The ER membrane protein complex is required to ensure correct topology and stable expression of flavivirus polyproteins
Figures
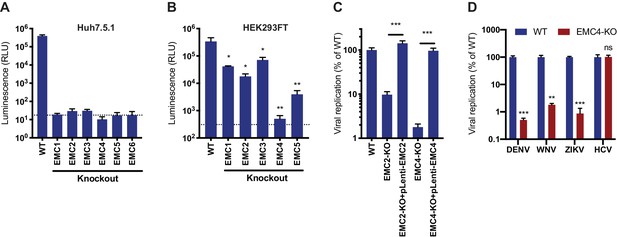
EMC is required for flavivirus infection.
(A) DENV infection of WT and EMC subunit KO Huh7.5.1 cells. Cells were infected with DENV-Luc, harvested at 30hpi and luminescence was measured. Dotted line indicates background from uninfected control cells. (B) DENV infection of WT and EMC subunit KO HEK293FT cells. Cells were infected with DENV-Luc, harvested at 48hpi and luminescence was measured. Dotted line indicates background from uninfected control cells. (C) Replication of DENV in WT, EMC2- and EMC4-KO, and cDNA complemented KO HEK293FT cells. Cells were infected with DENV-Luc, harvested at 48hpi and luminescence was measured. (D) Quantitative RT-PCR of DENV, WNV, ZIKV and HCV RNA in WT or EMC4-KO Huh7.5.1 cells. Cells were infected with an moi of 0.5 for all viruses and harvested at 30hpi for ZIKV, 48hpi for DENV and WNV, and 72hpi for HCV. Viral RNA levels were normalized to 18S levels and data is displayed relative to the respective WT condition. In all figures, values are shown as mean of three biological replicates with standard deviation in (A)-(C), and as mean with standard error of the mean in (D). t-tests were performed to determine statistical significance and p-values are defined as ns = non significant, *<0.05, **<0.01, ***<0.001.
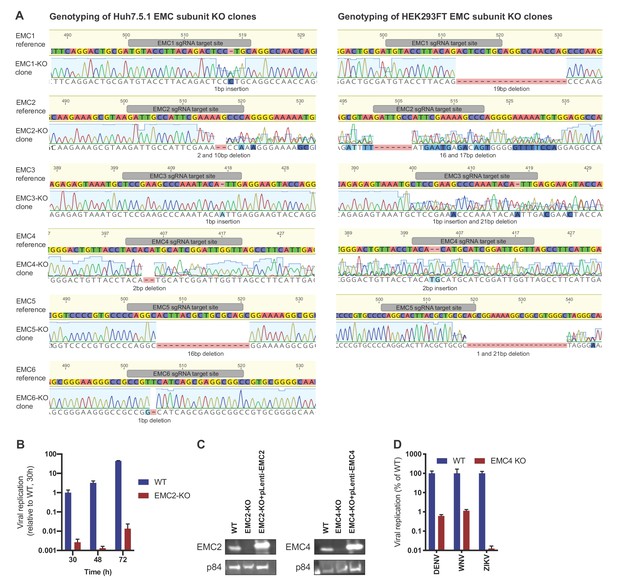
Genotyping and flavivirus infection of EMC KO cell lines.
(A) Sanger sequencing of clonal EMC subunit KO Huh7.5.1 and HEK293FT cells. sgRNA targeting site is highlighted in reference sequence (top) and indel mutation is shown below sequencing trace of KO cells. For heterozygous mutants, TIDE was used to determine indel mutations. (B) Timecourse of DENV infection in WT or EMC2-KO Huh7.5.1 at 30, 48 and 72 hr post infection. Cells were lysed and viral RNA was quantified by RT-PCR. Values are normalized to 18S RNA and shown relative to the WT, 30 hr condition. (C) Immunoblot analysis of EMC2 or EMC4 expression in WT, EMC2- and EMC4-KO, and cDNA complemented KO HEK293FT cells. p84 was used as loading control. (D) Quantitative RT-PCR of DENV, WNV and ZIKV RNA in WT or EMC4-KO HEK293FT cells. For QPCR experiments, values are shown as mean of three biological replicates with standard error of the mean.
