Interactions between Dpr11 and DIP-γ control selection of amacrine neurons in Drosophila color vision circuits
Figures
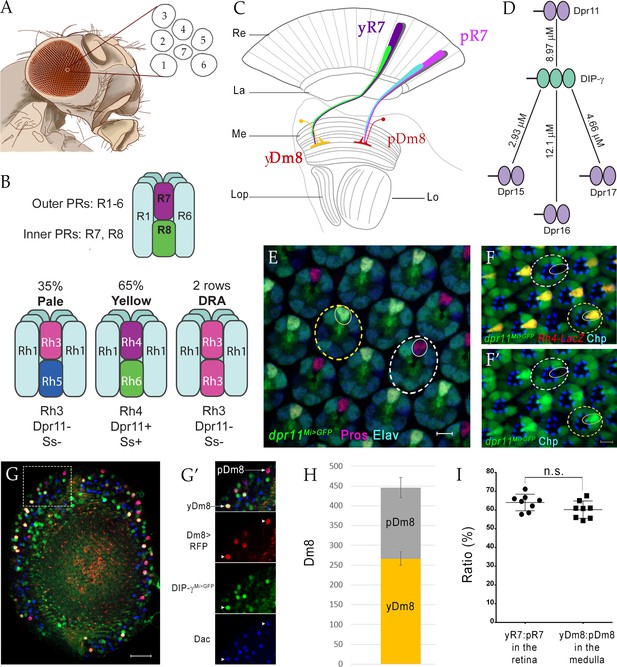
yDm8 and pDm8 populations are present in the same ratio as their presynaptic input yR7 and pR7 photoreceptors.
(A–C) Overview of the Drosophila visual system. (A) Compound eye. Each ommatidium contains 8 PRs. Rhabdomeres of 7 PRs are shown in diagram, because R7 is stacked on top of R8. (B) R1-6 are outer PRs, and R7 and R8 are inner PRs. Three ommatidial subtypes in the retina: yellow (y), pale (p) and Dorsal Rim Area (DRA). Each ommatidium is assigned as y or p based on Rh expression patterns in R7 and R8 PRs. y and p ommatidia are distributed randomly in a ~65y:35p ratio in wild-type. Rh4, Dpr11 and Spineless transcription factor (Ss) are expressed in R7 in y ommatidia only. (C) Schematic of the adult optic lobe. yR7 and pR7, and yR8 (green) and pR8 (cyan) project to M6 and M3 layers of the medulla, respectively. The axons of outer PRs (gray) terminate in the lamina. yR7 and pR7 synapse on yDm8 and pDm8 in M6. Re: Retina; La: Lamina; Me: Medulla; Lop: Lobula plate; Lo: Lobula. (D) The DIP-γ hub consists of Dprs 11, 15, 16 and 17. Dprs are 2-IgSF domain CSPs that interact with DIPs, which are 3-IgSF domain CSPs. KDs shown here are from Cosmanescu et al. (2018). Ig domains indicated by ovals. (E) dpr11Mi>GFP is expressed in R7 (small yellow circle) in select ommatidia (yellow dashed circle) and absent in others (white dashed circle) in the retina. R7 indicated by small circles. Mid-pupal retina labeled with anti-Pros for all R7 PRs (magenta), anti-GFP for dpr11Mi>GFP reporter (green) and anti-Elav for all neurons (blue). Maximum intensity projection; scale bar 5 µm. (F–F’) yR7 co-expresses dpr11Mi>GFP and Rh4-LacZ. Dpr11 is expressed in Rh4+ R7 in yellow ommatidia (yellow dashed circle) and absent in Rh3+ R7 in pale ommatidia (white dashed circle). Late pupal retina labeled with anti-β-galactosidase for Rh4-LacZ reporter (red), anti-GFP for dpr11Mi>GFP reporter (green), and anti-Chaoptin (Chp; blue) for all PRs. Single confocal slice; scale bar 5 µm. (G–G’) yDm8 and pDm8 cell bodies in adult medullary cortex. Adult optic lobes labeled with anti-RFP for pan-Dm8 driver >RFP (red), anti-GFP for DIP-γMi>GFP reporter (green) and anti-Dac for transcription factor Dachshund (blue). yDm8 expresses RFP, GFP and Dac and pDm8 expresses RFP and Dac. Inset in G shown in G’. yDm8 and pDm8 cell bodies indicated (arrows in merged, arrowheads in individual panels). Maximum intensity projection; scale bar 20 µm. (H) yDm8 and pDm8 populations in adult medullary cortex. The cell numbers of y and p Dm8 neurons determined with the pan-Dm8 driver and DIP-γMi>GFP are indicated on the y-axis. yDm8: 266.4+ /- 17.2, pDm8: 179.4+ /- 25.6 (n = 8–11 OLs; error bars indicate std. deviation). (I) Matching of yDm8:pDm8 ratio in the medulla with yR7:pR7 ratio in the retina. R7 and Dm8 ratios were determined as in F and G-H, respectively. Ratios (expressed as percentages) are shown on the y-axis. Graph shows mean + /- std. deviation and unpaired Student’s t-test p-values. yR7:pR7 63.9+ /- 4.4 (n = 8 retinas), yDm8:pDm8 60.2+ /- 4.6 (n = 8 OLs), not significant (n.s.). Complete genotypes in Table 2 of Materials and methods.
