Brain clusterin protein isoforms and mitochondrial localization
Figures
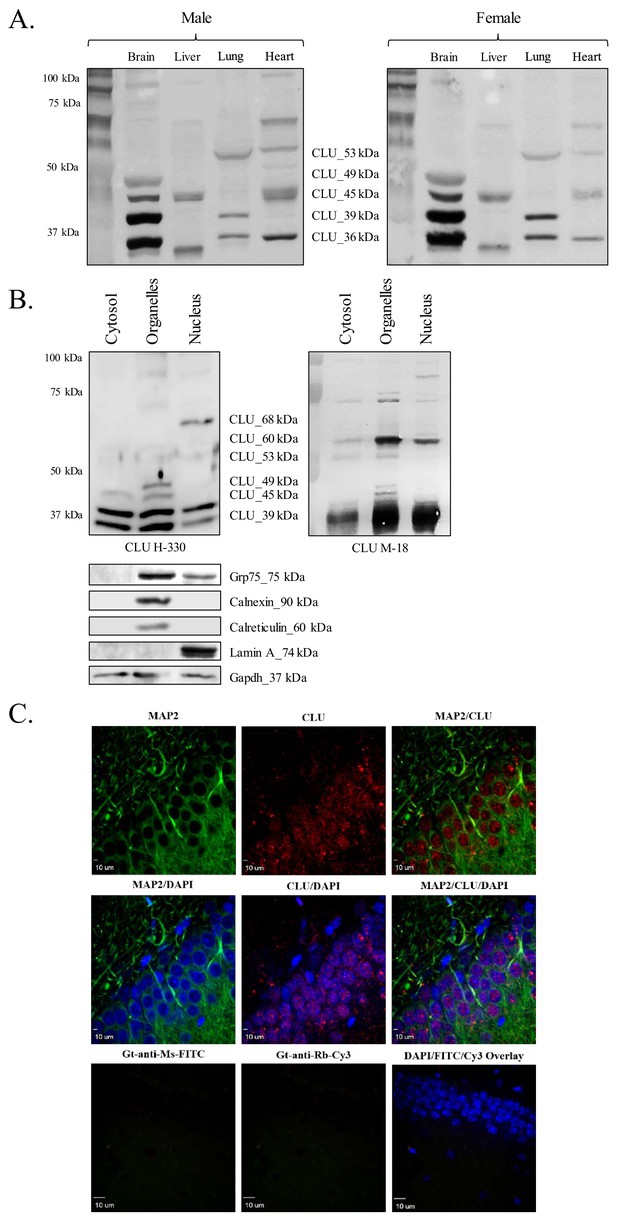
CLU protein is expressed in the brain as multiple protein isoforms that have distinct cellular localizations.
(A) Whole brain, liver, lung, and heart tissues were isolated from age-matched male (left panel) and female (right panel) wild-type (WT) mice and homogenized as indicated. Total protein was analyzed with SDS-PAGE and blots were probed for CLU immunoreactivity with anti-CLU H-330. (B) Cytosolic, organelle, and nuclear fractions were isolated from freshly harvested WT cortical tissue as indicated. Fractions were analyzed via SDS-PAGE and blots were probed for CLU immunoreactivity with anti-CLU H-330 (left panel) and anti-CLU M-18 (right panel). Blots were stripped and re-probed with fraction-/organelle-specific biochemical markers: Grp75 (mitochondria), calnexin and calreticulin (ER), lamin A (nucleus), and Gapdh (cytosol). (C) 40-µM-thick rodent brain sections were permeabilized and blocked as indicated and labeled with anti-MAP2 (green) or anti-GFAP (Figure 1—figure supplement 1C) and anti-CLU H-330 (red). Brain sections were then washed and probed with anti-mouse FITC (for MAP2) or anti-rat Cy5 (for GFAP) and pre-adsorbed anti-rabbit Cy3 (for CLU). To generate a secondary antibody control (bottom panel), one group of free-floating brain sections was incubated overnight in the same conditions without primary antibody. Brain sections from the hippocampal dentate gyrus were imaged at 4X (Figure 1—figure supplement 1B) and 40X using a customized Olympus IX81/spinning disk confocal inverted microscope and analyzed using the Slidebook Software.
