Long-term adult human brain slice cultures as a model system to study human CNS circuitry and disease
Figures

Electrophysiology of adult human pyramidal neurons.
(A) Example of biocytin filled pyramidal neuron (14 DIV) after streptavidin-Cy3 counterstaining, scale bar 150 µm. (B) Typical regular spiking pattern of human pyramidal neurons (acute slice, 3 DIV and 13 DIV) in response to +200 pA positive current injection. (C) Neurons recorded in acute slices (within 12 hr after surgery) and in brain slice cultures (2–3 DIV and 7–14 DIV) showed similar action potential firing frequencies and typical spike frequency adaptation upon 200 pA current injection. (D, E) Quantification of (from left to right) the AP half width, resting membrane potential, sag potential amplitude and input resistance revealed no significant correlation between these values and the days in vitro (DIV), (E) except membrane resting potential which slightly changed to more depolarized values within the first 2–3 days in culture and then stayed stable over the remaining time.
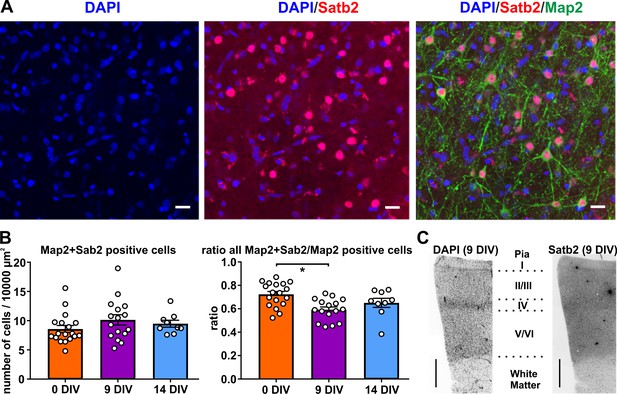
Satb2 positive neurons in human brain slice cultures.
(A) High-resolution image of a confocal z-stack collapsed into one optical plane processed for ICC of the glutamatergic neuronal marker Satb2 and neuron-specific cytoskeletal protein Map2 in layers 2/3, scale bar 20 µm. (B) Quantification of Map2/Satb2 double positive neurons normalized to 100 µm x 100 µm areas and the plot of the ratio of Map2+Satb2/Map2 positive cells. (C) The slices contained all six cortical layers, shown are representative examples with staining for DAPI and Satb2, scale bar 500 µm.
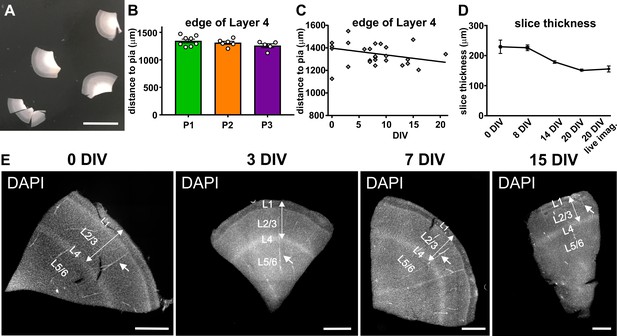
Stability of gross structural features of cortical slices between surgeries and over time in culture.
(A) On the day of the surgery slices are prepared from tissue blocks and carefully cut into even slices containing all six layers, scale bar 1 cm. (B) Comparison of slices originating from three independent surgeries of temporal cortex (P1-3) show a comparable distance from pia to the edge of layer 4, which was clearly visible upon DAPI staining. (C) This distance did not change significantly over time in culture up to the endpoint of analysis at 20 DIV. (D) Slice thickness slightly decreased throughout the analyzed three-week period in culture. (E) Examples of DAPI stainings of slices of the same surgery at different time points in culture (0 DIV - 15 DIV). Cortical thickness from pia to the edge of layer 4 (double arrow). Note blood vessels (arrows) running parallel to the slicing plane, indicating that sections indeed were prepared perpendicular to the cortical surface, scale bar 1000 µm.