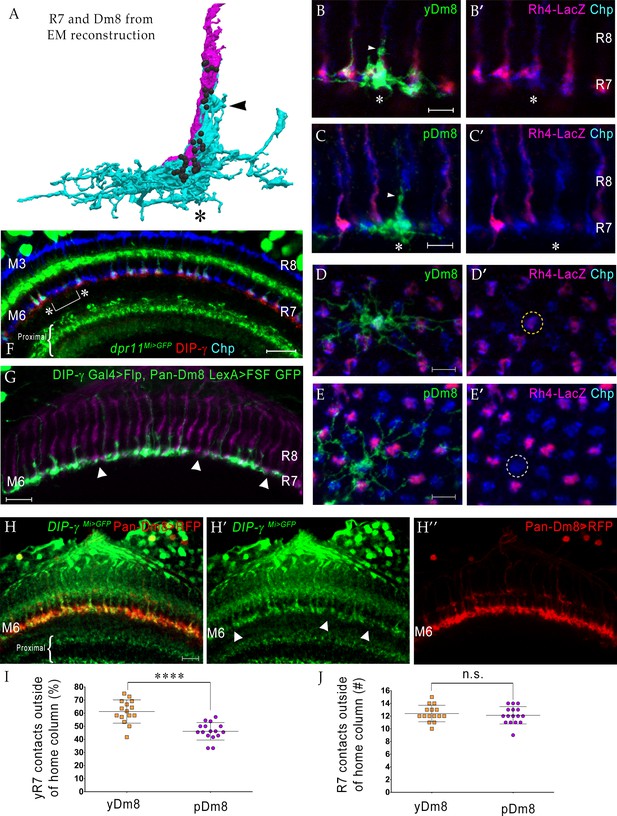
yDm8 neurons selectively innervate yR7 in their home columns and avoid pR7, and vice versa.
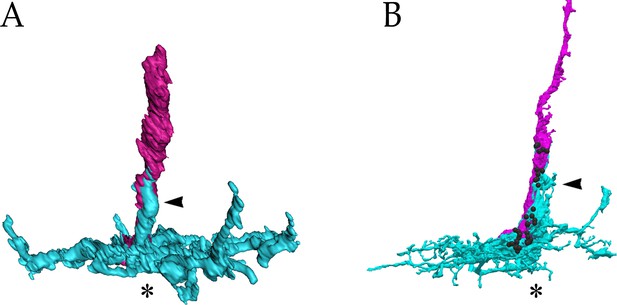
(A) Rendering of a yR7 terminal and a yDm8 arbor from an EM reconstruction, in a horizontal view. The distal dendritic projection (sprig; arrowhead) of Dm8 extensively contacts the R7 home column (asterisk). Dm8, cyan; R7, magenta. Black balls indicate R7 T-bars (output synapses). (B–E) Single flipout clones generated either with the DIP-γ split-Gal4 Dm8 driver (denoted as yDm8 split-Gal4 driver in Figure 2—figure supplement 1A) for yDm8 or with pan-Dm8 driver for pDm8. Adult optic lobes labeled with anti-GFP for flipout clone, anti-LacZ for yR7 reporter Rh4-lacZ and anti-Chp for all PRs. Medulla columns were identified as pale or yellow with yR7 reporter (magenta) and Chp (blue); pR7 columns were identified by the absence of Rh4-LacZ labeling. Maximum intensity projection; scale bar 5 µm. (B–B’) Horizontal view of a yDm8. The yDm8 distal dendritic projection (sprig; arrowhead) extends distally along the home column yR7 (asterisk) to the M4 layer. (C–C’) Horizontal view of a pDm8. pDm8 has a similar morphology to yDm8, with the sprig (arrowhead) in contact with a pR7 home column (asterisk). (D–D’) Cross-sectional (top-down) view of a yDm8. The dendritic arbor of this yDm8 contacts 13 columns (8y and 5 p). Home column yR7 indicated by yellow dashed circle. (E–E’) Cross-sectional view of a pDm8. The dendritic arbor of this pDm8 contacts 14 columns (8 p and 6y). Home column pR7 indicated by white dashed circle. (F) DIP-γ-expressing yDm8 neurons specifically contact Dpr11-expressing yR7 home columns. Horizontal view of mid-pupal medulla labeled with anti-DIP-γ (red), anti-GFP for dpr11Mi>GFP reporter (green) and anti-Chp for all PRs (blue). All yR7 PRs have DIP-γ labeling abutting the R7 (asterisks) and none of the pR7 PRs have any DIP-γ labeling apposed to them (bracket). Maximum intensity projection; scale bar 10 µm. (G) yDm8 and pDm8 populations have independent origins. The dendritic arbors of yDm8 neurons are labeled in flies carrying DIP-γ Gal4 >Flp and pan-Dm8 LexA >LexAop FRT-stop-FRT GFP transgenes. pDm8 that are not labeled appear as gaps (arrowheads) in the M6 layer, similar to the pattern of M6 layer in DIP-γMi>GFP where only yDm8 express DIP-γ (H’). Adult optic lobes were labeled with anti-GFP (green) and R7 and R8 PRs were labeled with anti-Chaoptin (magenta). Maximum intensity projection; scale bar 10 µm. (H–H”) Gaps representing pDm8 arbors are present in M6 layers labeled with the DIP-γMi>GFP reporter (H’), but not in M6 labeled with the pan-Dm8 reporter, which labels both Dm8 subtypes (H”). Adult optic lobes labeled with anti-GFP for DIP-γMi>GFP reporter (H’) and anti-RFP for pan-Dm8 Gal4 >RFP (H”). Gaps are marked (arrowhead in H’) in the M6 layer. The pan-Dm8 driver also labels lamina neuron L3, which is seen as