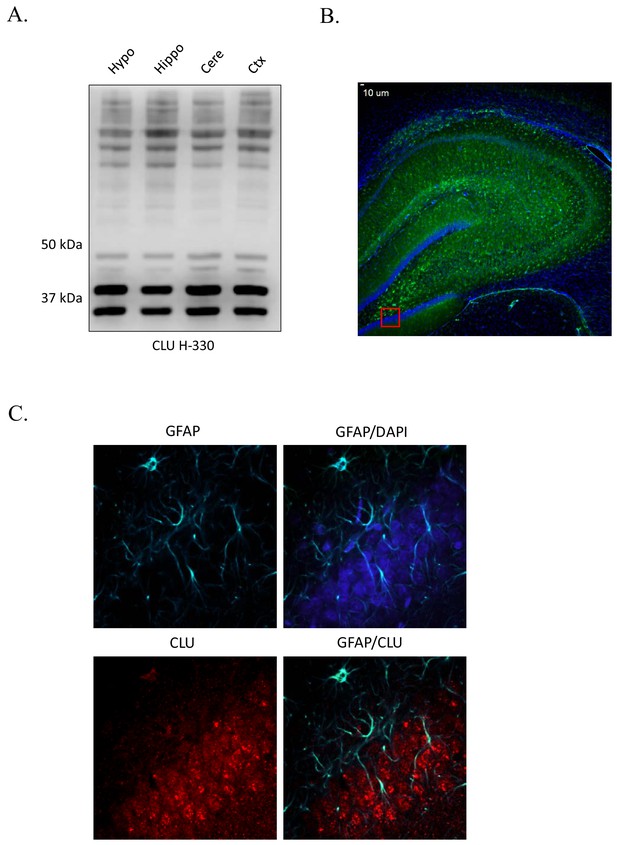
CLU protein expression in the rodent brain.
(A) CLU protein is expressed throughout the rodent brain. (B) 4X representation of the rodent brain used to generate Figure 1C. (C) CLU is minimally co-localized with GFAP. 40-µM-thick rodent brain sections were labeled and imaged as indicated in Figure 1C.
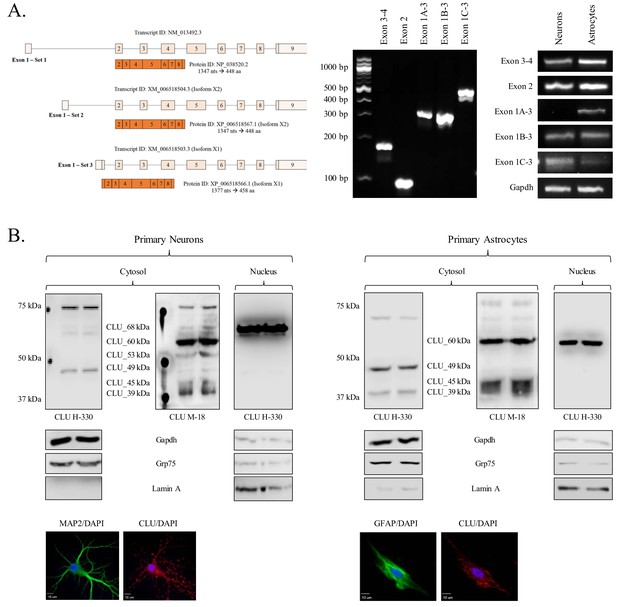
Characterization of neuronal and astrocytic CLU protein isoforms.
(A: left panel) Schematic of known murine CLU mRNA transcripts provided in the NCBI database. (A: right panel) Specific CLU gene transcript levels were examined using 25 ng cDNA isolated from DIV 9/16 neurons/astrocytes. Amplicons were visualized on a 4% agarose gel to ensure the appropriate size (left panel) and a comparison of CLU amplicon intensity was visualized (right panel). (B) Primary neurons (left panel) or astrocytes (right panel) were prepared as indicated. At DIV 9/16, cytosolic and nuclear fractions were isolated. 30 μg of each fraction was analyzed for CLU protein expression using anti-CLU H-330 and anti-CLU M-18. Fraction purity was analyzed using Gapdh (cytosol), Grp75 (mitochondria), and lamin A (nucleus). Isolation of cell type was confirmed by double labeling with either MAP2 (neurons) or GFAP (astrocytes) and CLU H-330, as indicated in the Materials and methods. Cells were imaged at 40X (air; neurons) and 60X (oil; astrocytes) using a customized Olympus IX81/spinning disk confocal inverted microscope and analyzed using the Slidebook Software.