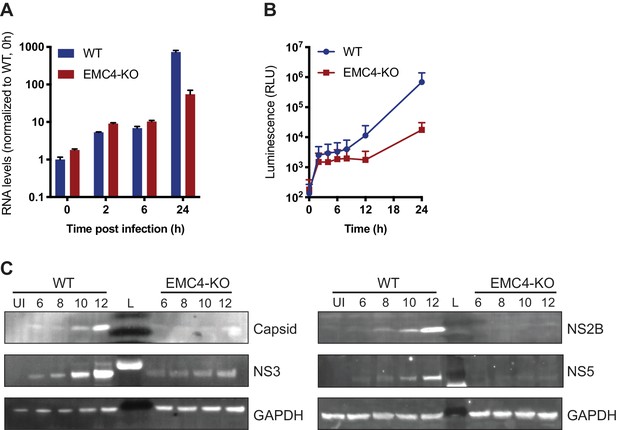
EMC is important for optimal viral protein expression.
(A) Quantitative RT-PCR of DENV RNA from cell-bound and internalized virions in WT and EMC4-KO Huh7.5.1 cells. Cells were infected on ice with an moi of 30, incubated for 1 hr, then moved to a 37C incubator, and finally lysed at indicated timepoints. The data are from three biological replicates and shown as mean with standard error of the mean. (B) Luminescence of WT and EMC4-KO HEK293FT cells, which were electroporated with DENV replicon expressing Renilla luciferase and collected at different times post-electroporation. The data are from three biological replicates for each timepoint and shown as mean with standard deviation. (C) Immunoblot analysis of DENV proteins from infected WT and EMC4-KO HEK293FT cells at different timepoints post-infection (6/8/10/12 hr). Lysates were blotted for different viral proteins (Capsid, NS2B, NS3 and NS5) and GAPDH was used as loading control. UI = uninfected control; L = ladder.

EMC directly affects viral protein expression and not genome replication.
(A) Luminescence of WT and EMC4-KO cells electroporated with DENV replicon expressing Renilla luciferase and treated with MK-0608 (a nucleoside analogue, which inhibits the DENV polymerase) or DMSO. Lysates were collected at different times post electroporation. The data are from three biological replicates and shown as mean with standard deviation. (B) Immunoblot analysis of DENV infected WT and EMC4-KO HEK293FT cells at different timepoints post-infection (6/8/10/12 hr) with treatment of MK-0608 to inhibit viral replication. Lysates were blotted for different viral proteins (Capsid, NS2B, NS3 and NS5) and GAPDH was used as loading control. UI = uninfected control; L = ladder.
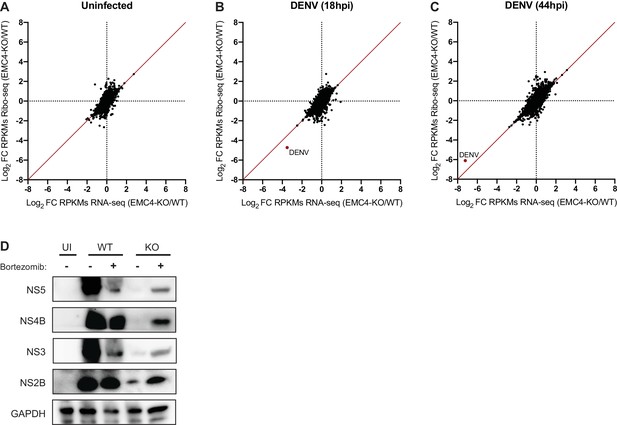
EMC knockout does not largely affect viral translation efficiency but leads to post-translational protein degradation.
(A) Ribosome Profiling (Ribo-seq) and RNA-seq were performed in uninfected WT and EMC4-KO HEK293FT cells to measure changes in translation efficiency (TE). Cycloheximide was added to stop translation and total RNA was isolated. RNA was treated with RNaseI and ribosome-protected footprints were purified, reverse transcribed and amplified for next-generation sequencing. Log2 fold-change (FC) of reads per kb of transcript, per million mapped reads (RPKMs) between EMC4-KO and WT cells for Ribosome Profiling and RNA-seq are displayed on the y- and x-axis, respectively. Each dot represents one RNA transcript. Fold-change in TE for a given transcript is defined as the change in Ribosome Profiling RPKMs normalized to the change in RNA-seq RPKMs. The red line represents fold-change of TE = 1. The Ribosome profiling experiments were performed once for each condition. (B) Ribosome Profiling (Ribo-seq) and RNA-seq were performed in DENV-infected WT and EMC4-KO HEK293FT cells 18 hr post-infection to measure changes in translation efficiency (TE). The DENV transcript is highlighted in red. The red line represents fold-change of TE = 1. (C) Ribosome Profiling (Ribo-seq) and RNA-seq were performed in DENV-infected WT and EMC4-KO HEK293FT cells 44 hr post-infection to measure changes in translation efficiency (TE). The DENV transcript is highlighted in red. The red line represents fold-change of TE = 1. (D) Viral protein expression from DENV infected WT and EMC4-KO HEK293FT cells with or without addition of bortezomib (50 nM), a proteasome inhibitor. Cells were harvested at 18hpi and lysates were analyzed by immunoblot using antibodies against dengue proteins. GAPDH was used as loading control. UI = uninfected. .