faint labeling above the Dm8 layer. Maximum intensity projection; scale bar 10 µm. (I) yDm8 have a bias for contacting yR7 outside of the home column, while pDm8 contact yR7 and pR7 equally. Single flipout clones were labeled as in Figure 2D–E and analyzed with Imaris software to analyze connections with neighboring columns. yR7 contacts made outside of the home column: yDm8 61.3+ /- 8.9 (n = 15), pDm8 46.2+ /- 6.7 (n = 16), ****p<0.0001 (J) The total number of R7 contacted outside of the home column by the two Dm8 subtypes are the same. Single flipout clones were labeled as in Figure 2D–E and analyzed with Imaris software to analyze connections with neighboring columns. Total R7 contacted: yDm8 12.4+ /- 1.3 (n = 15), pDm8 12.1+ /- 1.4 (n = 16), not significant (n.s.). Complete genotypes in Table 2 of Materials and methods.
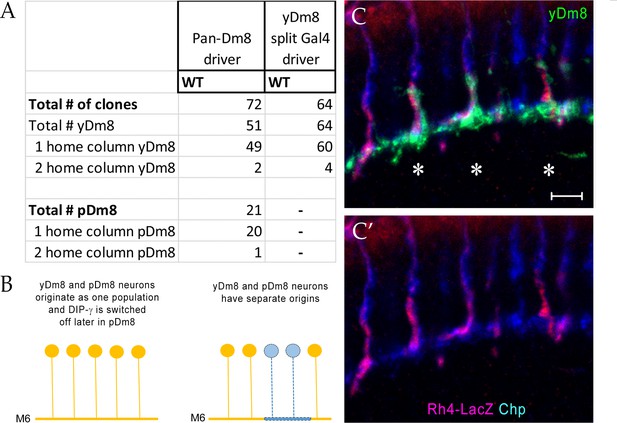
Quantitation of yDm8 and pDm8 flipout clones.
(A) Quantitation of yDm8 and pDm8 flipout clones in wild-type: Pan-Dm8 driver: R24F06-Gal4 (Nern et al., 2015). yDm8 split-Gal4 driver: R24F06-p65.AD; DIP-γ-Gal4-DBD (This study). With the pan-Dm8 driver, we examined 72 clones. 51/72 were yDm8 flipouts (identified by the home column R7 that was labeled with Rh4-LacZ for yR7) and 21/72 were pDm8 flipouts (identified by home column R7 labeled with Chp that did not show Rh4-LacZ signal, indicating a pR7). For both yDm8 and pDm8, we found clones with two home columns: 2/51 for yDm8 and 1/21 for pDm8. The two home columns were of the same identity: for yDm8, they were both yR7 and for pDm8, they were both pR7. With the yDm8 split Gal4 driver, we examined 64 yDm8 flipouts. We verified the identity of 43 clones as yDm8 by co-labeling with Rh4-LacZ and determining the identity of home column R7. 43/43 clones contacted yR7 home columns, confirming that the split Gal4 driver was an accurate yDm8 driver. 4/64 were two home column yDm8 flipouts. Complete genotypes in Table 3 of Materials and methods. (B) Schematic of origins of Dm8 subtypes. Two models are shown: yDm8 (express DIP-γ, indicated in yellow) and pDm8 (do not express DIP-γ, indicated in blue) originate as one population with DIP-γ switched off at a later developmental stage, or yDm8 and pDm8 populations originate independently. In our experiment, the dendritic arbors of DIP-γ expressing Dm8 neurons are labeled in flies carrying DIP-γ Gal4 >Flp and pan-Dm8-LexA > LexAop FRT-stop-FRT GFP transgenes. The first model predicts that the M6 layer will be labeled as a continuous line, whereas the second model predicts that the M6 layer will have gaps that are occupied by the pDm8 arbor. Dash indicates gaps (pDm8 arbors) that are not labeled. (C–C’) Horizontal view of a dense flipout showing 3 yDm8 clones (asterisks) with yR7 home columns labeled with Rh4-LacZ (red) and Chp (blue). Note the absence of sprig labeling on a pR7 column labeled with Chp only (blue). Maximum intensity projection; scale bar 5 µm.
