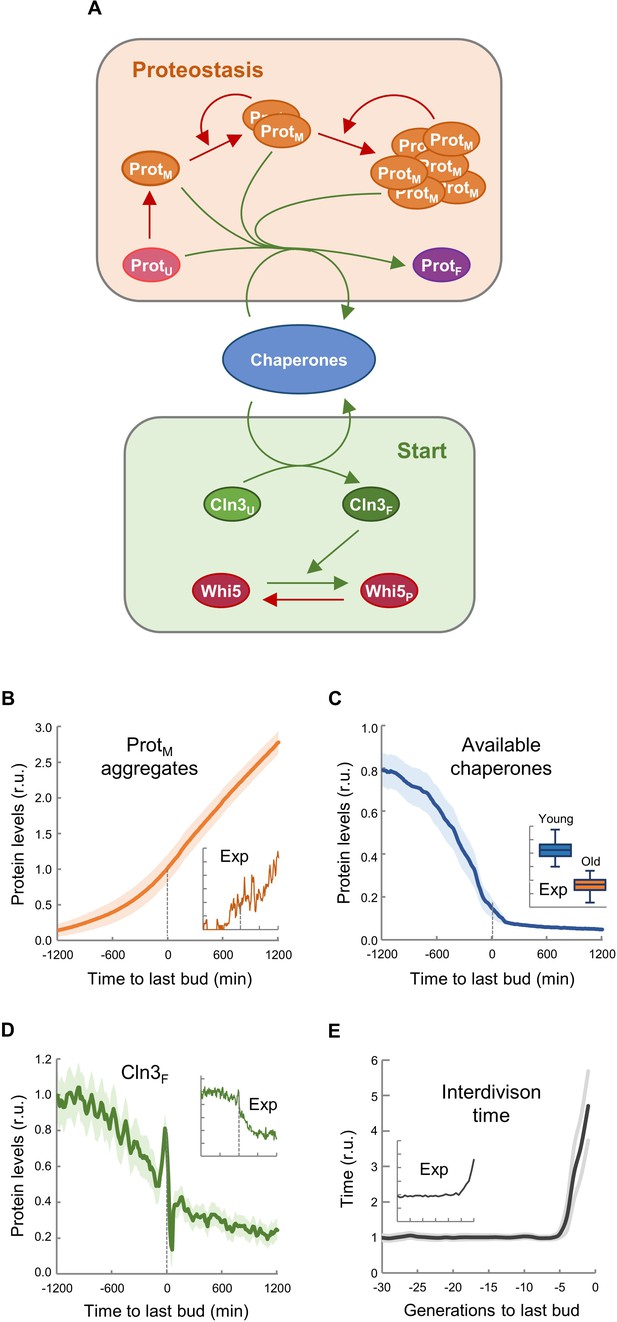
Asymmetric aggregate inheritance predicts a decrease in chaperone availability and a G1 arrest in aging cells.
(A) Scheme of the integrative mathematical model with chaperones playing concurrent roles in proteostasis and Start. (B–E) Predicted aggregate protein (B), available chaperone (C), and free folded Cln3 (D) levels and interdivision times in aging cells aligned at the last budding event. Values (mean ±CL, n = 75) are plotted as lines. Experimental (Exp) data from Figures 1B, H, 2F and 3E are also shown as insets for direct comparison.
-
Figure 4—source data 1
Asymmetric aggregate inheritance predicts a decrease in chaperone availability and a G1 arrest in aging cells.
- https://doi.org/10.7554/eLife.48240.018
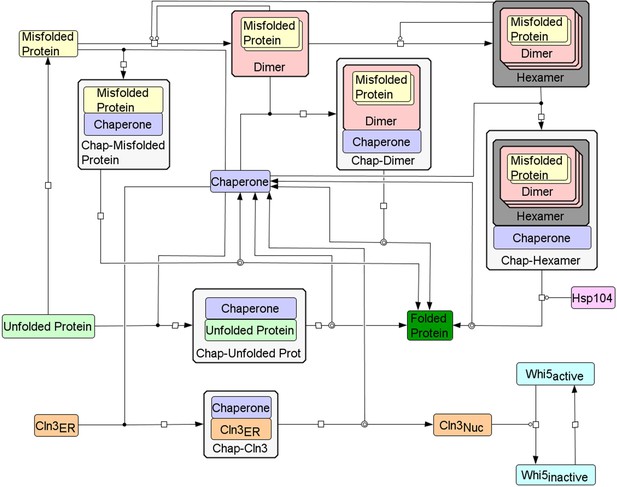
Wiring diagram of the integrative mathematical model.