-
Figure 3—source data 1
Ribosome Profiling data including read counts and fold changes for ribosome footprints and RNA-seq used to generate Figure 3A–C.
- https://doi.org/10.7554/eLife.48469.009
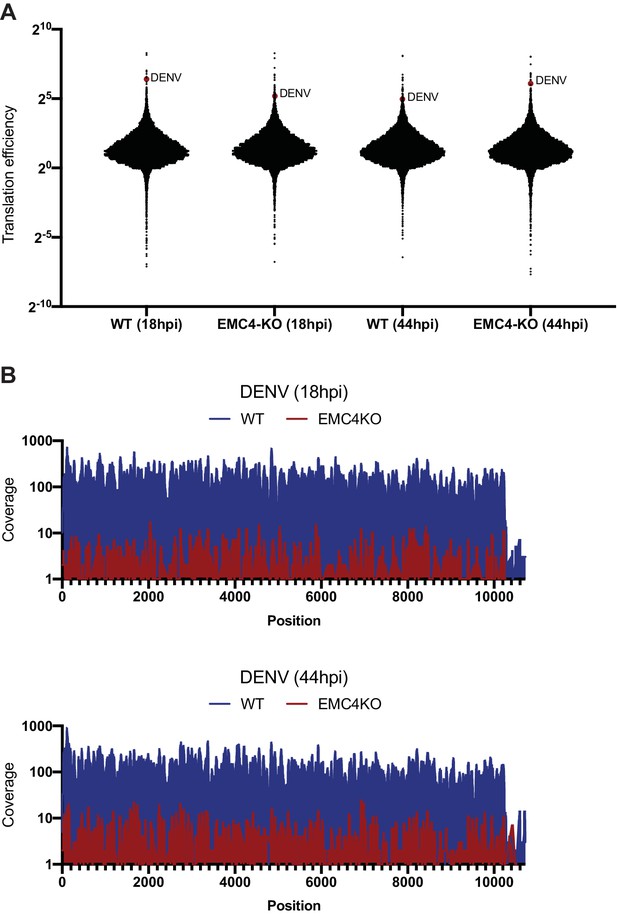
DENV efficiently translates and produces full-length polyproteins in EMC KO cells.
(A) Translation efficiencies (number of ribosomal footprints normalized to the number of RNAseq reads) of cellular mRNA and DENV RNA in WT and EMC4-KO cells at 18 and 44hpi. DENV is highlighted as red dot. (B) Footprint distribution of Ribosome Profiling reads mapped to the DENV genome in WT and EMC4-KO cells at 18 and 44hpi. Nucleotide position of DENV genome is indicated on x-axis. .
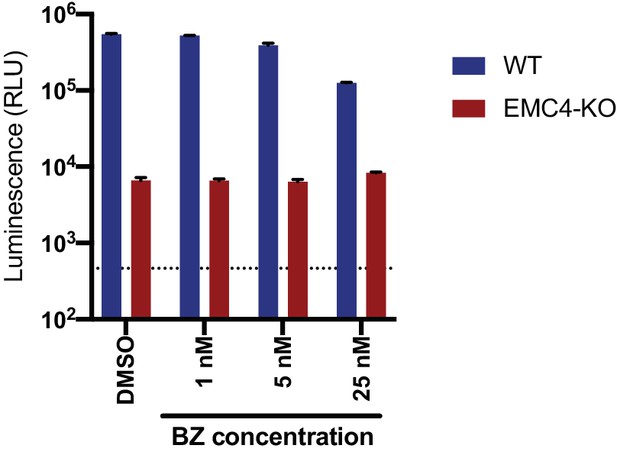
Bortezomib-recovered viral proteins do not restore viral replication.