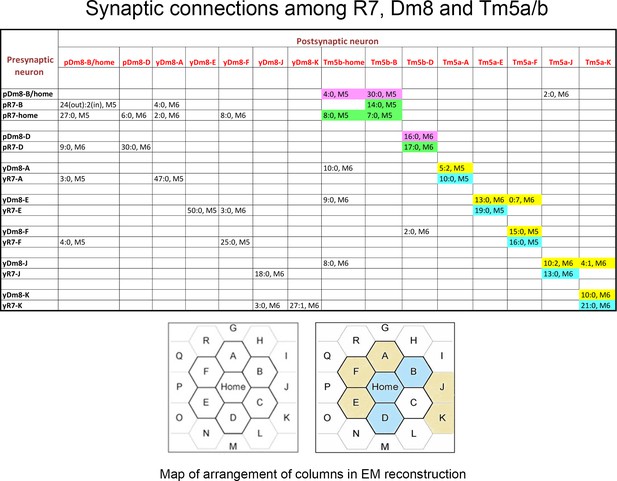
Synaptic connections among R7, Dm8, and Tm5a/b from the EM reconstruction.
The entries in column one indicate the EM column and y/p identities for each R7 and Dm8. The entries in row one indicate the column and y/p identities for the Dm8, Tm5a, and Tm5b neurons that are postsynaptic (and sometimes presynaptic) to the R7 and Dm8 neurons. In each box, the x:y nomenclature indicates the numbers of output synapses and input synapses. The layer in which most of these synapses are located is also indicated (M5 or M6). Classes of synapses are indicated in different colors, as follows: pR7→Tm5b, green; pDm8→Tm5b, purple; yR7→Tm5a, aqua; yDm8→Tm5a, yellow. At the bottom are maps of the arrangement of columns in the reconstruction, from Takemura et al. (2015), with y columns indicated in gold and p columns in blue in the right-hand map. We could not assign y/p identities to columns C, P, or Q, and there are no R7 outputs listed for the other columns. Tm5-C has an ambiguous morphology and could be either a Tm5a or a Tm5b, and R7-P and R7-Q have no listed synapses onto Tm5a or b. Figure 3—source data 1 contains additional information for non-home-column Dm8 inputs, and synapses onto Tm5c and Dm9.
-
Figure 3—source data 1
This is a table listing non-home column R7 synapses, and R7/Dm8 synapses with Tm5c and Dm9, in addition to the information in Figure 3.
Column two indicates the identities of the input synapses from R7 onto each Dm8. The reconstructed volume is unlikely to cover the entire arbor of these Dm8 neurons, since each Dm8 can contact 12–16 R7 (Gao et al., 2008). However, column two shows that within the reconstructed columns there is no apparent specificity in synaptic connectivity between y and p R7 and Dm8 neurons outside of their home columns. For example, pDm8-B/home receives synapses from four non-home column R7, at least 2 of which are yR7, while yDm8-A receives synapses from two non-home column pR7 and a yR7. Even the Dm8 at the center of the reconstruction (Dm8-B/home) only receives synapses from four non-home column R7 PRs, so many Dm8 contacts with R7 columns may not be associated with input synapses from R7. The total number of non-home column R7 PRs that synapse onto the Dm8 is indicated in the parentheses in column 2. Synapses labeled as ? in column two are from R7 PRs in columns that cannot be assigned as y or p. Also note that (Karuppudurai et al., 2014) found that Tm5a dendrites specifically associate with yR7 axons, but they stated that Tm5b dendrites have no specificity for association with y vs. p R7 axons. Our analysis of the EM reconstruction, however, shows that all 3 pR7 PRs synapse only onto Tm5b and not Tm5a. The conclusion of Karuppudurai et al. (2014) likely arises from the fact that Tm5b usually has two distal dendritic projections. One of these always arborizes with a pR7 axon and receives the pR7 input. The other one can arborize with either a y or a p R7, but does not receive yR7 input, at least not for this set of columns. Four of the 7 Dm8, and 3 of the 8 R7 PRs, also have synapses onto Tm5c neurons. There is no specificity for y vs. p in the connections to Tm5c. Both R7 classes have input and output synapses with Dm9, and both classes of Dm8 have output synapses onto Dm9.