Wiring diagram of the model to describe the interaction between a minimal Start network and the protein folding/aggregation pathway. Chaperones can bind to all forms of unfolded and misfolded proteins, but misfolded dimers and hexamers require two or six chaperones, respectively, to properly refold all monomers. Although not shown in the wiring diagram, all species are subject to degradation reactions, and Cln3, chaperone and unfolded protein synthesis is set as a function of growth rate. Created using CellDesigner (Funahashi et al., 2008).
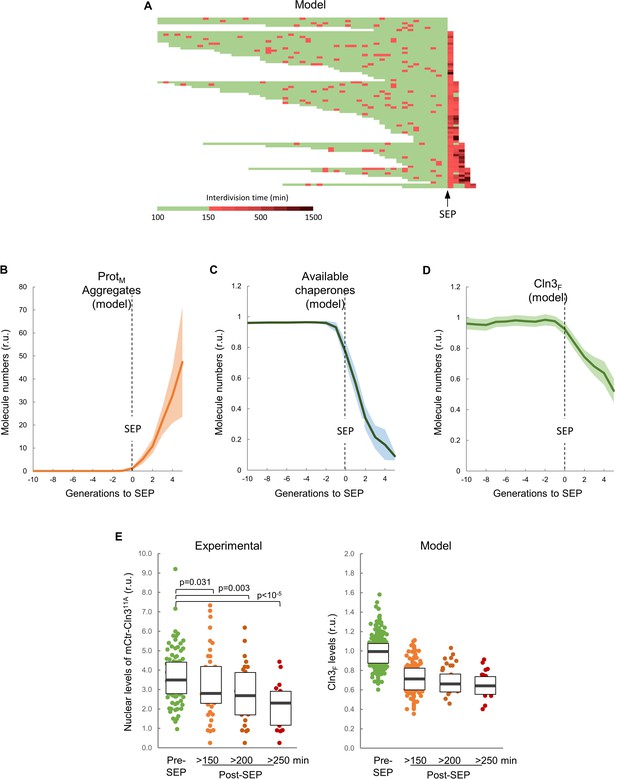
The integrative model predicts a decrease in chaperone availability and free Cln3 at the first generation after the SEP.
(A) Interdivision times obtained by simulations. Independent runs were aligned at the generation in which the interdivision time was maintained over a fixed threshold afterwards so as to simulate the SEP. (B–D) Predicted aggregate protein (B), available chaperone (C), and free folded Cln3 (D) levels in aging cells aligned at the SEP as shown in panel A. Values (mean ±CL, n = 75) are plotted as lines. (E) Nuclear levels of mCitrine-Cln311A in cells (n = 150) before or at the indicated times after the SEP. Median ±Q values are also plotted. The plot on the right shows the predicted values of free folded Cln3 (Cln3F) obtained from independent simulations (n = 75) for the corresponding times. Shown p-values were obtained using a Mann-Whitney U test.
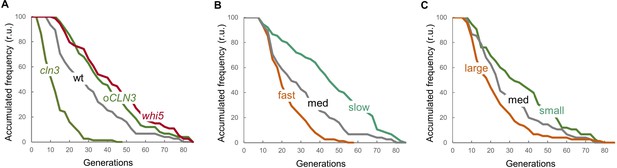
Lifespan of mutants and growth conditions as predicted by the integrative mathematical model.
(A) Survival curves predicted for the indicated genotypes of cells (n = 75). (B) Survival curves predicted for wild-type fast-, medium- and slow-growing cells (n = 75). (C) Survival curves predicted for wild-type cells with initial large, medium and small cell sizes (n = 75). Simulations were obtained as shown in Supplementary file 3.