DENV-Luc infection of WT and EMC4 KO HEK293FT cells treated with DMSO control or different concentrations of BZ. Cells were harvested at 24hpi and luminescence was measured. Dotted line indicates background signal from uninfected cells. .
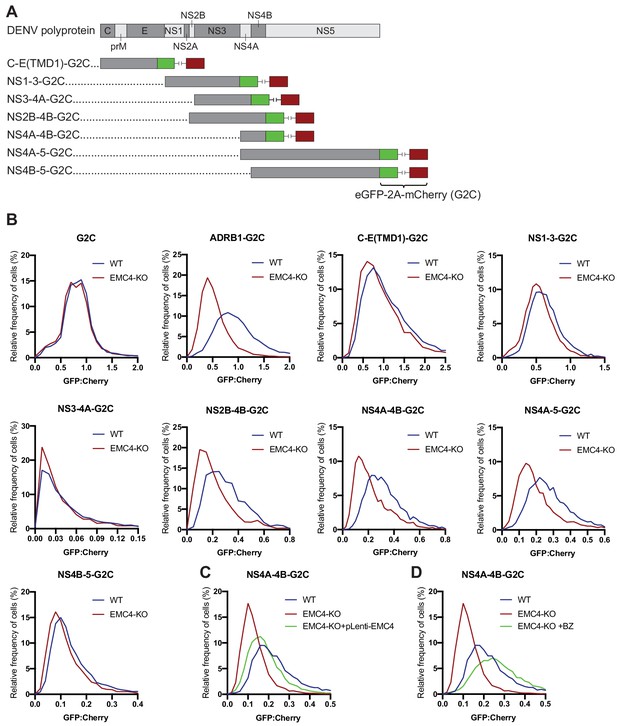
EMC is required for proper biogenesis of the NS4A-4B region of the DENV polyprotein.
(A) Overview of dual-fluorescence reporter constructs of DENV polyprotein for analysis of protein stability. Segments of the DENV genome encoding parts of the polyprotein were cloned with a C-terminal fusion to eGFP-T2A-mCherry (G2C). The T2A peptide leads to peptide bond skipping during translation resulting in two protein products, DENV protein fused to eGFP and mCherry. The GFP:mCherry fluorescence ratio reflects changes in DENV protein stability. (B) GFP:mCherry fluorescence ratio for G2C constructs in WT and EMC4-KO Huh7.5.1 cells. Cells were transfected with G2C construct and analyzed by flow cytometry after 24–48 hr. For each individual cell the GFP:mCherry ratio was calculated and the fluorescence ratios are depicted as histograms. ADRB1=β1-adrenergic receptor. (C) Measurement of GFP:mCherry fluorescence ratios of NS4A-4B-G2C construct in WT, EMC4-KO and EMC4 cDNA complemented EMC4-KO HEK293FT cells. (D) Measurement of GFP:mCherry fluorescence ratios of NS4A-4B-G2C construct in WT, EMC4-KO and bortezomib-treated EMC4-KO HEK293FT cells. Note that (C) and (D) use same WT and EMC4-KO data as experiments were performed in parallel.
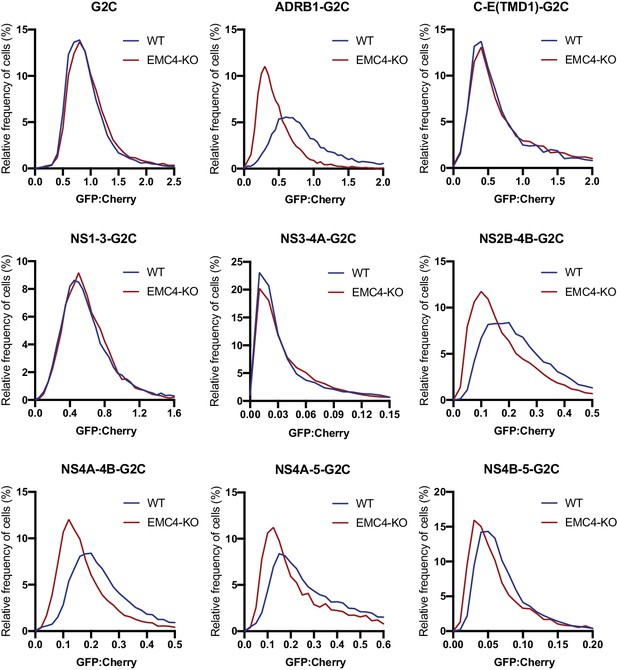
EMC is required for proper biogenesis of the NS4A-4B region of the DENV polyprotein in HEK293 cells.