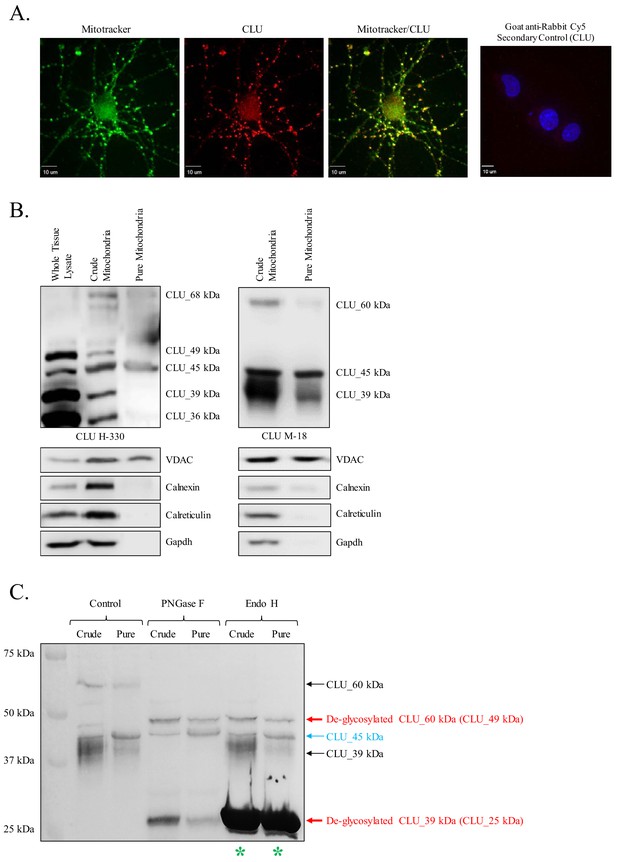
Identification of a mitochondrial CLU protein isoform.
(A) DIV 9 Mitotracker-stained (green) primary neurons were probed for CLU immunoreactivity using anti-CLU H-330 (red) and visualized using 40X confocal microscopy. (B) Pure cortical mitochondria were isolated as indicated. Equal concentrations of whole tissue lysate, crude mitochondria, and pure mitochondria were analyzed via SDS-PAGE and probed for CLU immunoreactivity using anti-CLU H-330 (left panel) and anti-CLU M-18 (right panel) (n = 3 independent isolations). Biochemical characterization of isolated fractions was performed using a panel of organelle-specific antibodies: voltage-dependent anion channels (VDAC) (mitochondria), calnexin and calreticulin (ER), and Gapdh (cytosol). (C) Crude and pure mitochondria were isolated and subjected to endoglycosidase treatment using PNGase F and Endo H. Deglycosylated mitochondrial lysates were then analyzed for CLU immunoreactivity using anti-CLU M-18. Red font: deglycosylated protein isoforms; blue font: isoforms that were unaffected by glycosidase treatment; green asterisk: excess Endo H enzyme.
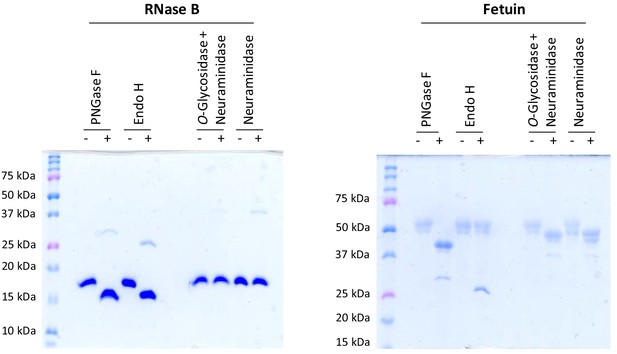
Positive controls for deglycosylation studies.
RNase B, a high mannose glycoprotein, has a single N-linked glycosylation site and was used as a positive control for endoglycosidases that cleave N-linked carbohydrates. Fetuin, a glycoprotein containing sialylated N-linked and O-linked glycans, was used as a positive control for endoglycosidases that cleave both N-linked and O-linked carbohydrates.