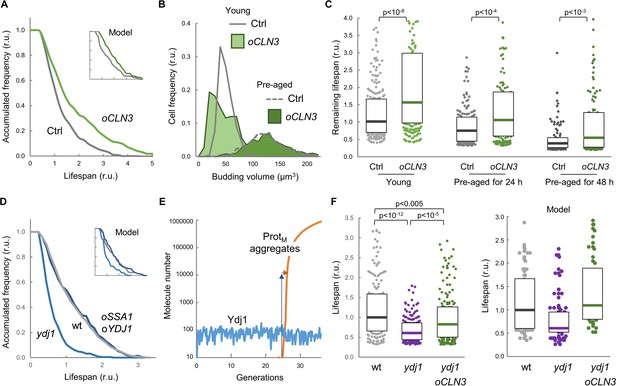
Enforced expression of Cln3 increases lifespan in a chaperone-dependent manner.
(A) Survival curves of control and CLN3 overexpressing cells (n > 300). Curves predicted by the integrative model in Figure 4A are also shown (inset). (B) Budding volume distributions (n > 250) of young or old cells pre-aged for 24 hr before induction of CLN3 expression. (C) Lifespan effects of CLN3 overexpression in young (n = 200) or old cells pre-aged for 24 hr (n = 150) and 48 hr (n = 100). Median ±Q values are also plotted. (D) Survival curves of wild-type, Ydj1-deficient and SSA1 YDJ1 overexpressing cells (n > 300). Curves predicted by the integrative model in Figure 4A are also shown (inset). (E) Simulation of free Ydj1 and ProtM aggregate numbers during successive replicative cycles in wild-type cells by the integrative model. The results of a representative run in stochastic mode are shown. A 2-order of magnitude increase in Ydj1 levels (blue arrow) would only cause a very small delay (orange arrow) in protein aggregation and, as a consequence, in lifespan. (F) Lifespan of wild-type (wt) and Ydj1-deficient cells with empty vector (ydj1) or overexpressing CLN3 (ydj1 oCLN3) (n = 150). Median ±Q values are also plotted. The plot on the right shows the predicted lifespan obtained from independent simulations (n = 75) for the corresponding genotypes. Shown p-values were obtained using a Mann-Whitney U test. Results shown in this figure are representative of at least two replicate experiments.
-
Figure 5—source data 1
Enforced expression of Cln3 increases lifespan in a chaperone-dependent manner.
- https://doi.org/10.7554/eLife.48240.021
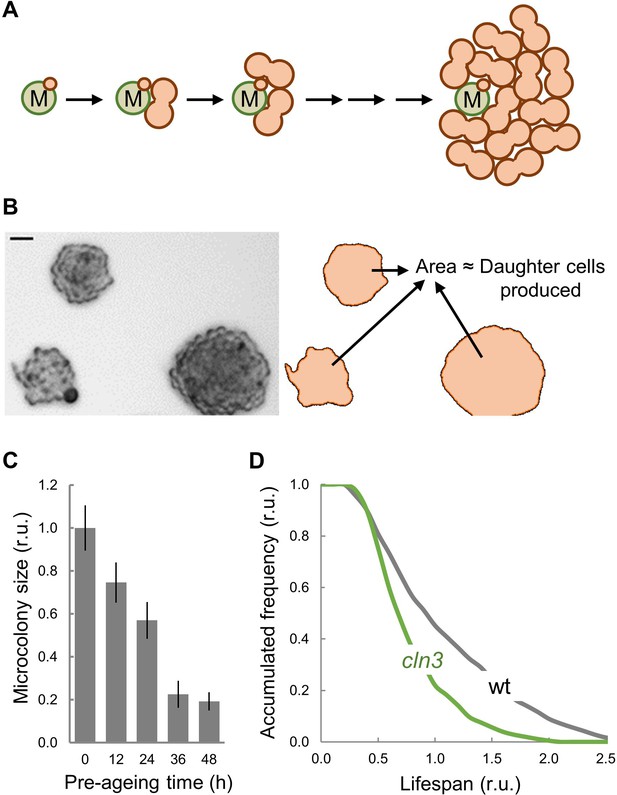
Lifespan analysis by MEP-induced microcolony size.