- https://cdn.elifesciences.org/articles/48935/elife-48935-fig3-data1-v2.xlsx
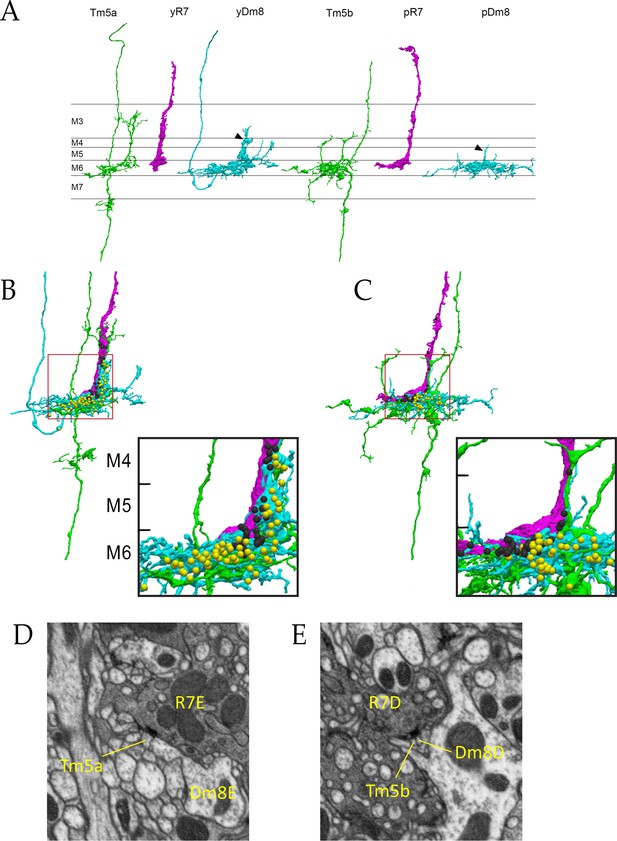
Electron microscopic reconstruction of wavelength discrimination circuits.
Renderings of separated and combined cells from the EM reconstruction (Takemura et al., 2015) are shown in (A-D). R7, magenta; Dm8, cyan; Tm5a/b, green. (A) Separated cells in a yellow circuit, from column E, and a pale circuit, from column D. yR7-E, yDm8-E, and Tm5a-E pR7-D, pDm8-D, and Tm5b-D are shown. yDm8-E and pDm8-D sprigs indicated by arrowheads. (B-C) Renderings of R7-Dm8-Tm5a/b circuits. The insets show the home column region where most synapses are located. Black balls, R7 T-bars; yellow balls, Dm8 T-bars. The borders of M6, M5, and M4 are indicated. (B) The column E circuit. (C) The column D circuit. See also associated Figure 4—videos 5 and 6 (vertical and horizontal rotation of each of these circuits, as well as of the two-home column B/home circuit, which is shown in Figure 4—figure supplement 1). For a comparison of an ExM image of a wild-type yR7 and yDm8 to yR7-E and yDm8-E from the EM reconstruction, see Figure 6—figure supplement 1. (D) A section from column E, showing a polyadic synapse of yR7-E onto yDm8-E and Tm5a-E. (E) A section from column D, showing a polyadic synapse of pR7-D onto pDm8-D and Tm5b-D. In (D) and (E), the R7 T-bars are the black shapes on the R7 membranes where they are apposed to both postsynaptic cells.
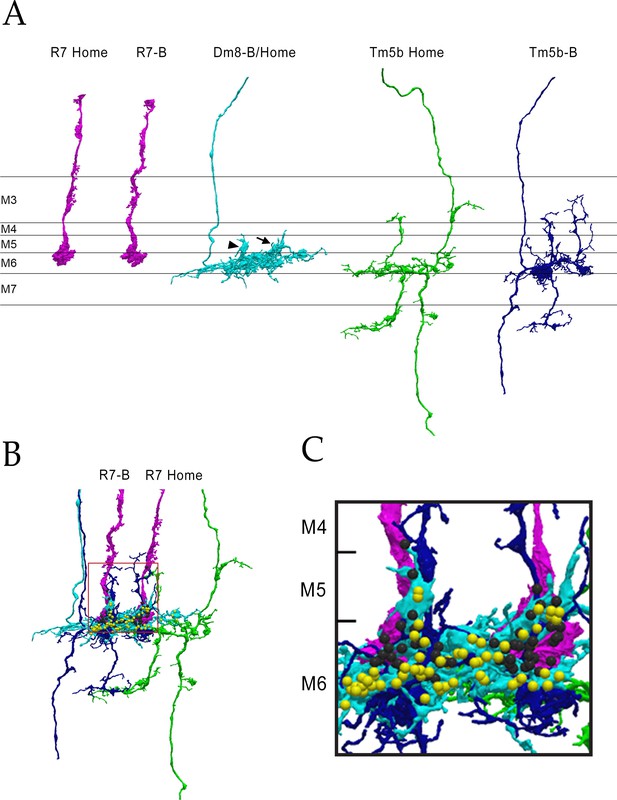
The columns B and home circuit.
