A toolbox of nanobodies developed and validated for use as intrabodies and nanoscale immunolabels in mammalian brain neurons
Figures
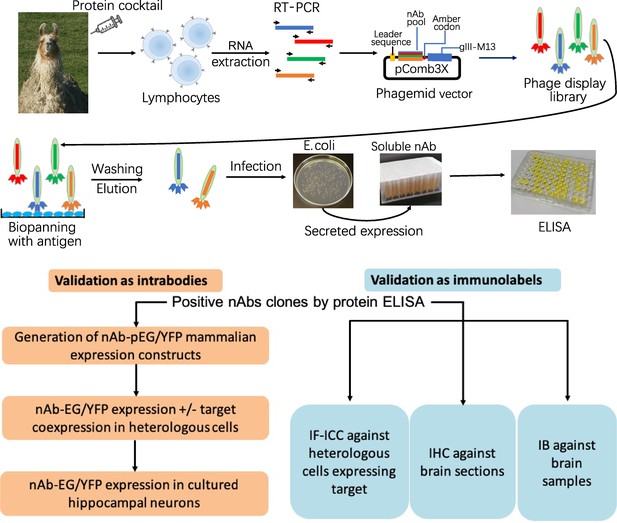
Schematic of nAb generation and validation pipeline.
(A) Schematic of nAb generation pipeline. See text for details. (B) Schematic of pipeline for validating ELISA-positive nAbs for utility as intrabodies and immunolabels.
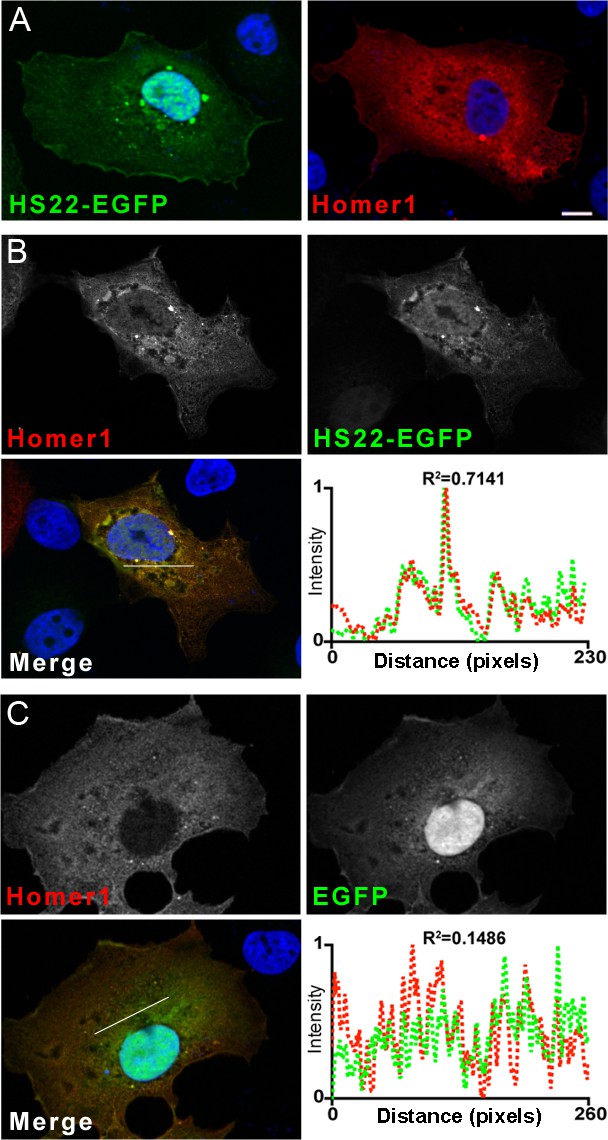
Anti-Homer1 nAbs that function as intrabodies colocalize with exogenously expressed Homer1 when coexpressed in heterologous COS-1 cells.