Cells were transfected with G2C construct and analyzed by flow cytometry after 24–48 hr. For each individual cell the GFP:mCherry ratio was calculated and the fluorescence ratios are depicted as histograms. ADRB1=β1-adrenergic receptor.
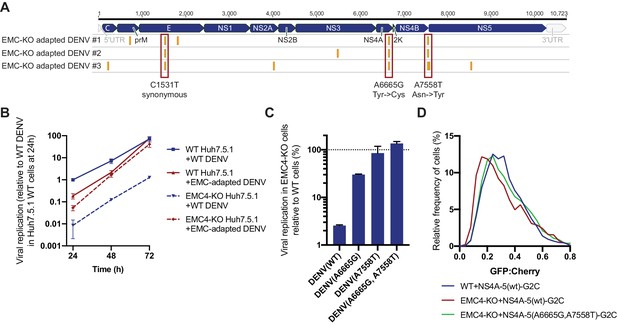
Adaptation of DENV to EMC KO cells reveals non-synonymous point mutations in NS4A and NS4B.
(A) Identification of SNPs in EMC4-KO adapted DENV relative to DENV 16681 reference genome by next-generation sequencing of supernatant-extracted viral RNA. DENV was serially passaged on EMC4 KO Huh7.5.1 cells every 3–4 days. After 20–22 passages supernatant was collected and RNA extracted from three independent passaging experiment. Truseq total RNA sequencing libraries were generated and reads were mapped to the reference genome. Mutations common across all three replicates are highlighted by red boxes and their nucleotide and amino acid changes are shown below. A table of all identified SNPs can be found in Figure 5—source data 1). (B) Viral growth curves of WT and EMC-adapted DENV in WT and EMC4 KO cells measured by quantitative RT-PCR. Cells were infected with WT or EMC-adapted virus at an moi of 0.5 and harvested at indicated timepoints. The data are from three biological replicates and shown as mean with standard error of the mean. (C) Viral replication of WT DENV-Luc or mutant DENV-Luc (containing A6665G and A7558T either individually or in tandem) in EMC4 KO cells relative to WT cells. Lysates were collected 72hpi and luminescence was measured. The data are from three biological replicates and shown as mean with standard deviation. (D) Measurement of GFP:mCherry fluorescence ratios of WT NS4A-5-G2C and NS4A-5(A6665G,A7558T)-G2C constructs in WT and EMC4-KO Huh7.5.1.
-
Figure 5—source data 1
Identified SNPs in EMC KO adapted DENV isolates relative to the DENV 16681 reference genome.
A minimum variant frequency cutoff of 80% was used.
- https://doi.org/10.7554/eLife.48469.013
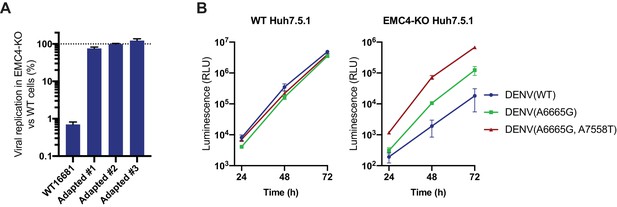
DENV containing EMC KO adapted mutations efficiently replicates in EMC deficient cells.
(A) Quantitative RT-PCR of DENV RNA from WT DENV 16681 and the three isolated EMC-adapted DENV in WT and EMC4 KO Huh7.5.1. Cells were infected with an moi of 0.5 and harvested 48hpi. Viral RNA was normalized to 18S RNA and values are displayed relative to the respective WT cell condition. The data are from three biological replicates and shown as mean with standard error of the mean. (B) Timecourse infection experiment with WT, A6665G and A665G/A7558T DENV-Luc in WT or EMC4 KO Huh7.5.1. Cells were infected, lysates harvested at 24, 48 and 72 hpi and luminescence was measured. The data are from three biological replicates and shown as mean with standard deviation.