This is a two-home column circuit, including pR7-B, pR7-home, pDm8-B/home, Tm5b-B, and Tm5b-home. Videos of this circuit are linked to Figure 4, as Figure 4—videos 5 and 6. (A) Separated cells for the circuit. pDm8-B/home sprigs indicated: B sprig, arrowhead; home sprig, arrow. (B) Combined images for the 5 cells. Boxed region is magnified in (C). (C) Magnified image of B and Home columns. Black balls, R7 output synapses; yellow balls, Dm8 output synapses.
Horizontal rotation of column E yellow circuit Colors as in Figure 4.
Vertical rotation of column E yellow circuit.
Horizontal rotation of column D pale circuit.
Vertical rotation of column D pale circuit.
Horizontal rotation of column B/home pale circuit.
Vertical rotation of column B/home pale circuit.
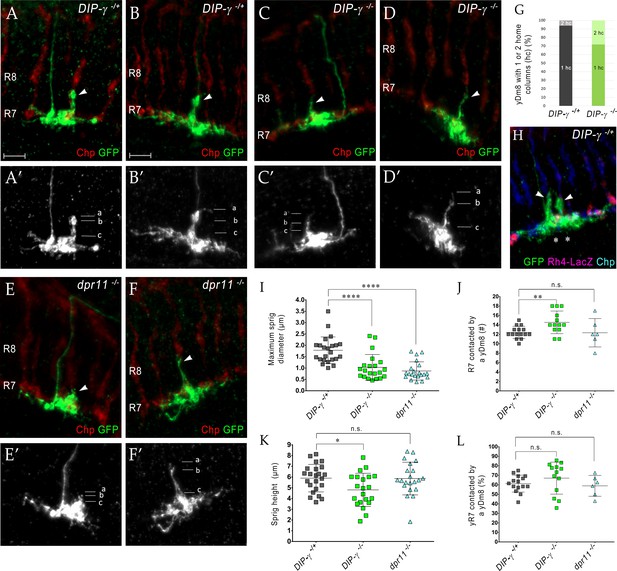
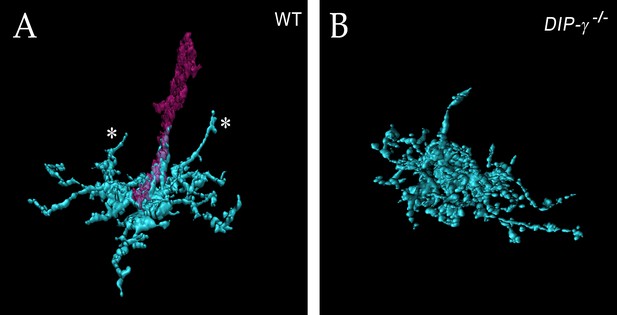
Loss of DIP-γ or Dpr11 alters morphology of yDm8 distal dendrites.