Representative images of the intrabody positive anti-Homer1 nAb HS22 validated in the heterologous COS-1 cell intrabody assay. (A) Images of HS22-EGFP (green) and Homer1 (red) expressed alone in COS-1 cells. The scale bar on the top left panel is 10 μm and holds for all panels in the figure. (B) Images of HS22-EGFP (green) and Homer1 (red) coexpressed in COS-1 cells and demonstrating the loss of HS22-EGFP nuclear localization and its cytoplasmic colocalization with coexpressed Homer1. The graph shows the normalized fluorescence intensity values across the line scan depicted by the white line in the merged image, with the corresponding R2 value. (C) Images of EGFP (green) and Homer1 (red) coexpressed in COS-1 cells and demonstrating the lack of ran effect of Homer1 coexpression on the nuclear localization of EGFP. The graph shows the normalized fluorescence intensity values across the line scan depicted by the white line in the merged image, with the corresponding R2 value. Note the concordance of the red and green intensity values for HS22-EGFP and Homer1 but not EGFP and Homer1.
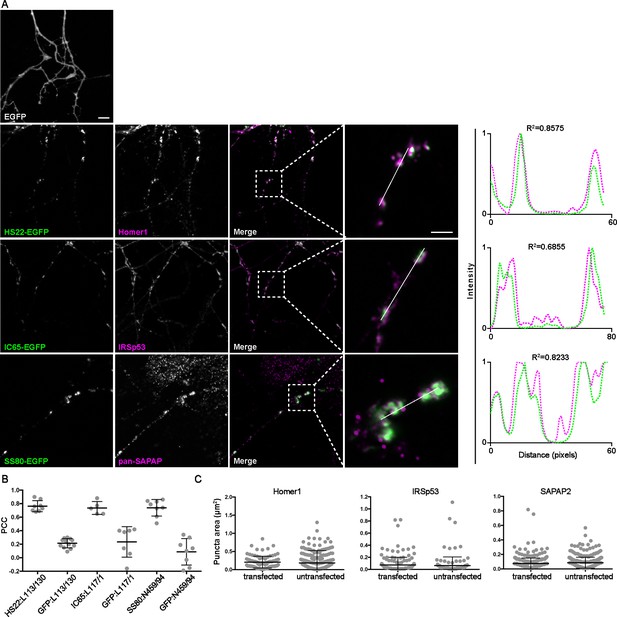
nAbs that function as intrabodies against excitatory synaptic target proteins colocalize with the endogenously expressed target proteins in cultured hippocampal neurons.
Representative images of intrabody-positive nAbs bound to endogenous excitatory synapse targets after expression in cultured hippocampal neurons. (A) The top left panel shops the pattern of EGFP expression in the dendrites of a transfected neuron. The subsequent rows show neurons expressing nAb-EGFP fusions and showing EGFP fluorescence (green), target-specific mAb labeling (magenta), and the merged image, with an adjacent panel showing a magnified image of the inset marked by the dashed box. Panels to the far right of each row are the normalized fluorescence intensity values across the individual line scans from the magnified inset, with the corresponding R2 values. Note the concordance of the magenta and green intensity values for the target and the positive nAb-EGFP fusions, but not for EGFP itself. The scale bar in the top left panel is 5 μm and holds for all panels in figure except the magnified insets, for which the scale bar is 2 μm. (B) Graph of Pearson’s Correlation Coefficient (PCC) values between nAb-EGFP and anti-target mAb labeling (Homer1: L113/130; IRSp53: L117/1; pan-SAPAP:N459/94). PCC values of Homer1 and IRSp53 mAb immunolabeling with EGFP fluorescence in cells expressing EGFP are also shown. (C) Size analysis of mAb-labeled puncta of target proteins between nAb-EGFP transfected and untransfected cells. Homer1: ns, p=0.3828; IRSp53: ns, p=0.5410; pan-SAPAP: ns, p=0.0706; two-tailed unpaired t-tests. Bars on all graphs are mean ± SD.