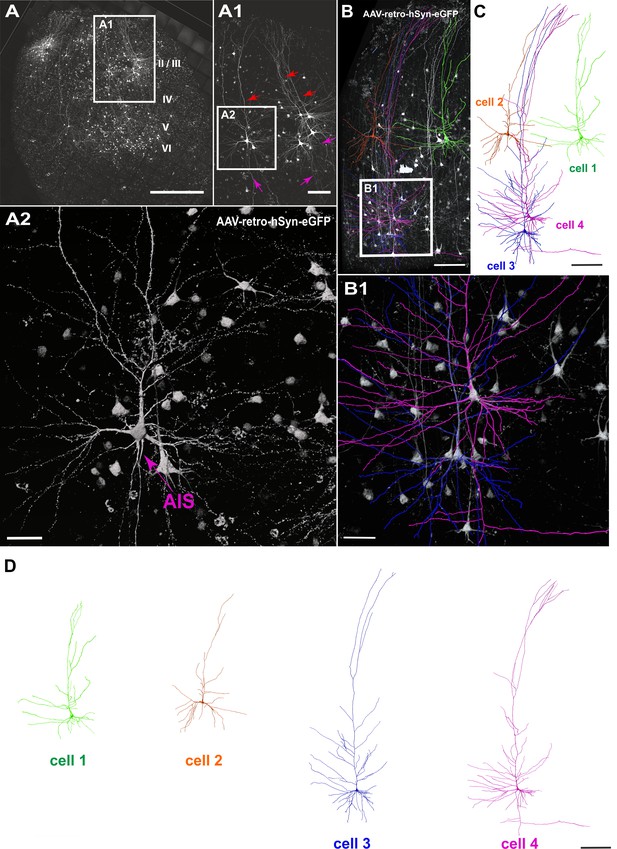
Viral transduction in human brain slice cultures and 3D reconstruction of GFP-labeled pyramidal neurons.
(A) Representative example of a human brain slice after viral transduction with AAVrg-hSyn-GFP at 9 DIV, scale bar 1000 µm. (A1) Enlarged confocal image from A: layers 2/3 pyramidal neurons with intact apical dendrites (red arrows) and axons (magenta arrows), scale bar 200 µm. (A2) The soma and axon initial segment (AIS) are clearly visible in the virally transduced neurons, scale bar 50 µm. (B) 3D reconstructions of four GFP transduced pyramidal neurons were performed from confocal z-stack tile scans, scale bar 200 µm. The cells were individually traced and pseudo colored (C). (B1) Example of two neurons within close proximity of each other, which could still be clearly separated for further analysis, scale bar 50 µm. (D) Separation of the four distinct GFP-labeled pyramidal cells for further analysis, scale bar 200 µm.

3D reconstruction of biocytin filled pyramidal neurons.
All 24 biocytin labeled reconstructed pyramidal neurons sorted by their soma distance to the pia ranging from 385 to 2400 µm represented in the colors red (apical dendrites), blue (basal dendrites) and magenta (axons).
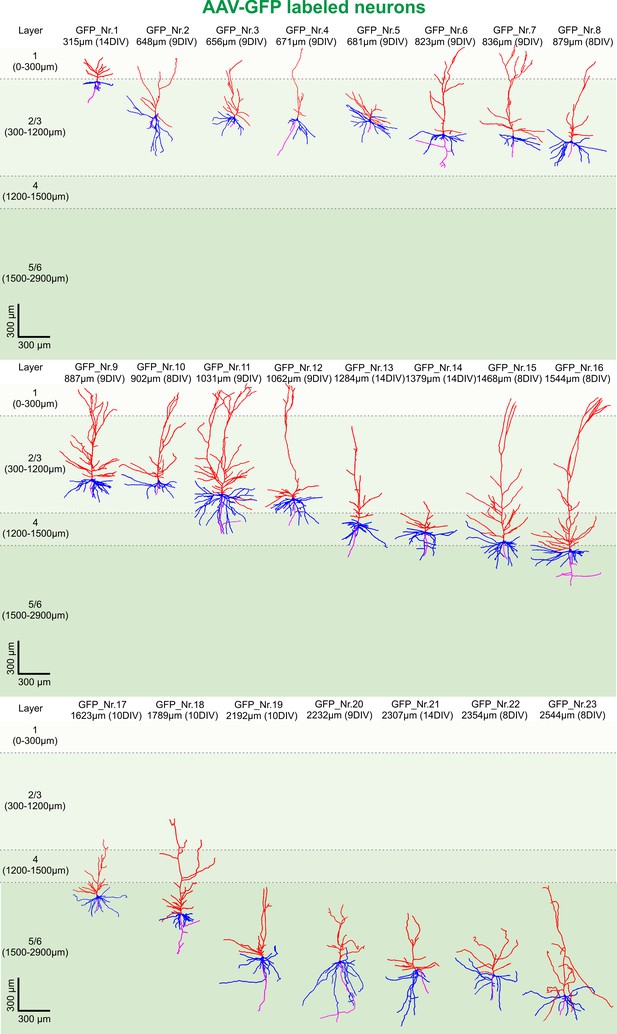
3D reconstruction of GFP-labeled pyramidal neurons.