(A–F) yDm8 flipouts generated in (A–B) wild-type, (C–D) DIP-γ mutant and (E–F) dpr11 mutant. Sprigs are marked with arrowheads and PRs labeled with anti-Chp (red). (A’–F’) Positions of the measurements used in graphs I and K are indicated. Height of the sprig was determined by measuring distance from ‘a’ to ‘c’ and sprig diameter was measured at position marked ‘b’. Frequency of clones obtained for all three genotypes in Figure 5—source data 1. Flipouts in wild-type and DIP-γ mutants were generated with the yDm8 split-Gal4 driver; dpr11 mutant flipouts were generated with the pan-Dm8 driver and scored as yDm8 using Rh4-LacZ labeling. Complete genotypes in Table 2 of Materials and methods (Table 1). Maximum intensity projection; scale bar 5 µm. (G–H) Two-home column yDm8 flipouts were observed more frequently (4.5-fold increase) in DIP-γ mutant (11/39 clones) than in control (4/64 clones). Panel H shows a two home column yDm8 with both sprigs (arrowheads) located on yR7 columns (asterisks). (I, K) Maximum sprig diameter is reduced significantly in both dpr11 and DIP-γ mutants. Sprig height was slightly affected in DIP-γ mutants but not in dpr11 mutants. Graph shows mean + /- std. deviation and unpaired Student’s t-test p-values. Sprig diameter: DIP-γ -/+ 1.8+ /- 0.6 (n = 23), DIP-γ -/-1.0+ /- 0.6 (n = 21), dpr11 -/- 0.9+ /- 0.4 (n = 21), ****p<0.0001 for both mutants. Sprig height: DIP-γ -/+ 5.9+ /- 1.3 (n = 23), DIP-γ -/-4.8+ /- 1.5 (n = 21), dpr11 -/- 5.9+ /- 1.5 (n = 21), from left to right *p=0.014, not significant (n.s.) (J, L) Contact with neighboring columns is affected in DIP-γ mutants. (J) Total number of R7 columns contacted by a yDm8: DIP-γ -/+ 12.4+ /- 1.3 (n = 15), DIP-γ -/-14.5+ /- 2.4 (n = 13), dpr11 -/- 12.33+ /- 3.0 (n = 6), from left to right **p=0.0061, not significant (n.s.) (L) Percentage of yR7 columns contacted by a yDm8: DIP-γ -/+ 61.3+ /- 8.9 (n = 15), DIP-γ -/-67.03+ /- 16.8 (n = 13), dpr11 -/- 58.9+ /- 11.0 (n = 6), from left to right, not significant (n.s.).
-
Figure 5—source data 1
Frequency of two-home column yDm8 clones is increased in DIP-γ mutants.
The percentage of two-home column yDm8 clones in DIP-γ -/- mutants is 4.5-fold higher than in wild-type. Quantitation of flipout clones in wild-type, DIP-γ and dpr11 mutants shown. Flipouts were generated with either pan-Dm8 driver R24F06-Gal4 or the yDm8 split-Gal4 driver R24F06-p65.AD; DIP-γ Gal4-DBD. Complete genotypes in Table 3 of Materials and methods.
- https://cdn.elifesciences.org/articles/48935/elife-48935-fig5-data1-v2.xlsx
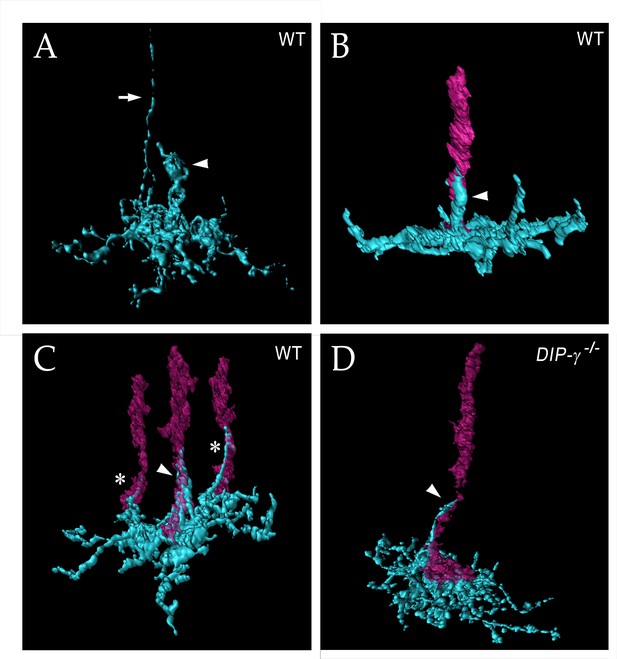
yDm8 arbor morphology in wild-type and DIP-γ mutant visualized with expansion microscopy.
yDm8 dendritic arbors (cyan) visualized with expansion microscopy and surface rendered with Imaris software. (A-C) Wild-type; (D) DIP-γ -/-. Arrowheads, sprigs; arrow in (A), axon. In (B-D), one or more R7 terminals/axons are included in the rendering. Two different views of the same flipout clone are shown in panels (B) and (C). The R7 terminals/axons are semi-transparent in (C) and (D). The home column R7 located at the center of the arbor makes extensive contacts with the sprig as well as with the base of the dendritic arbor in M6 in (B) and (C). Two thinner dendritic processes positioned on the edges of the arbor (asterisks) contact two non-home column R7 in (C). The yDm8 in the DIP-γ -/- mutant has a much thinner sprig as compared to wild-type (D). See associated Figure 6—videos 1–4 (vertical and horizontal rotations) and Figure 6—figure supplement 2 for additional views of the wild-type clone in (C) and the mutant clone in (D). ExM analysis of yDm8 in wild-type (n = 6) and DIP-γ -/- (n = 5) genotypes.