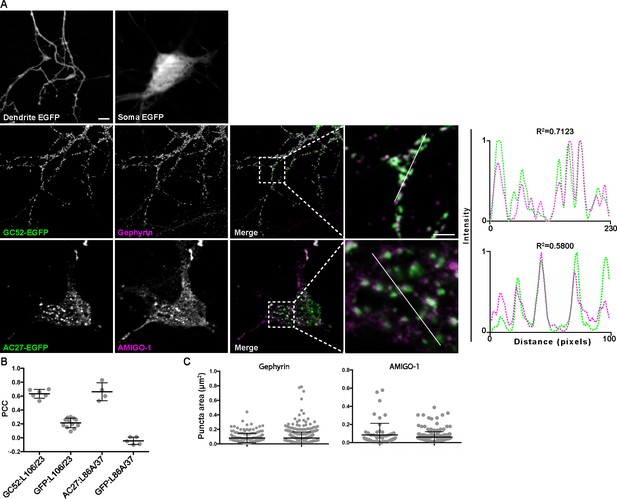
nAbs that function as intrabodies against inhibitory synaptic and ER-PM junction target proteins colocalize with endogenously expressed target proteins in cultured hippocampal neurons.
Representative images of intrabody-positive nAbs bound to endogenous targets after expression in cultured hippocampal neurons. (A) The top row shows the diffuse localization of EGFP in the dendrite (left) and soma (right) of an EGFP-transfected neuron. The subsequent rows show neurons expressing nAb-EGFP fusions and showing EGFP fluorescence (green), target-specific mAb labeling (magenta), and the merged image, with an adjacent panel showing a magnified image of the inset marked by the dashed box. The scale bar in the top left panel is 5 μm and holds for all panels in figure except the magnified insets, for which the scale bar is 2 μm. Panels to the far right of each row are the normalized fluorescence intensity values across the individual line scans from the magnified inset, with the corresponding R2 values. Note the concordance of the nAb (green) and anti-target mAb (magenta) intensity values. (B) Graph of PCC values between nAb-EGFP fusions and anti-target mAb labeling (Gephyrin: L106/23; AMIGO-1: L86A/37). PCC values of Gephyrin and AMIGO-1 mAb immunolabeling with EGFP fluorescence in cells expressing EGFP are also shown. (C) Size analysis of mAb labeled puncta of target proteins between nAb-EGFP transfected and untransfected cells. Gephyrin: ns, p=0.8758; AMIGO-1: ns, p=0.1091; two-tailed unpaired t-tests. Bars on all graphs are mean ± SD.
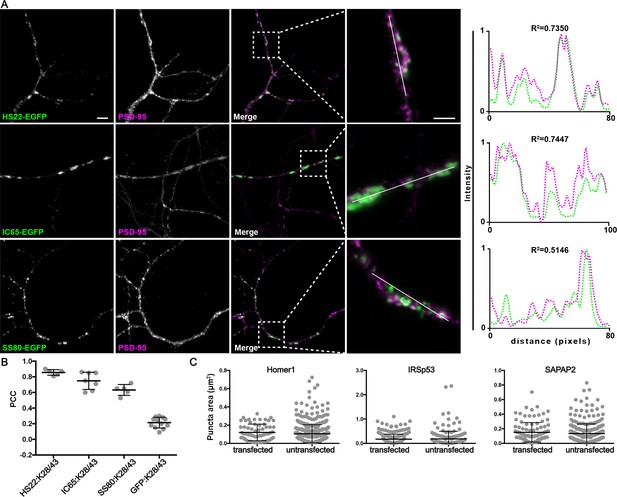
nAbs that function as intrabodies against excitatory synaptic target proteins colocalize with the excitatory synaptic marker PSD-95 in cultured hippocampal neurons.
(A) Each row shows representative images of a nAb against an excitatory synaptic protein (Homer1L, IRSp53, SAPAP2) expressed in neurons as nAb-EGFP fusions (green), and immunolabeling for a mAb (K28/43) against the synaptic marker PSD95 (magenta). The scale bar in the top left panel is 5 μm and holds for all panels in figure except the magnified insets, for which the scale bar is 2 μm. Panels to the far right of each row are the normalized fluorescence intensity values across the individual line scans from the magnified inset, with the corresponding R2 values. Note the concordance of the intensity values for the nAb-EGFP fusions (green) with PSD-95 labeling (magenta). (B) Graph of PCC values between the different nAb-GFP fusions and EGFP with immunolabeling for anti-PSD-95 mAb K28/43. (C) Size analysis of mAb labeled puncta of PSD-95 between nAb-EGFP transfected and untransfected cells. Homer1: ns, p=0.2093; IRSp53: ns, p=0.7063; pan-SAPAP: ns, p=0.2683; two-tailed unpaired t-tests. Bars on all graphs are mean ± SD.