All 23 GFP-labeled reconstructed pyramidal neurons sorted by their soma distance to the pia ranging from 315 to 2544 µm represented in the colors red (apical dendrites), blue (basal dendrites) and magenta (axons).

Quantification of the total length of apical and basal dendrites of biocytin filled and GFP-labeled neurons.
(A) Upper panels: Neurons that were patched and filled in layers 2/3 are represented by red circles, neurons in layer four by red squares and neurons in layers 5/6 by red upward triangles. Bottom panels: Each biocytin filled neuron is represented by a red circle. (B) GFP-labeled layers 2/3 neurons are represented by green circles, neurons in layer four by green squares and neurons in layers 5/6 by green upward triangles. (C) Left: Analysis of the distribution of biocytin filled (red) and GFP-labeled (green) pyramidal neurons in the different layers. (C) Middle: Total dendritic length of biocytin filled (red) and GFP-labeled (green) pyramidal neurons plotted against DIV. (C) Right: Reconstructions of pyramidal neurons based on GFP expression slightly underestimated the total dendritic length compared to classic biocytin fillings (n = 22 for biocytin, n = 23 for GFP-labeled, **p<0.01).
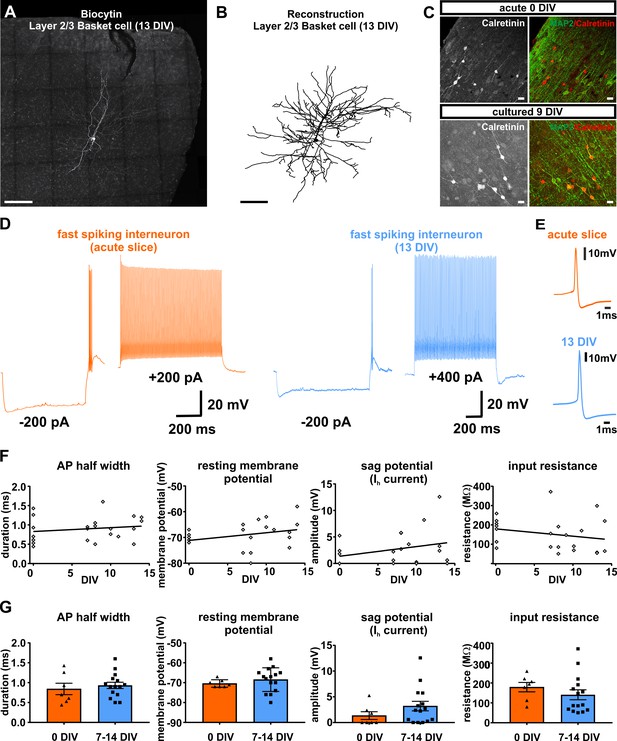
Presence and functionality of interneurons in human slice cultures.
(A) Example of Layers 2/3 basket cell labled with biocytin and (B) after reconstruction, both scale bars 200 µm. (C) Staining of calretinin revealed presence of a subpopulation of inhibitory interneurons in acute (0 DIV) and late in culture (9 DIV), scale bar 20 µm. (D) Example of fast spiking interneuron firing in acute slices (orange) and late (blue) in culture (13 DIV). (E) Examples of interneuron APs in acute slice (orange) and late in culture (13 DIV, blue) reveal comparable AP half width. (F) Quantification and plotting of basic properties of IN in relation to the DIV. (G) Group comparison between the properties in acute slice and late in culture measured interneurons revealed no significant differences (Mann-Whitney test, p>0.05).