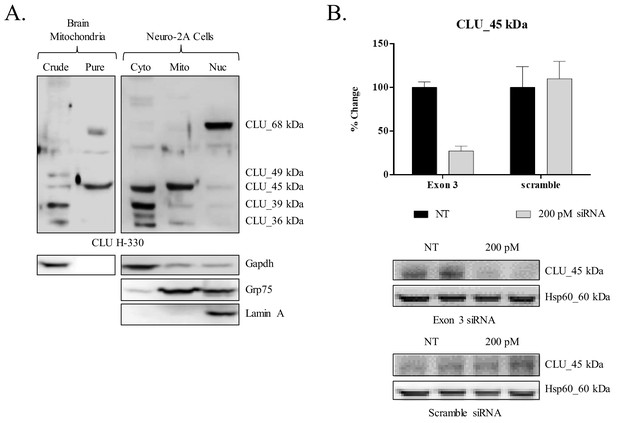
siRNA-mediated knockdown of CLU_45 kDa in mouse neuroblastoma cells.
(A) Mouse neuroblastoma cells (Neuro-2a) were cultured and subjected to a subcellular fractionation to isolate the cytosolic, crude mitochondrial, and nuclear fractions. 30 μg of each fraction was analyzed for CLU protein expression using anti-CLU H-330. Fraction purity was analyzed using fraction-specific markers: Gapdh (cytosol), Grp75 (mitochondria), and lamin A (nucleus). (B) Neuro-2a cells were transfected with 200 pM of Exon-3-targeting siRNA or non-specific scramble for 18 hr followed by 72 hr incubation. Whole cell lysates were analyzed for CLU protein expression using anti-CLU H-330. To ensure consistent loading, blots were stripped and re-probed for Hsp60 immunoreactivity. The relative intensity of CLU_45 kDa was normalized to the relative intensity of Hsp60 and is plotted as % change compared to the non-transfected cells. Values represent the mean of two individual experiments, with error bars representing the standard error.
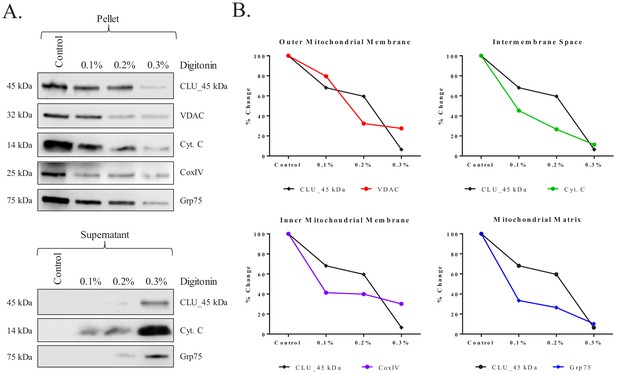
Submitochondrial localization of CLU_45 kDa in rodent brain mitochondria.
Pure cortical mitochondria were isolated using a discontinuous percoll gradient and subjected to a mitochondrial subfractionation using increasing concentrations of digitonin (0%–0.3%). Following permeabilization, mitochondrial samples were separated to yield the mitochondrial pellet (retained proteins) and the mitochondrial supernatant (released proteins). (A) Samples were probed for CLU immunoreactivity with CLU H-330 and mitochondrial-compartment-specific markers: VDAC (outer mitochondrial membrane), cytochrome C (Cyt. C; intermembrane space), CoxIV (inner mitochondrial membrane), and Grp75 (Matrix). (B) Percent change was calculated by setting each protein control sample to 100%. CLU expression changes in the pellet were compared to the expression changes in each fraction-specific marker. In the provided plots, the y-axis represents % change in protein expression and the x-axis represents % digitonin from 0% (Control) to 0.3%. The black lines in each of the four plots represent CLU % change while the colored lines represent the % change of the respective mitochondrial marker.
