Asexual reproduction reduces transposable element load in experimental yeast populations
Figures
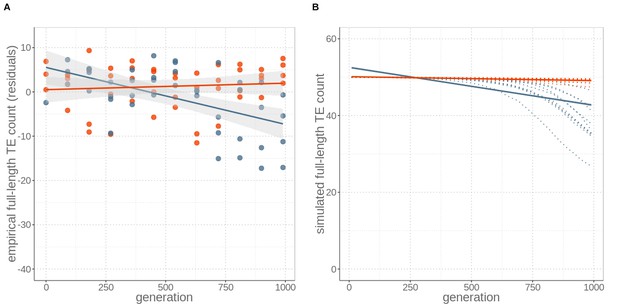
Sex maintains constant TE loads through time, while its absence leads to TE copy number reductions, for both (A) empirical data and (B) simulations including an allele modifying TE activity rates.
(A) Number of full-length TE copies inserted in genomes of four replicates of otherwise identical occasionally sexual (red) and wholly asexual (blue) yeast strains over 1000 generations of experimental evolution. Numbers are expressed as residuals, since the TE detection probability depends on sequencing coverage (Figure 1—figure supplement 2). (B) Individual-based simulations for studying the TE load dynamics expected under sexual and asexual reproduction with ten replicates (red and blue dotted lines). The simulations are parameterised with yeast-specific values and include a modifier alleles. For both (A) empirical and (B) simulation data, asexuals lost about nine active, full-length TEs by generation 1000. Lines represent linear regression for sexuals (red) and asexuals (blue) and the grey areas represent 95% CI.
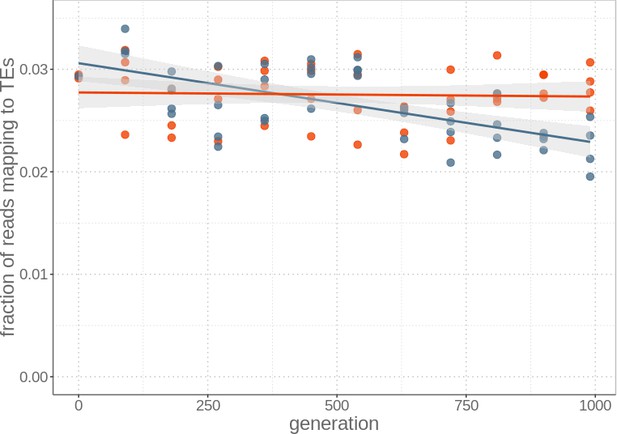
Overall transposable element load remains stable in sexual strains, but is reduced in asexual strains after 1000 generations.
Read fraction mapping to TEs relative to the sum of reads mapping to the genome and/or the TE library for each of the four replicate sexual (red) and asexual (blue) strains sequenced every 90 generations (from generation 0 to 990). Lines represent linear regression for sexuals (red) and asexuals (blue) and the grey areas represent 95% CI..
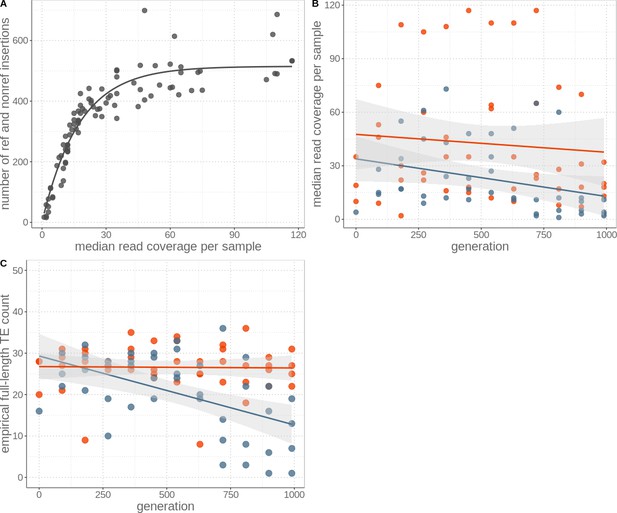
Identification of TE insertions depends on the sequencing coverage.
(A) TE insertions (including those present in the reference genome and de novo insertions) vs. median sequencing coverage from paired reads. Coverage influences the ability to detect TE insertions (Wilcoxon signed-rank test V = 4095, p-value<0.001). (B) Median read coverage per sample for sexual (red) and asexual (blue) strains over 1000 generations. Data from asexual strains had lower coverage, but were not different to sexuals through time (generation effect p=0.096, reproductive mode effect p=0.002, and interaction between generation and mode p=0.588; permutation ANOVA). Lines represent linear regression and the grey areas represent 95% CI. (C) Subsampling to the mean asexual read coverage per generation for all samples results in similar findings (generation effect p=0.012, reproductive mode effect p=0.302, and interaction between generation and mode p=0.004; permutation ANOVA).