(A) Schematic of a microcolony formed by the accumulation of G2-arrested daughter cells (orange) produced after MEP induction by a single mother cell (M, green) during successive division cycles. (B) A representative image (left) of microcolonies of different size formed under MEP-inducing conditions. ImageJ-delimited microcolony areas used to estimate the number of daughter cells produced and, hence, the number of generations, are also shown (right). (C) Microcolony size (mean ±CL) as a function of pre-aging time before plating. (D) Survival curves of wild-type and cln3 mutant cells estimated from MEP-induced microcolony sizes (n > 250). Bar = 20 µm.
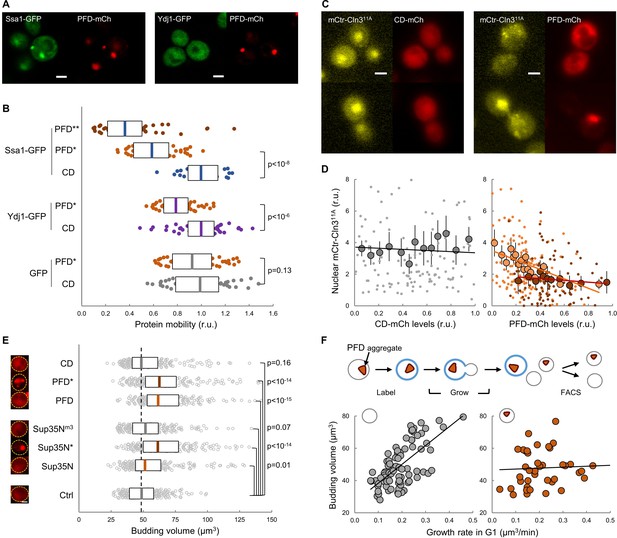
Protein aggregation hinders chaperone mobility and nuclear accumulation of Cln3 in young cells.
(A) Representative young cells expressing the prion-forming domain (PFD)-mCh and either Ssa1-GFP or Ydj1-GFP. (B) Mobility of Ssa1-GFP, Ydj1-GFP and GFP in young cells expressing control CD or displaying PFD aggregates (*). Ssa1-GFP mobility was also analyzed within PFD aggregates (**). Median ±Q (n = 50) values are also plotted. (C) Representative images of young cells expressing mCtr-Cln311A and either CD or PFD. (D) Nuclear levels of mCtr-Cln311A in young cells as a function of CD (left) or PFD (right) expression levels. Single-cell (small circles), binned (mean ±CL, n = 10) data and the corresponding linear regression lines are plotted. Cells with PFD aggregates are indicated (red circles). (E) Budding volume of young cells expressing the indicated protein domains. Cells with PFD or Sup35N aggregates are indicated (*). Median ±Q (n = 200) values are also plotted. (F) Budding volume of newborn daughter cells in the absence (left, n = 82) or presence (right, n = 42) of PFD aggregates after FACS selection as a function of growth rate in G1. Shown p-values were obtained using a Mann-Whitney U test. Bar = 2 µm. Results shown in this figure are representative of at least two replicate experiments.
-
Figure 6—source data 1
Protein aggregation hinders chaperone mobility and nuclear accumulation of Cln3 in young cells.
- https://doi.org/10.7554/eLife.48240.024
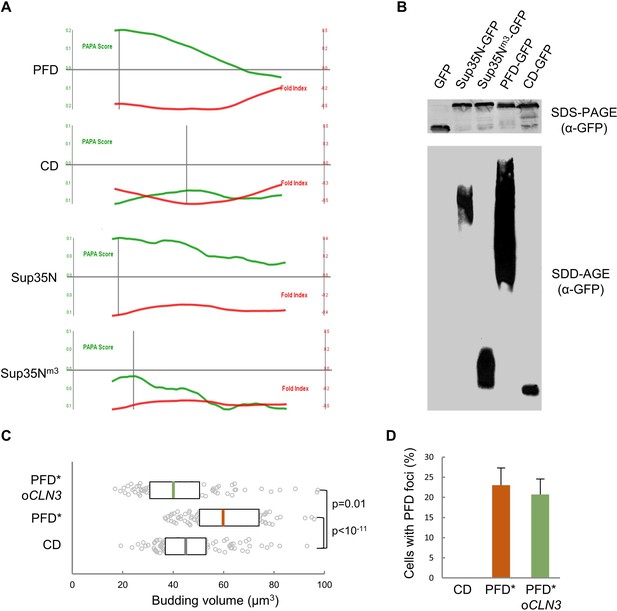
Enforced protein aggregation and CLN3 overexpression effects in young cells.