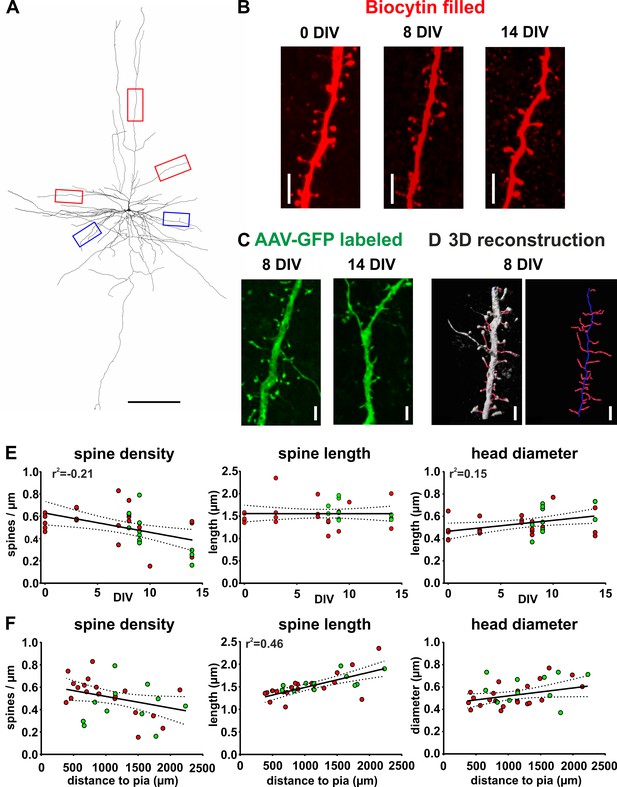
Spine measurements of human cortical pyramidal neurons in acute and cultured slices filled with biocytin and labeled with GFP.
(A) 3D reconstruction of a typical layers 2/3 pyramidal neuron (biocytin filled, 7 DIV) in human brain slice cultures, scale bar 300 µm. Of each neuron five dendritic regions were chosen (three of the apical dendritic compartment, red boxes, and two of the basal dendritic tree, blue boxes). (B) Typical examples of spines localized on apical dendrites of representative layers 2/3 pyramidal neurons recorded and biocytin filled at 0 DIV (acute slice), at 8 DIV and at 14 DIV, scale bar 5 µm. (C) Typical examples of spines of GFP-labeled neurons at 8 and 14 DIV, scale bar 5 µm. (D) For quantitative assessment, the z-stacks were 3D reconstructed and analyzed using NeuronStudio and Imaris software, scale bar 5 µm (see Materials and methods for details). (E, F) Neurons that were patched and filled with biocytin are represented by red circles and transduced GFP positive neurons by green circles. (E) Data reveal a moderate negative correlation of spine density with DIV and a moderate positive correlation of spine head diameter with DIV. (F) Plots of the same parameters versus the distance of the somata of the analyzed neurons to the pia revealed a strong positive correlation for spine length, but not the spine density or the spine head diameter.
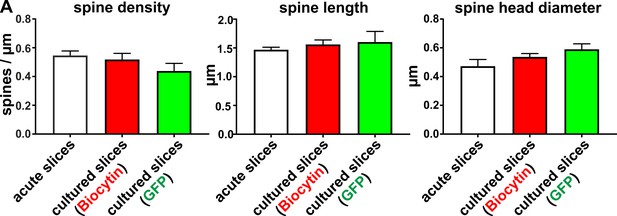
Analysis of mean values of spine density, length and head diameter of biocytin filled and virally transduced GFP positive pyramidal neurons.
Mean values for spine density, spine length and spine head diameter of pyramidal neurons were not significantly different in acute slices (0 DIV) compared to cultured slices (2–14 DIV).
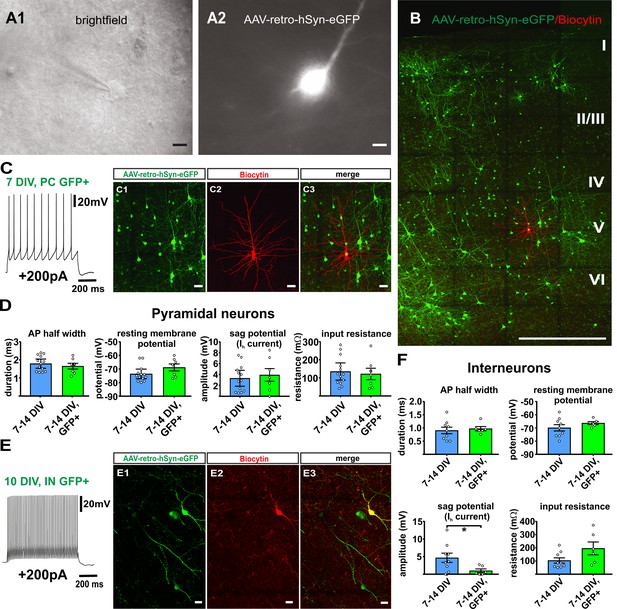
Virally transduced neurons show no major changes of basic properties.