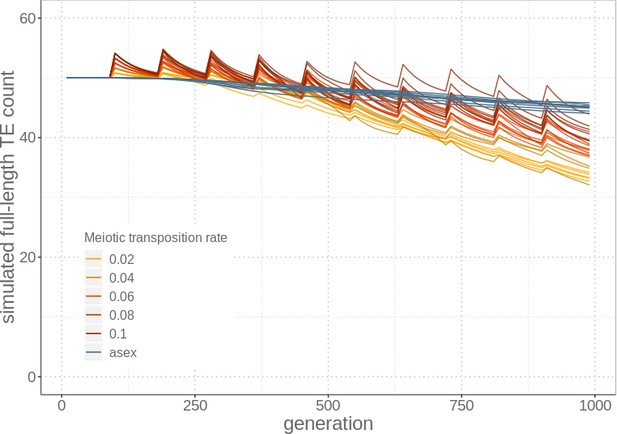
Simulations with higher transposition rates during meiosis than mitosis.
Meiosis generates the TE load spikes following events of sexual reproduction, but allows for selection to effectively remove genotypes with high TE loads by generating fitness variation among genotypes. Parameters used in the simulations are indicated in Supplementary file 2A (bold values).
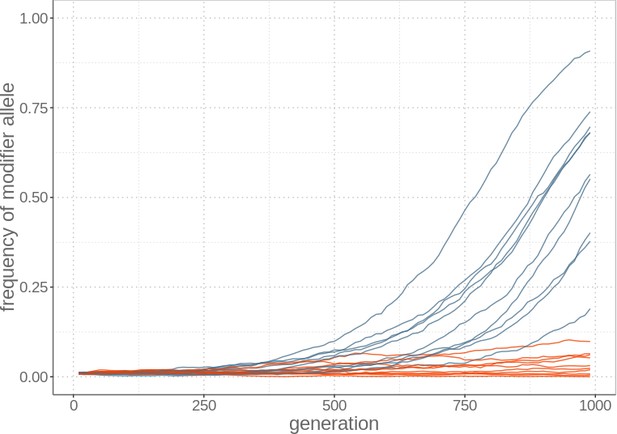
In the simulations, the spread of a modifier of excision rates is faster in asexual than sexual populations because it remains linked to genomes that have few TE copies and therefore a high relative fitness.
The modifier allele frequency is shown over time for simulations under sexual (red) and asexual (blue) reproduction, with ten replicates.
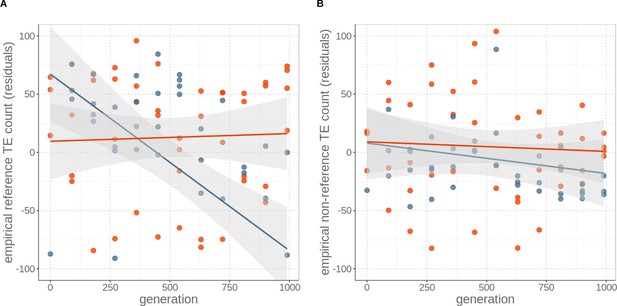
Decrease of insertions in asexuals over time is largely due to loss of ‘ancestral’ reference insertions (A) rather than novel insertions (B).
Count of all TE insertions, irrespective whether full-length TE, solo LTR, truncated elements or other types in genomes of four replicates of sexual (red) and asexual (blue) yeast strains over 1000 generations of experimental evolution. Numbers are expressed as residuals, since TE detection probability depends on sequencing coverage. Lines represent linear regression for sexuals (red) and asexuals (blue) and the grey areas represent 95% CI.
Additional files
-
Supplementary file 1
S. cerevisiae TY elements and the sizes (in bp) of internal regions and LTRs and the size boundaries used for filtering.
- https://doi.org/10.7554/eLife.48548.009
-
Supplementary file 2
(A) Explored parameter space of the simulations as pertinent for yeast (empirically determined values in bold). Selection_a and selection_b are selection coefficients for linear fitness effects and epistasis, respectively. Lost_TEs refers to the total number of TE lost after 1000 generations (averaged over ten replicates). (B) Explored parameter space for simulations including a modifier allele. Highlighted is the simulation closest to empirical observations. Init_f is the frequency of the modifier at the start of the simulations. Selection_a and selection_b are selection coefficients for linear fitness effects and epistasis, respectively. Lost_TEs refers to the total number of TE lost after 1000 generations (averaged over ten replicates). The bold lines refer to parameter combinations that generate results close to the observed empirical values.
- https://doi.org/10.7554/eLife.48548.010
-
Transparent reporting form
- https://doi.org/10.7554/eLife.48548.011