(A) PAPA-score and fold-index plots as a function of amino acid position in the indicated peptides. (B) Western blot analysis of the indicated protein fusions after gel electrophoresis under full (SDS-PAGE, top) or semi (SDD-AGE, bottom) denaturing conditions. (C) Budding volume of young cells expressing the indicated protein domains combined or not with CLN3 overexpression. Cells with PFD aggregates are indicated (*). Median ±Q (n = 100) values are also plotted. (D) Percentage (±CL, n = 100) of cells with PFD foci in cells as in panel C. Shown p-values were obtained using a Mann-Whitney U test.
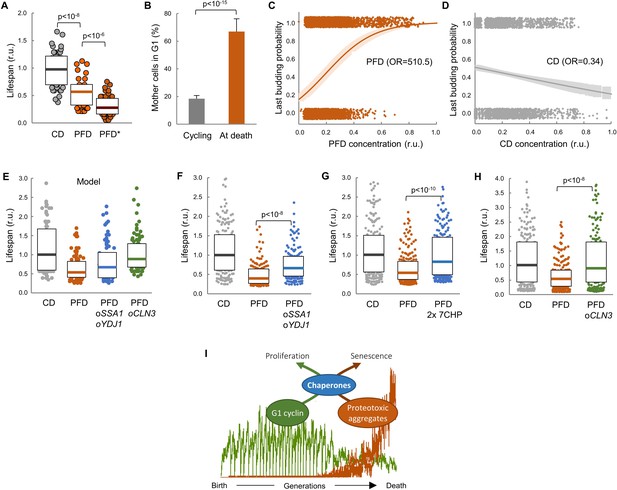
Lifespan shortening by protein aggregation can be overcome by enforced expression of chaperones or Cln3.
(A) Lifespan effects of CD or PFD expression in young cells in the CLiC microfluidics chamber (n = 50). Cells with PFD aggregates at the initial time point are indicated (*). Median ±Q values are also plotted. (B) Percentage (±CL, n = 57) of PFD expressing cells in G1 in young cycling cultures or at death. (C–D) Last budding probability within the following 180 min (twice the generation time of young mother cells) after reading PFD (C) or CD (D) cell concentration. Sampled single-cell data (closed circles) and the logistic regression lines (mean ±CL) are plotted. Odds ratios are also indicated. (E) Predicted lifespan effects of SSA1 YDJ1 overexpression and CLN3 overexpression in PFD expressing cells (n = 75). Predicted values for control CD-expressing cells are shown as reference. Median ±Q values are also plotted. (F–H) Lifespan effects of concerted SSA1 YDJ1 overexpression (F), duplication of seven chaperone genes (2 × 7CHP: SSA1, YDJ1, HSP82, CDC37, CDC48, UFD1 and NPL4) (G) or CLN3 overexpression (H) in PFD expressing cells (n > 150). Values of control CD-expressing cells are shown as reference. Median ±Q values are also plotted. (I) By compromising chaperone availability, proteostasis deterioration would exclude cyclin Cln3 from the nucleus and, as a direct consequence, drive the cell into senescence. Shown p-values were obtained using a Mann-Whitney U test. Results shown in this figure are representative of at least two replicate experiments.
-
Figure 7—source data 1
Lifespan shortening by protein aggregation can be overcome by enforced expression of chaperones or Cln3.
- https://doi.org/10.7554/eLife.48240.027
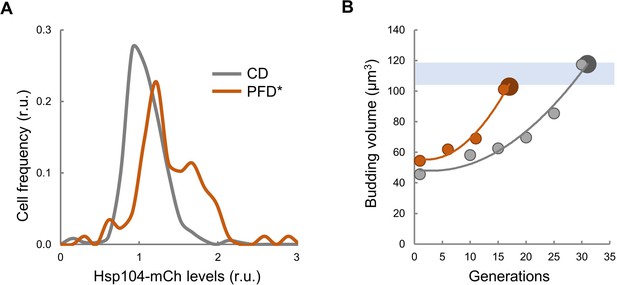
Enforced protein aggregation increases levels of Hsp104 and budding size during aging.