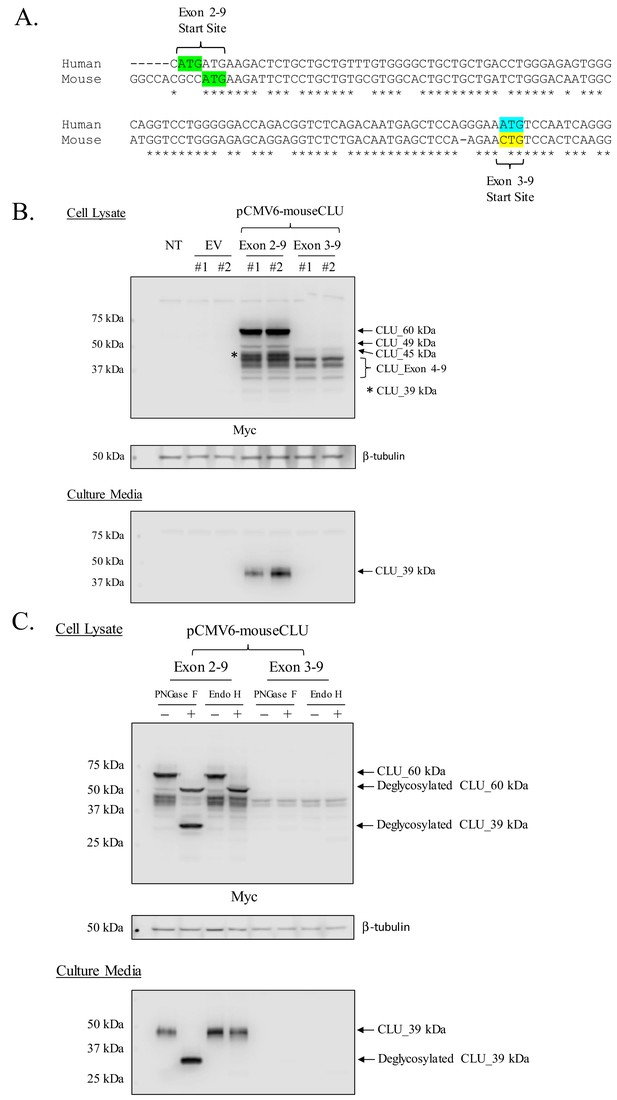
Mouse CLU_45 kDa is translated from a non-canonical CUG start site in Exon 3.
(A) Sequential alignment of human and mouse CLU: DNA of Exons 2 and 3. Green sequences represent the Exon 2 translational start site (ATG) in both mouse and human CLU. Blue and yellow sequences represent the Exon 3 translational start site for human CLU (ATG) and the proposed Exon 3 translational start site for mouse CLU_45 kDa (CTG), respectively. (B) Empty vector (pCMV6), Myc-tagged pCMV6-mouse CLU Exon 2–9, or Exon 3–9 constructs were transfected into Neuro-2a cells. Cell lysates and culture media were harvested and probed for CLU immunoreactivity using the anti-Myc antibody. (C) pCMV6-mouse CLU Exon 2–9 or Exon 3–9 constructs were transfected into Neuro-2a cells. Cell lysates and culture media were harvested and treated with endoglycosidases. The resulting samples were analyzed for CLU immunoreactivity using the anti-Myc antibody.
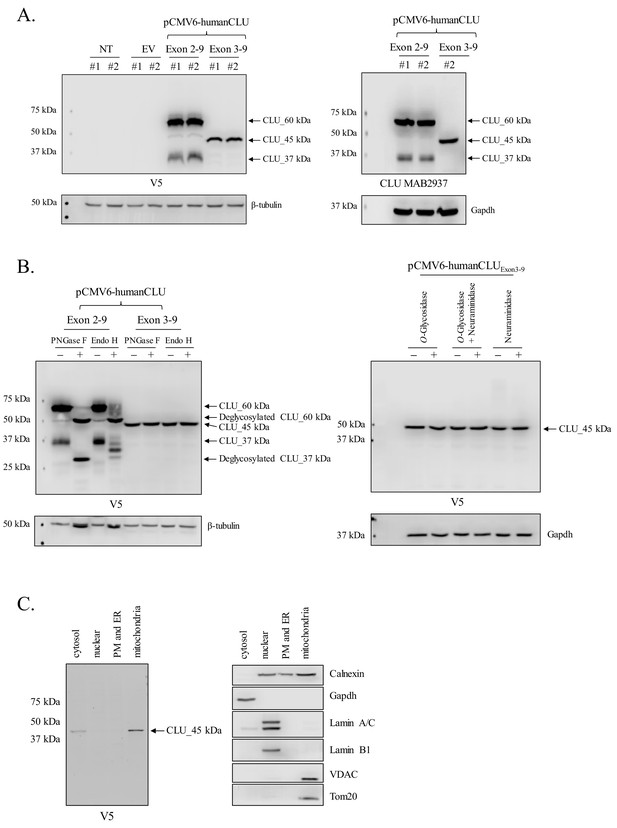
Human CLU_45 kDa is translated from an AUG start site in Exon 3.