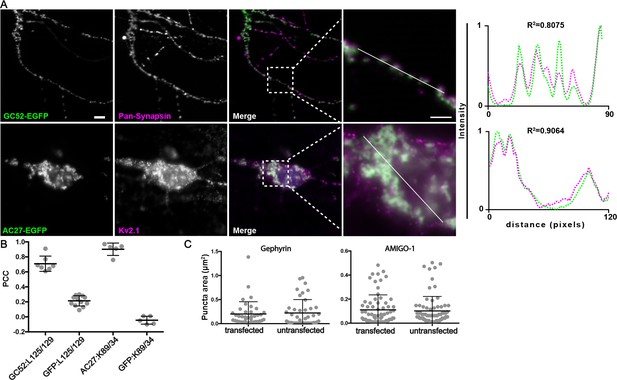
nAbs that function as intrabodies against inhibitory synaptic and ER-PM junction target proteins localize to their respective subcellular domains in cultured hippocampal neurons.
Representative images of the colabeling for proteins related to targets of nAbs and expressed nAbs-EGFP in neurons. (A) Each row shows representative images of a nAb-EGFP fusions (green) against the inhibitory synaptic protein Gephyrin (top row) or the ER-PM junction protein AMIGO-1 (second row) and in magenta immunolabeling with mAbs that label synapses (the pan-synapsin mAb L125/129 that sees all synapsin isoforms,) or Kv2 channel-containing ER-PM junctions (mAb K89/34 against Kv2.1 to label. The scale bar in the top left panel is 5 μm and holds for all panels in figure except the magnified insets, for which the scale bar is 2 μm. Panels to the far right of each row are the normalized fluorescence intensity values across the individual line scans from the magnified inset, with the corresponding R2 values. Note the concordance of the intensity values for the nAb-EGFP fusions (green) with labeling for the compartment-specific markers (magenta). (B) Graph of PCC values between the different nAb-GFP fusions and EGFP with immunolabeling for synapses (L125/129) or ER-PM junctions (K89/34). (C) Size analysis of mAb labeled puncta. Left: synapses (labeled with anti-pan-synapsin mAb L125/126) in anti-Gephyrin nAb-EGFP transfected and untransfected cells: ns, p=0.7063. Right: Kv2.1-containing ER-PM junctions (labeled with anti-Kv2.1 mAb K89/34) in anti-AMIGO-1 nAb-EGFP transfected and untransfected cells: ns, p=0.6513. Two-tailed unpaired t-tests. Bars on all graphs are mean ± SD.
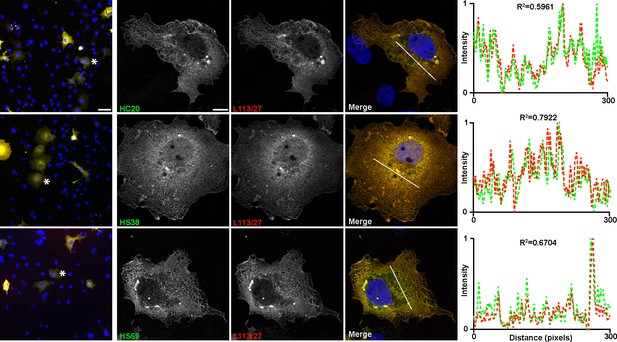
Anti-Homer1 nAbs exhibit labeling of exogenously expressed Homer1 in heterologous COS-1 cells.