(A) Distribution by Hsp104 levels of young cells displaying PFD aggregates or expressing CD as control. (B) Mean budding volume (n0 = 50) during the lifespan of cells expressing PFD or CD.
Videos
Movie of a representative Whi5-GFP (green) Hsp104-mCh (red) cell in the CLiC microfluidic chamber.
Images were taken every 10 min. The frame where the last budding event takes place is indicated.
Movie of a representative mCitrine-Cln311A (yellow) cell in the CLiC microfluidic chamber.
Images were taken every 10 min. The frame where the last budding event takes place is indicated.
Tables
| Reagent type or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Saccharomyces cerevisiae) | BY4741 | Lab stock | MATa his3-Δ1 leu2Δ0 met15Δ0 ura3Δ0, from S288C | |
| Strain, strain background (S. cerevisiae) | MAG248 | This work | MATa his3-Δ1 leu2Δ0 met15Δ0 ura3Δ0 NAT::TEFp-GFP, from S288C | |
| Strain, strain background (S. cerevisiae) | MAG261 | (Moreno et al., 2019) | MATa his3-Δ1 leu2Δ0 met15Δ0 ura3Δ0 YDJ1-GFP-FS::HIS3, from S288C | |
| Strain, strain background (S. cerevisiae) | MAG1078 | This work | MATa his3-Δ1 leu2Δ0 met15Δ0 ura3Δ0 YDJ1-GFP-FS::HIS3 SSA1-mCherry::HYG, from S288C | |
| Strain, strain background (S. cerevisiae) | MAG1689 | This work | MATa his3-Δ1 leu2Δ0 met15Δ0 ura3Δ0 SSA1-GFP::HIS3 OLE1-mCherry::GEN, from S288C | |
| Strain, strain background (S. cerevisiae) | YOR083W-GFP | Lab stock | MATa his3-Δ1 leu2Δ0 met15Δ0 ura3Δ0 WHI5-yGFP::HIS3, from S288C | |
| Strain, strain background (S. cerevisiae) | CML128 | (Gallego et al., 1997) | MATa leu2-3,112 ura3-52 trp1-1 his4-1 canr, from 1788 | |
| Strain, strain background (S. cerevisiae) | MAG1077 | This work | MATa leu2-3,112 ura3-52 trp1-1 his4-1 canr WHI5-sGFP::GEN HSP104-mCherry::HYG, from 1788 | |
| Strain, strain background (S. cerevisiae) | MAG1512 | (Moreno et al., 2019) | MATa leu2-3,112 ura3-52 trp1-1 his4-1 canr NAT::TEF1p-mCherry, from 1788 | |
| Strain, strain background (S. cerevisiae) | MAG1767 | This work | MATa leu2-3,112 ura3-52 trp1-1 his4-1 canr mCitrine-CLN3(11A)::NAT, from 1788 | |
| Strain, strain background (S. cerevisiae) | MAG1767 | This work | MATa leu2-3,112 ura3-52 trp1-1 his4-1 canr HSP104-mCherry::HYG, from 1788 | |
| Strain, strain background (S. cerevisiae) | UCC5179 | (Lindstrom and Gottschling, 2009) | MATa ade2::hisG his3 leu2 lys2 ura3Δ0 trp1Δ63 hoΔ::SCW11pr-Cre-EBD78-NatMX loxP-UBC9-loxP-LEU2 loxP-CDC20-intron-loxP-HPHMX, from S288C | |
| Strain, strain background (S. cerevisiae) | MAG1013 | This work | MATa ade2::hisG his3 leu2 lys2 ura3Δ0 trp1Δ63 hoΔ::SCW11pr-Cre-EBD78-NatMX loxP-UBC9-loxP-LEU2 loxP-CDC20-intron-loxP-HPHMX (ARS-CEN URA3 HSP104 SSA1 YDJ1 HSC82 CDC37 CDC48 UFD1 NPL1), from S288C | |