(A) Empty vector (pCMV6), V5-tagged pCMV6-human CLU Exon 2–9, or Exon 3–9 constructs were transfected into human SH-SY5Y cells. Cell lysates were harvested and probed for CLU immunoreactivity using anti-V5 (left panel) or anti-Clusterin (right panel). (B: left panel) pCMV6-human CLU Exon 2–9 or Exon 3–9 constructs were transfected into human SH-SY5Y cells. The resulting cell lysates were treated with PNGase F or Endo H and probed for CLU immunoreactivity using anti-V5. (B: right panel) Cell lysates harvested from pCMV6-human CLU Exon 3–9-transfected SH-SY5Y cells were treated with O-glycosidase, neuraminidase, or a combination of the two and probed for expression of CLU_45 kDa protein using anti-V5. (C) pCMV6-human CLU Exon 3–9-transfected SH-SY5Y cells were subjected to cellular fractionation using the Qiagen Qproteome Mitochondrial Isolation Kit and analyzed for CLU immunoreactivity using anti-V5. Fraction isolation was confirmed using calnexin (ER), Gapdh (cytosol), lamin A/C and lamin B1 (nucleus), and VDAC and Tom20 (mitochondria).
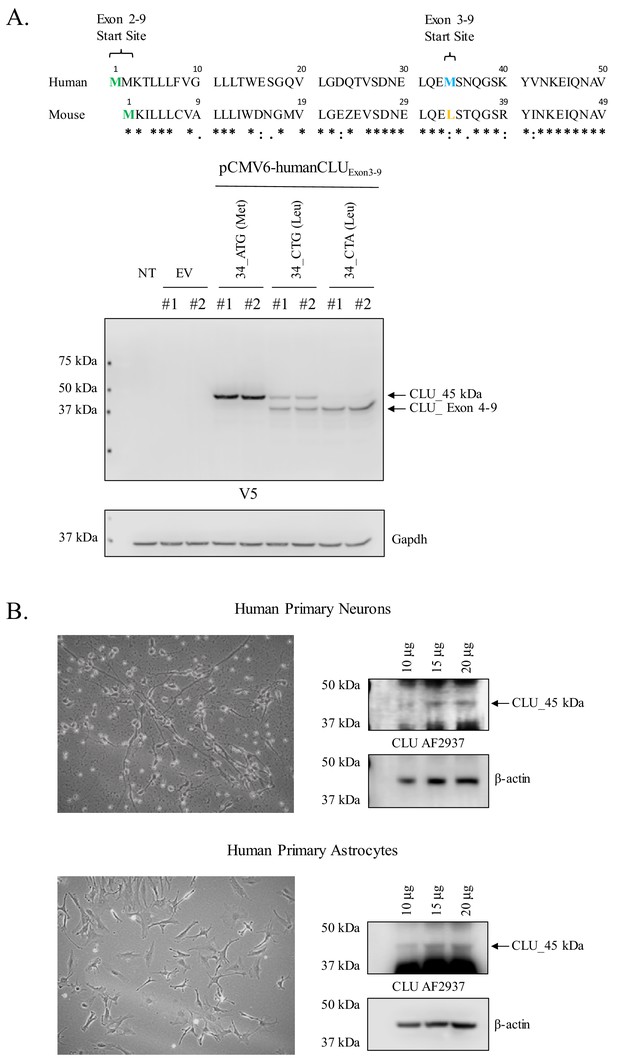
Human CLU_45 kDa is translated from an AUG in Exon 3 and is present in human primary neurons and astrocytes.
(A: upper panel) Alignment of human and mouse CLU: amino-acid sequences of Exons 2 and 3. Green represents the Exon 2 start site in both human and mouse CLU (Met), blue represents the Exon 3 start site for human CLU (Met), and yellow represents the Exon 3 start site for mouse CLU (Leu). (A: lower panel) SH-SY5Y cells were transfected with empty vector (pCMV6) or pCMV6-human CLU Exon 3–9 amino-acid 34 variants: 34_ATG (wild-type), 34_CTG (mimics mouse CLU), or 34_CTA (negative control). Cell lysates were probed for CLU immunoreactivity using the anti-V5 antibody. (B) Human primary cortical neurons and astrocytes were cultured and imaged to demonstrate cell type visually. Increasing concentrations of whole cell lysate were probed for CLU_45 kDa immunoreactivity using anti-CLU AF2937.