Representative images of anti-Homer1 nAb labeling (green) of transiently transfected COS-1 cells expressing Homer1L double-labeled with the anti-Homer1 mAb L113/27 (red). The left column shows a mosaic of transfected and untransfected cells, nuclei are stained with Hoechst dye in blue. Note overlap of nAb and mAb labeling at both the cellular and subcellular level. The scale bar in the top left panel is 50 μm and holds for all panels in column. The three panels to the right show higher magnification views of the cells labeled with the asterisk in the left panels. The scale bar in the top left panel is 5 μm and holds for all panels in column. The graph to the right shows the normalized fluorescence intensity values across the line scans depicted by the white line in the merged images, with the corresponding R2 values.
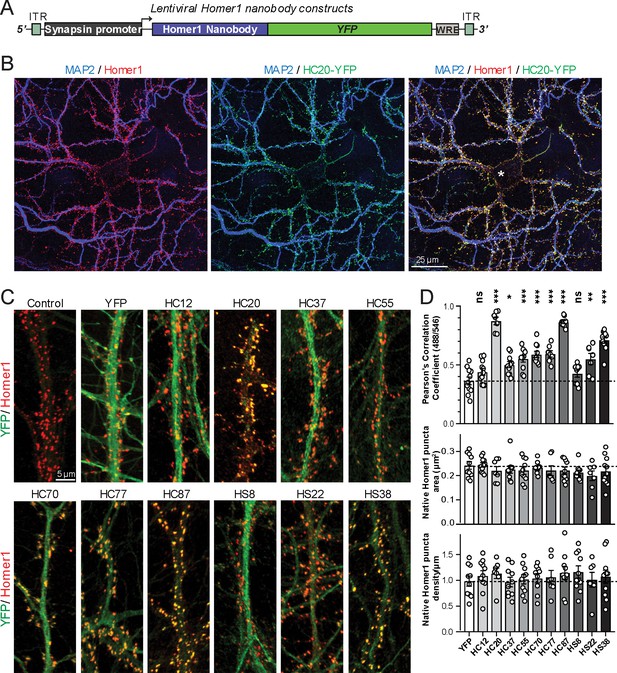
Anti-Homer nAbs expressed as intrabodies from recombinant lentivirus target to dendritic spines in cultured hippocampal neurons.
(A) Schematic of the lentivirus targeting construct. (B) Image shows a representative field of a CHN culture infected at 5 days in vitro (DIV) with recombinant lentivirus encoding the anti-Homer1 HC20-YFP nAb fusion (green) and imaged at 14 DIV after immunolabeling for endogenous Homer1 (red) and the dendritic marker MAP2 (blue). (C) Images show representative fields of dendrites of infected CHNs expressing different nAb-YFP fusions as indicated (green) and immunolabeled for endogenous Homer1 (red). The scale bar in the top left Control panel is 5 μm and holds for all panels in C. (D) The top graph shows Pearson’s Correlation Coefficient values between YFP or the different nAb-YFP fusions and anti-Homer1 immunolabeling. *p<0.01; **p<0.001; ***p<0.0001 for values of different anti-Homer1 nAb-YFP fusions versus for YFP alone. ns = not significant versus YFP alone. Values were analyzed by a one-way ANOVA followed by a Dunnett's post hoc test. The middle graph shows a size analysis of anti-Homer1 Ab labeled synaptic puncta in CHNs expressing YFP or the different anti-Homer1 nAb-YFP fusions. The bottom graph shows the density of anti-Homer1 Ab labeled synaptic puncta in CHNs expressing YFP or the different anti-Homer1 nAb-YFP fusions. Values for the size and density of anti-Homer1 Ab labeled synaptic puncta in CHNs expressing different anti-Homer1 nAb-YFP fusions are not significantly different than in CHNs expressing YFP alone. Bars on all graphs are mean ± S.E.M.