| Strain, strain background (S. cerevisiae) | MAG1095 | This work | MATa ade2::hisG his3 leu2 lys2 ura3Δ0 trp1Δ63 hoΔ::SCW11pr-Cre-EBD78-NatMX loxP-UBC9-loxP-LEU2 loxP-CDC20-intron-loxP-HPHMX YDJ1-GFP-FS::HIS3, from S288C | |
| Strain, strain background (S. cerevisiae) | MAG1096 | This work | MATa ade2::hisG his3 leu2 lys2 ura3Δ0 trp1Δ63 hoΔ::SCW11pr-Cre-EBD78-NatMX loxP-UBC9-loxP-LEU2 loxP-CDC20-intron-loxP-HPHMX SSA1-GFP::HIS3, from S288C | |
| Strain, strain background (S. cerevisiae) | MAG1578 | This work | MATa ade2::hisG his3 leu2 lys2 ura3Δ0 trp1Δ63 hoΔ::SCW11pr-Cre-EBD78-NatMX loxP-UBC9 -loxP-LEU2 loxP-CDC20-intron-loxP-HPHMX ydj1Δ::GEN, from S288C | |
| Strain, strain background (S. cerevisiae) | MAG1745 | This work | MATa ade2::hisG his3 leu2 lys2 ura3Δ0 trp1Δ63 hoΔ::SCW11pr-Cre-EBD78-NatMX loxP-UBC9-loxP-LEU2 loxP-CDC20-intron-loxP-HPHMX YDJ1-GFP-FS::HIS3 SSA1-mCherry::KAN, from S288C | |
| Strain, strain background (S. cerevisiae) | MAG1952 | This work | MATa ade2::hisG his3 leu2 lys2 ura3Δ0 trp1Δ63 hoΔ:: SCW11pr-Cre-EBD78-NatMX loxP-UBC9-loxP-LEU2 loxP-CDC20-intron-loxP-HPHMX Hsp104-mCherry::GEN, from S288C | |
| Strain, strain background (S. cerevisiae) | MAG2060 | This work | MATa ade2::hisG his3 leu2 lys2 ura3Δ0 trp1Δ63 hoΔ::SCW11pr-Cre-EBD78-NatMX loxP-UBC9-loxP-LEU2 loxP-CDC20-intron-loxP-HPHMX GAL1p-CLN3 URA3::TRP1, from S288C | |
| Strain, strain background (S. cerevisiae) | MAG1253 | This work | MATa ade2::hisG his3 leu2 lys2 ura3Δ0 trp1Δ63 hoΔ::SCW11pr-Cre-EBD78-NatMX loxP-UBC9-loxP-LEU2 loxP-CDC20-intron-loxP-HPHMX trp1Δ63::SCW11pr-Cre-EBD78 -KanMX4, from S288C | |
| Strain, strain background (S. cerevisiae) | MAG1569 | This work | MATa ade2::hisG his3 leu2 lys2 ura3Δ0 trp1Δ63 hoΔ:: SCW11pr-Cre-EBD78-NatMX loxP-UBC9-loxP-LEU2 loxP-CDC20-intron-loxP-HPHMX trp1Δ63::SCW11pr-Cre-EBD78- KanMX4 CLB2-GFP::HIS3MX, from S288C | |
| Strain, strain background (S. cerevisiae) | MAG1576 | This work | MATa ade2::hisG his3 leu2 lys2 ura3Δ0 trp1Δ63 hoΔ::SCW11pr-Cre-EBD78-NatMX loxP-UBC9-loxP-LEU2 loxP-CDC20-intron-loxP-HPHMX trp1Δ63::SCW11pr-Cre-EBD78-KanMX4 CLB2-GFP::HIS3 GALp-CLN3-URA3::TRP1, from S288C | |
| Strain, strain background (S. cerevisiae) | MAG1795 | This work | MATa ade2::hisG his3 leu2 lys2 ura3Δ0 trp1Δ63 hoΔ::SCW11pr-Cre-EBD78-NatMX loxP-UBC9-loxP-LEU2 loxP-CDC20-intron-loxP-HPHMX trp1Δ63::SCW11pr-Cre-EBD78-KanMX4 CLB2-GFP::HIS3MX Δcln3::URA3MX, from S288C | |
| Strain, strain background (S. cerevisiae) | W303-1A | Lab stock | MATa ade2-1 trp1-1 leu2-3,111 his3-11,75 ura3 can1-100, from W303 | |
| Strain, strain background (S. cerevisiae) | KSY083-5 | (Schmoller et al., 2015) | MATa ADE2 trp1-1 leu2-3,111 his3-11,75 ura3 can1-100 mCitrine-CLN3-11A::NAT | |
| Strain, strain background (S. cerevisiae) | MAG876 | This work | MATa ade2-1 trp1-1 leu2-3,111 his3-11,75 ura3 can1-100 SSA1-GFP::HIS3, from W303 | |