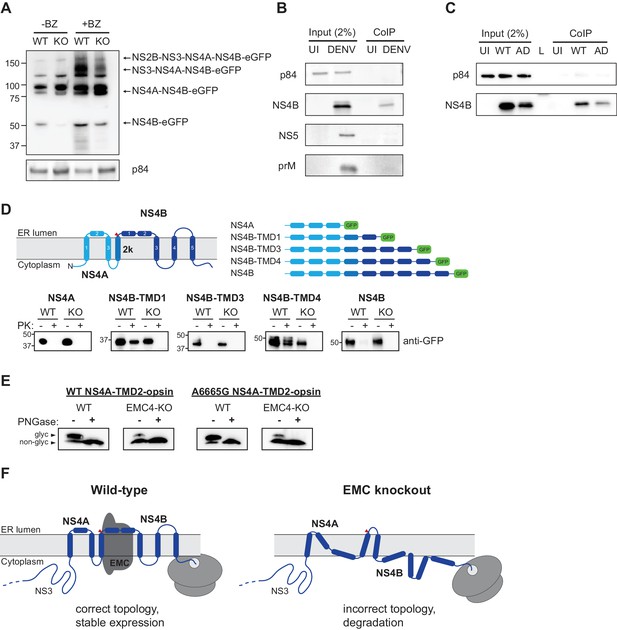
EMC physically interacts with DENV NS4B post-cleavage and knockout of EMC leads to an aberrant topology of NS4A and NS4B.
(A) Immunoblot for processed products of transfected NS2B-4B-GFP construct in WT and EMC4 KO cells in presence or absence of BZ. Anti-GFP antibody was used to detect processed viral proteins and p84 was used as loading control. (B) Co-immunoprecipitation of EMC4-FLAG from lysates of uninfected (UI) or DENV infected HEK293FT EMC4-FLAG cells. Cells were harvested after 96 hr and lysates were incubated with anti-FLAG magnetic beads. Eluates were immunoblotted for different viral proteins (prM, NS4B, NS5). p84 was used as loading and non-specific binding control. (C) Co-immunoprecipitation of EMC4-FLAG from lysates of uninfected cells (UI), cells infected with WT DENV (WT) or infected with EMC adapted DENV (AD). (D) Protease protection assay for different NS4A-4B-eGFP fusion constructs. Experimentally determined topology according to Miller et al. (2006); Miller et al. (2007) and generated constructs are displayed. Different shades of blue indicate NS4A, 2K and NS4B in the model and red triangle points to signal peptidase cleavage site. Constructs were transfected into WT or EMC4-KO cells. After 24–48 h cell pellets were resuspended in buffer with digitonin, and subsequently treated with proteinase K (PK) or left untreated. Finally, samples were lysed and immunoblotted using anti-GFP antibody. (E) Deglycosylation assay using PNGase F to probe luminal localization of NS4A TMD2 using constructs containing the NS4A WT sequence or A6665G mutation. Opsin tags containing a N-K-T sequon were inserted downstream of NS4A TMD2. WT or EMC4-KO cells were transfected with WT or mutant construct and harvested after 24 hr. Lysates were treated with PNGase F to remove N-linked oligosaccharides or left untreated following analysis by immunoblot using anti-FLAG antibody. Upper bands represent NS4A with glycosylated and thus luminally localized opsin tag while lower bands are non-glycosylated forms. (F) Possible model for the role of EMC in flavivirus infection. In infected WT cells, the EMC co-translationally engages with NS4A and NS4B TMDs to facilitate or maintain correct insertion and topology of TMDs , thus ensuring stable expression. In the absence of the EMC, an aberrant topology is obtained during biogenesis, leading to misfolding and degradation of the NS4A-4B viral proteins.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (human) | HEK293FT | ThermoFisher | R70007 | |
| Cell line (human) | EMC1-KO HEK293FT | This study | See Material and Methods, ‘Generation of knockout cell lines’ | |
| Cell line (human) | EMC2-KO HEK293FT | This study | See Material and Methods, ‘Generation of knockout cell lines’ | |
| Cell line (human) | EMC3-KO HEK293FT | This study | See Material and Methods, ‘Generation of knockout cell lines’ | |
| Cell line (human) | EMC4-KO HEK293FT | This study | See Material and Methods, ‘Generation of knockout cell lines’ | |
| Cell line (human) | EMC5-KO HEK293FT | This study | See Material and Methods, ‘Generation of knockout cell lines’ | |
| Cell line (human) | Huh7.5.1 | PMID: 15939869 | RRID: CVCL_E049 | |