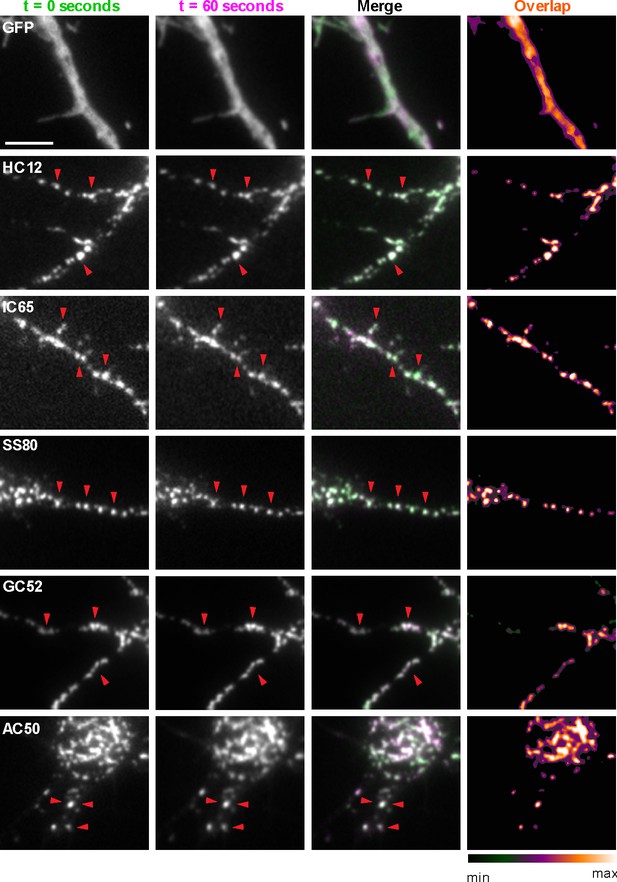
nAbs that function as intrabodies localize to immobile structures in cultured rat hippocampal neurons.
TIRF images of live cultured rat hippocampal neurons transfected with GFP (shown in A), or nAb-GFP fusions against Homer1 (clone HC12, shown in B), IRSp53 (clone IC65, shown in C), SAPAP2 (clone SS80, shown in D), Gephyrin (clone GC52, shown in E), or AMIGO-1 (clone AC50, shown in F). For each, two images (of the same field of view) taken one min apart are shown. To the right is an overlay of the initial image (in green) and the subsequent image (in magenta). Overlap of green and magenta yields a white signal. Arrows point to punctate structures. The column to the far right shows an analysis of the extent of overlap of pixels between the initial and subsequent images. The scale bar in the top left panel is 5 μm and holds for all panels in figure.
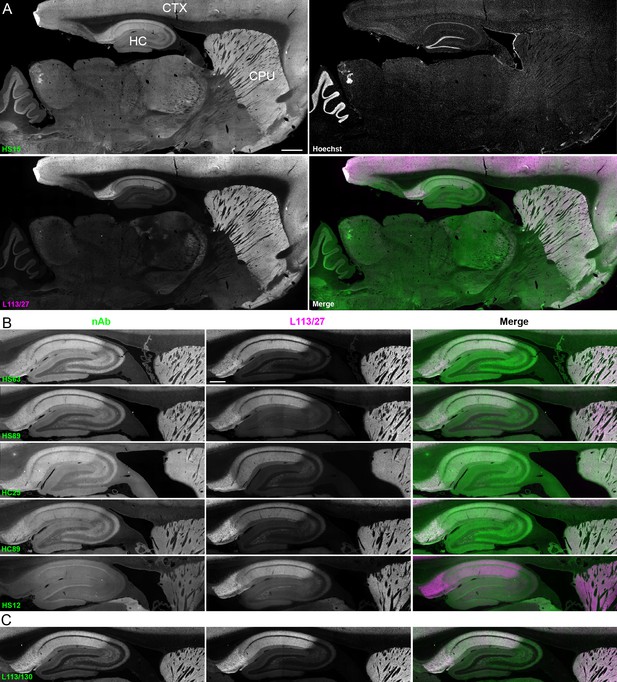
nAbs against Homer1 label rat brain sections through fluorescent immunohistochemistry.