(A) GFP positive neurons were visualized according to their GFP fluorescence (A2) and targeted for whole-cell patch clamp recordings (A1), scale bars 10µm. (B) Overview image of a double positive pyramidal neuron (GFP and Biocytin) after whole cell recording and post-hoc staining, scale bar 500 µm; (C1–C3) and in detail, scale bar 20 µm. (C) Example of a representative regular firing pattern of a GFP positive pyramidal neuron (+200 pA). (D) Quantification of basic properties of GFP negative and GFP positive human pyramidal neurons. (E) Example of a GFP positive fast spiking interneuron. (E1-E3) GFP signal, biocytin filling and merge of the same neuron, scale bar 20 µm. (F) Quantification of basic properties of GFP negative and GFP positive human interneurons, *p<0.05, Mann Whitney test.
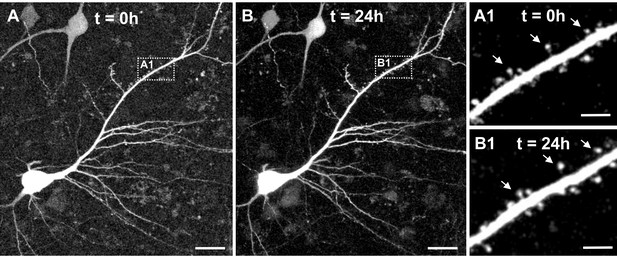
Two-photon live imaging of human neurons over 24 hr.
(A) Maximum projection of a GFP positive human pyramidal neuron in slice culture at the start of the measurement and (B) after 24 hr life cell imaging with acquisition of image stacks every 30 min. (A1) and (B1) inserts are zoomed in images of the same branch of the apical dendrite at 0 and 24 hr of imaging. Arrows point to the same spines. Scale bars 20 µm (A, B) and 5 µm (A1, B1).
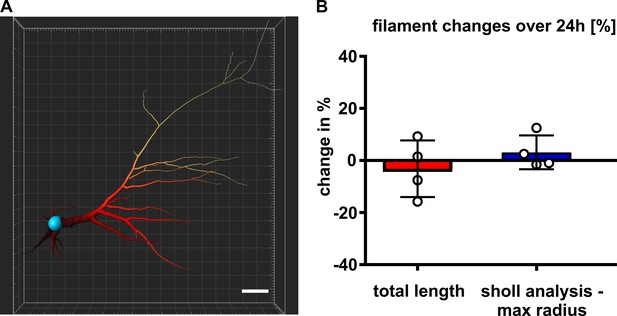
Filament measurements reveal stability over the time of 24 hr.
(A) Filament reconstruction for quantification, scale bar 20 µm. (B) Slight changes of the filament total length and maximal sholl radius at 24 hr in % of the measurement at 0 hr. Mean values ± standard deviation (SD).
Tables
Tissue samples included in this study.
https://doi.org/10.7554/eLife.48417.015| Age at surgery | Gender | Resected brain area | Number and type of cells included in this study |
|---|---|---|---|
| 30 | M | temporal lobe | 3 PC |
| 30 | M | temporal lobe | 2 PC |
| 10 | F | temporal lobe | 3 PC + 3 IN |
| 38 | F | temporal lobe | 2 IN |
| 16 | M | temporal lobe | 1 PC |
| 18 | M | frontal lobe | 6 PC |
| 40 | F | temporal lobe | 4 PC + 3 IN |
| 40 | F | frontal lobe | 1 IN |
| 29 | M | temporal lobe | 5 PC |
| 56 | F | temporal lobe | 3 PC |
| 31 | M | temporal lobe | 3 PC + 2 IN |
| 44 | M | temporal lobe | 5 PC + 3 IN |
| 46 | F | temporal lobe | 2 PC + 1 IN |
| 67 | M | temporal lobe | 2 PC + 1 IN |
| 57 | M | frontal lobe | 6 PC + 6 IN |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.48417.016