| Cell line (human) | EMC1-KO HUH7.5.1 | This study | See Material and Methods, ‘Generation of knockout cell lines’ | |
| Cell line (human) | EMC2-KO HUH7.5.1 | This study | See Material and Methods, ‘Generation of knockout cell lines’ | |
| Cell line (human) | EMC3-KO HUH7.5.1 | This study | See Material and Methods, ‘Generation of knockout cell lines’ | |
| Cell line (human) | EMC4-KO HUH7.5.1 | This study | See Material and Methods, ‘Generation of knockout cell lines’ | |
| Cell line (human) | EMC5-KO HUH7.5.1 | This study | See Material and Methods, ‘Generation of knockout cell lines’ | |
| Cell line (human) | EMC6-KO HUH7.5.1 | This study | See Material and Methods, ‘Generation of knockout cell lines’ | |
| Biological sample (virus) | DENV serotype 2 strain16681 | PMID: 9143286 | NCBI Reference Sequence: NC_001474.2 | Generated from infectious clone obtained from Karla Kirkegaard |
| Biological sample (virus) | DENV-Luc | PMID: 27383987 | ||
| Biological sample (virus) | WNV Kunjin strain CH16532 | Other | GenBank: JX276662.1 | Source: John F. Anderson |
| Biological sample (virus) | ZIKV PRVABC59 | BEI Resources | NR-50240 | |
| Biological sample (virus) | HCV JFH1 | PMID: 11424123 | GenBank: AB047639.1 | |
| Antibody | P84 (mouse, monoclonal) | Genetex | GTX70220 | 1:2000 |
| Antibody | GAPDH (mouse, monoclonal) | SCBT | sc-32233 | 1:2000 |
| Antibody | EMC2/TTC35 (rabbit, polyclonal) | Proteintech | 25443–1-AP | 1:750 |
| Antibody | EMC4 (rabbit, polyclonal) | Bethyl | A305-752A | 1:500 |
| Antibody | Dengue virus prM protein (rabbit, polyclonal) | Genetex | GTX128093 | 1:1000 |
| Antibody | Dengue virus capsid protein (rabbit, polyclonal) | Genetex | GTX103343 | 1:1000 |
| Antibody | Dengue virus NS2B (rabbit, polyclonal) | Genetex | GTX124246 | 1:1000 |
| Antibody | Dengue virus NS3 protein (mouse, monoclonal) | Genetex | GTX629477 | 1:1000 |
| Antibody | Dengue virus NS4B protein (rabbit, polyclonal) | Genetex | GTX124250 | 1:1000 |
| Antibody | Dengue virus Type 2 NS5 protein (mouse, monoclonal) | Genetex | GTX629447 | 1:1000 |
| Antibody | GFP (rabbit, polyclonal) | Genetex | GTX113617 | 1:5000 |
| Antibody | FLAG (mouse, monoclonal) | Sigma | F1804 | 1:2000 |
| Recombinant DNA reagent | EMC2 cDNA | GenScript | clone OHu30604 | |
| Recombinant DNA reagent | EMC4 cDNA | GenScript | clone OHu00964 | |
| Commercial assay or kit | Renilla Luciferase Assay system | Promega | E2810 | |
| Commercial assay or kit | Power SYBR Cells-to-CT kit | ThermoFisher | 4402953 | |
| Chemical compound, drug | MK-0608 (2'-C-Methylcytidine) | Carbosynth | NM07918 | working concentration: 25 μM |
| Chemical compound, drug | Bortezomib | Selleckchem | S1013 | working concentration: 1–50 nM |
| Chemical compound, drug | Cycloheximide | Sigma | C7698 | working concentration: 100 μg/ml |
| Software, algorithm | Prism 8 | GraphPad | ||
| Software, algorithm | FlowJo | FlowJo |
Additional files
-
Supplementary file 1
Table of oligonucleotides used in this study: sgRNA oligos, genotyping primers, qPCR primers, primers to generate G2C constructs.
- https://doi.org/10.7554/eLife.48469.016
-
Transparent reporting form
- https://doi.org/10.7554/eLife.48469.017