(A) Representative images of a whole brain section labeled with a nAb BC supe against Homer1 (HS15, in green), a mouse mAb against Homer1 (L113/27, in magenta) and a nuclear label (Hoechst, in grayscale) showing brain regions with high Homer1 labeling. Merge is of antibody labeling only. CPu = caudate putamen; CTX = cerebral cortex; HC = hippocampus. The scale bar in the HS15 panel is 1 mm and holds for all four panels. (B) Representative images of HC and CPu from brain sections labeled with several anti-Homer1 nAb BC supes (green) and a mouse mAb against Homer1 (L113/27, in magenta). Note that nAb HS12 was scored as negative in this application. The scale bar in the top L113/27 panel is 500 μm and holds for all panels in B and C. (C) Representative images from brain sections labeled with mouse mAb L113/130 that recognizes both the long and short splice variants of Homer1 (green), and mouse mAb L113/27 that recognizes only the long splice variants of Homer1 (magenta).
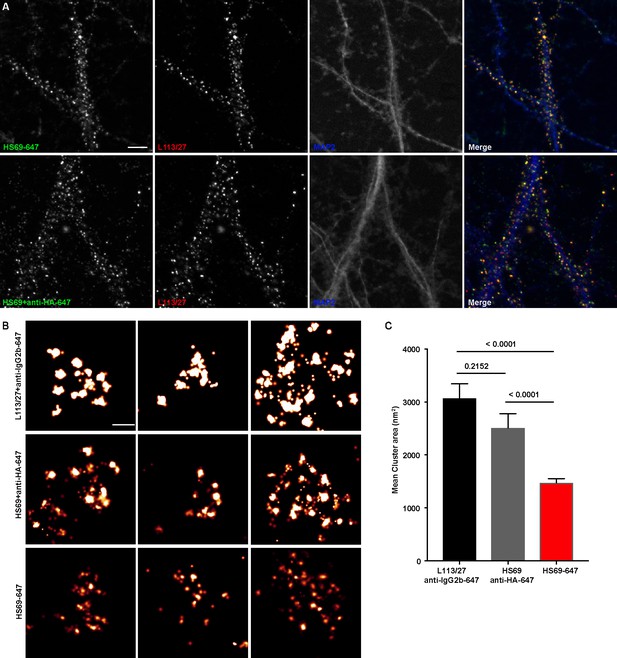
Immunolabeling with nAbs enhances spatial resolution.
(A) Representative TIRF images of CHNs. Neurons were immunolabeled with anti-Homer1 nAb plus a mouse mAb against Homer1 (L113/27, in red), and an antibody against MAP2 (blue). Top row: Dendrites of a CHN immunolabeled with nAb HS69 directly conjugated to Alexa647 (HS69-647, in green). Bottom row: Dendrites of a CHN immunolabeled with HA-tagged HS69 nAb plus Alexa647 conjugated anti-HA mAb (HS69 + anti-HA-647, in green). Scale bar is 5 μm and holds for all panels in A. (B) Representative super-resolution Homer1 localization maps of neurons immunolabeled for Homer1 with a mouse mAb plus Alexa647-conjugated secondary Ab (L113/27 + anti IgG2a-647), with HS69 nAb plus Alexa647-conjugated anti-HA antibody (HS69 + anti-HA-647), or with directly Alexa647-conjugated HS69 nAb (HS69-647). Scale bar is 200 nm and holds for all images in panel B. (C) Bar plot of the mean Homer1 cluster area ± SD for the three different labeling groups described in B (n = 227, 403, and 593 clusters respectively from five different cells per group). p values were calculated using a one-way ANOVA and a Bonferroni’s multiple comparisons test.