| Recombinant DNA reagent | YCplac22 | Lab stock | Centromeric TRP1 vector | |
| Recombinant DNA reagent | YCplac33 | Lab stock | Centromeric URA3 vector | |
| Recombinant DNA reagent | YCpGAL | Lab stock | GAL1/10 p in YCplac22 | |
| Recombinant DNA reagent | p425MET25-FFL-GFP | (Abrams and Morano, 2013) | MET25p-FFL-GFP in pRS425 | |
| Recombinant DNA reagent | pCYC87 | Lab stock | GAL1/10p-CLN3-3HA in YCplac33 | |
| Recombinant DNA reagent | pMAG438 | (Moreno et al., 2019) | SSA1 YDJ1 HSC82 CDC37 CDC48 UFD1 NPL4 in YAC URA3 | |
| Recombinant DNA reagent | pMAG600 | This work | GAL1/10p-Sup35Nm3-GFP in YCplac22 | |
| Recombinant DNA reagent | pMAG602 | This work | GAL1/10p-PFD-GFP in YCplac22 | |
| Recombinant DNA reagent | pMAG604 | This work | GAL1/10p-CD-GFP in YCplac22 | |
| Recombinant DNA reagent | pMAG605 | This work | GAL1/10p-Sup35N-GFP in YCplac22 | |
| Recombinant DNA reagent | pMAG610 | This work | GAL1/10p-GFP in YCplac22 | |
| Recombinant DNA reagent | pMAG633 | This work | GAL1/10p-PFD-mCh in YCplac33 | |
| Recombinant DNA reagent | pMAG634 | This work | GAL1/10p-CD-mCh in YCplac33 | |
| Recombinant DNA reagent | pMAG1182 | This work | GAL1p-SSA1 GAL10p-YDJ1 in YCplac33 | |
| Recombinant DNA reagent | pMAG1228 | Lab stock | TE1Fp-GFP in YCplac33 | |
| Antibody | αGFP (Mouse monoclonal) | Merck | G1546 | |
| Chemical compound, drug | Azetidine-2-carboxilic acid | Sigma-Aldrich | A0760 | |
| Chemical compound, drug | β-estradiol | Sigma-Aldrich | E2758 | |
| Software, algorithm | MODEL1901210001 | This work | BioModels database | |
| Software, algorithm | ImageJ | Wayne Rasband, NIH | imagej.nih.gov/ij/download.html | |
| Software, algorithm | BudJ | (Ferrezuelo et al., 2012) | ibmb.csic.es/groups/spatial-control-of-cell-cycle-entry | |
| Software, algorithm | CoinRICSJ | (Moreno and Aldea, 2019) | ibmb.csic.es/groups/spatial-control-of-cell-cycle-entry | |
| Software, algorithm | RICS analysis plugins | Jay Unruh, Stowers Institute | research.stowers.org/imagejplugins | |
| Software, algorithm | Microcolony_size.ijm | This work | ibmb.csic.es/groups/spatial-control-of-cell-cycle-entry | |
| Software, algorithm | PAPA | (Toombs et al., 2012) | combi.cs.colostate.edu/supplements/papa |
Additional files
-
Supplementary file 1
Chemical reactions of the integrative mathematical model.
- https://doi.org/10.7554/eLife.48240.028
-
Supplementary file 2
Parameter set of the integrative mathematical model.
- https://doi.org/10.7554/eLife.48240.029
-
Supplementary file 3
Parameter modifications to simulate different genotypes or relevant physiological conditions.
- https://doi.org/10.7554/eLife.48240.030
-
Transparent reporting form
- https://doi.org/10.7554/eLife.48240.031