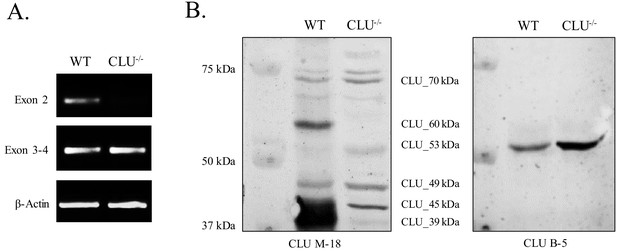
Mitochondrial CLU_45 kDa is present in the CLU–/–animal model.
(A) Total RNA was isolated from WT and CLU–/– cortical tissue. 1.5 µg RNA was reverse transcribed and 50 ng cDNA was analyzed for Exon 2- and Exon 3–4-containing mRNA. To confirm equal loading, β-actin mRNA was amplified using both WT and CLU-/- cDNA. Following amplification, amplicons were run on a 2% agarose gel. (B) 30 µg cortical tissue lysate isolated from WT and CLU–/– mice was analyzed by immunoblotting and probed with anti-CLU M-18 (left panel) or anti-CLU B-5 (right panel) overnight at 4°C. Blots were washed and probed with species-specific HRP-conjugate secondary antibodies.
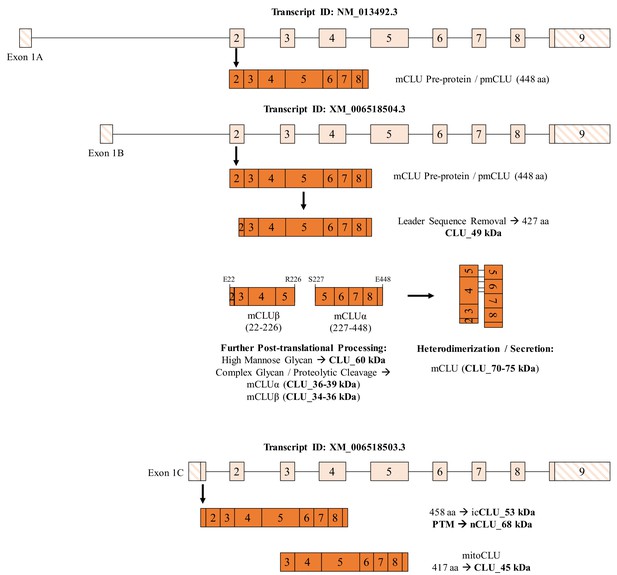
Summary of CLU mRNA transcripts and translated protein isoforms in the brain.
Three previously described and one newly hypothesized CLU mRNA transcript result in the production of five distinct protein isoforms, including one mature isoform with associated intermediates and three intracellular isoforms. The mCLU pre-protein is generated via translation of Exon-1B-containing mRNA (astrocytes and neurons; middle panel) and Exon-1A-containing mRNA (astrocytes only; upper panel). Once the mCLU pre-protein is translated, the 21-amino-acid ER-targeting sequence (majority of Exon 2) is cleaved and the resultant 427-amino-acid protein (CLU_49 kDa) is glycosylated, first by high mannose glycans (CLU_60 kDa) and then by complex glycans, followed by proteolytic cleavage to form mCLUβ (CLU_34–36 kDa) and mCLUα (CLU_36–39 kDa). Heterodimerization by five disulfide bonds results in the formation of mature CLU (mCLU; CLU_70–75 kDa), which can then be secreted into the extracellular space. In addition, on the basis of our data, we hypothesize that the Exon -C-containing mRNA transcript results in the production of the intracellular CLU protein isoform, icCLU_53 kDa, which is then post-translationally modified to form nCLU_68 kDa. The spliced transcript containing Exons 3–9 results in the production of mitoCLU_45 kDa, which is present in both human and rodent brain mitochondria and is translated from a non-canonical CUG start site in rodents.
Additional files
-
Supplementary file 1
Cell line authentication certificate.
- https://cdn.elifesciences.org/articles/48255/elife-48255-supp1-v1.pdf
-
Supplementary file 2
Mycoplasma testing of cell lines at different passages.
- https://cdn.elifesciences.org/articles/48255/elife-48255-supp2-v1.pdf
-
Transparent reporting form
- https://doi.org/10.7554/eLife.48255.014