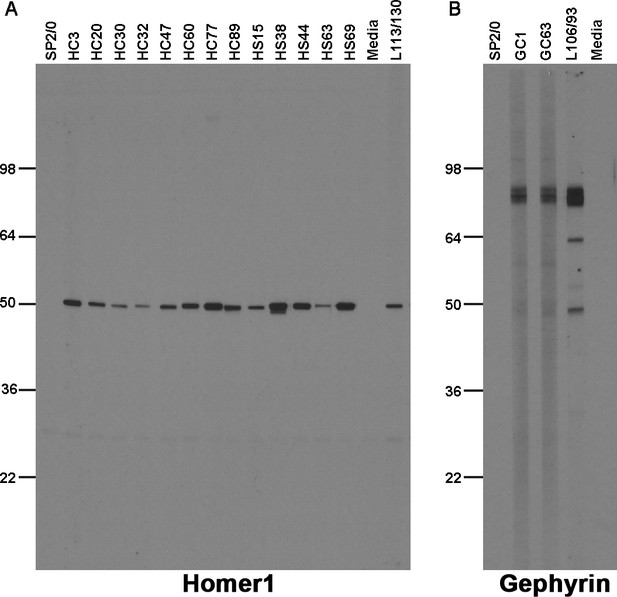
Immunolabeling with nAbs on immunoblots against a crude rat brain membrane fraction.
(A) Positive immunoblot labeling with anti-Homer1 nAbs expressed from E. coli Top10F’ cells; (B) Positive immunoblot labeling with anti-Gephyrin nAbs expressed from E. coli Top10F’ cells. Control lanes are the respective positive control monoclonal antibodies, and negative controls the SB bacterial culture medium (‘media’) and conditioned medium from culture of the non-secreting SP2/0 myeloma cell line ("SP2/0").
Tables
Summary of nanobody generation and validation.
https://doi.org/10.7554/eLife.48750.009| Primary selection and validation | Validation as intrabodies | Validation as immunolabels | ||||||
|---|---|---|---|---|---|---|---|---|
| Target | Phage clones selected | ELISA positives | Unique ELISA positives | COS-1 intrabody positives | Neuron intrabody positives | COS-1 IF-ICC positives | Brain IHC positives | Brain IB positives |
| Homer1 | 180 | 135 | 39 | 32 | 12 | 33 | 25 | 13 |
| IRSp53 | 160 | 33 | 17 | 8 | 3 | 0 | 0 | 0 |
| SAPAP2 | 172 | 32 | 15 | 7 | 2 | 0 | 4 | 0 |
| Gephyrin | 182 | 78 | 24 | 9 | 5 | 0 | 1 | 2 |
| AMIGO-1 | 173 | 38 | 18 | 13 | 5 | 0 | 0 | 0 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Cercopithecus aethiops) | COS-1 | ATCC Cat # CRL-1650, Lot # 59102713; PMID: 6260373 | RRID:CVCL_0223 | |
| Antibody | numerous | See Supplementary file 4 | ||
| Recombinant DNA reagent | pComb3XSS | PMID: 10986398 | Addgene #63890 (Addgene RRID:SCR_002037) | |
| Software algorithm | Photoshop | Adobe Systems | RRID:SCR_014199 | |
| Software algorithm | Axiovision | Carl Zeiss MicroImaging | RRID:SCR_002677 | |
| Software algorithm | Fiji | PMID: 22743772 | RRID:SCR_002285 |
Additional files
-
Supplementary file 1
Summary of corresponding mouse monoclonal antibody and nanobody immunolabeling results.
Table lists the screening results from prior monoclonal antibody (mAb) projects and the results from the corresponding nanobody screens. The numbers in parentheses in the final three columns of the nanobody rows represent the number of expected application-positive nanobodies obtained from the population of unique ELISA-positive nanobodies tested, based on percentages in mouse mAb projects.
- https://doi.org/10.7554/eLife.48750.015
-
Supplementary file 2
Summary of immunogens used for llama immunization.
Table lists nanobody project target, fragment used for llama immunization and tag.
- https://doi.org/10.7554/eLife.48750.016
-
Supplementary file 3
Primers used for heavy chain repertoire cloning.
Table lists primers used to amplify llama heavy chain variable regions.
- https://doi.org/10.7554/eLife.48750.017
-
Supplementary file 4
Non-nanobody antibodies used in this study.
Table lists non-nanobody antibodies used throughout this study, the immunogen used in their development, the species and IgG subclass, the manufacturer and Antibody Registry/RRID information, and the figure in which each antibody was used.
- https://doi.org/10.7554/eLife.48750.018
-
Transparent reporting form
- https://doi.org/10.7554/eLife.48750.019