A comparison of the (A) ExM rendering of the wild-type yDm8 and yR7 shown in Figure 6B, and the (B) EM reconstruction of yDm8-E and yR7-E shown in Figure 2A.
yDm8, cyan; yR7, magenta. Black balls indicate yR7 T-bars. Home column positions and sprigs are indicated by asterisks and arrowheads, respectively. Based on the comparison with B, we can infer that there are likely to be many R7-Dm8 synapses on the sprig in A.
Additional views of expanded yDm8 in (A) wild-type and (B) DIP-γ -/-.
(A) The wild-type rendering is the same column as in Figure 6C, but with only the home column yR7 included. This allows clearer visualization of the other two dendritic projections (asterisks). (B) The mutant rendering is without the R7, allowing clearer visualization of sprig morphology.
Horizontal rotation of an expanded yDm8 in wild-type.
Horizontal rotation of a yDm8 (cyan) with Chp labeled R7 PRs (magenta). A single yDm8 wild-type clone was expanded and surface rendered with Imaris software. Note that in addition to the major dendritic process (sprig) wrapping around the home column R7, two thinner dendritic processes extend distally along other R7.
Vertical rotation of an expanded yDm8 in wild-type.
Vertical rotation of the same yDm8 clone.
Horizontal rotation of an expanded yDm8 in DIP-γ mutant.
Horizontal rotation of a yDm8 (cyan) and a Chp labeled home column yR7 (magenta). A single yDm8 DIP-γ mutant clone was expanded and surface rendered with Imaris software. The thin process that is not in contact with the R7 is the yDm8 axon.
Vertical rotation of an expanded yDm8 in DIP-γ mutant.
Vertical rotation of the same mutant yDm8 clone.
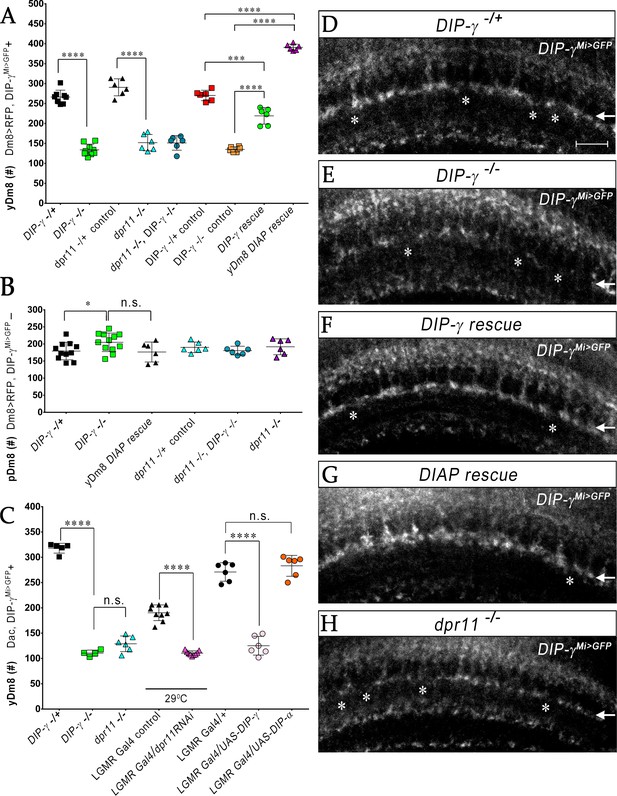
yR7 PRs provide survival signals to yDm8 neurons through Dpr11-DIP-γ interactions.
(A–B) yDm8 and pDm8 cell numbers were determined using pan-Dm8 driver >RFP and DIP-γMi>GFP reporter. yDm8 neurons express both RFP and GFP, and pDm8 neurons express only RFP. (C) yDm8 cell number determined using anti-GFP for DIP-γMi>GFP reporter and anti-Dac. The DIP-γMi>GFP reporter (indicated on the x-axis in all graphs as DIP-γ-/+) is an insertion in the 5’ UTR intron that has no detectable protein expression. Thus, this line serves as a mutant as well as a reporter of DIP-γ transcript expression. Graphs show mean + /- std. deviation and unpaired Student’s t-test p-values. Complete genotypes in Table 2 of Materials and methods (Table 1). (A) Both DIP-γ and dpr11 mutants show ~50% loss of yDm8 neurons, and this is rescued in DIP-γ mutants by inhibiting cell death with DIAP. yDm8 cell numbers in heterozygous controls, DIP-γ and dpr11 mutants, a double mutant of both genes, and DIP-γ and DIAP rescues in DIP-γ mutant are shown. DIP-γ -/+ 266.4+ /- 17.2 (n = 8), DIP-γ -/-134+ /- 14.3 (n = 9), ****p<0.0001; dpr11 -/+ control 290.7+ /- 21.3 (n = 6), dpr11 -/- 152+ /- 20.4 (n = 6), ****p<0.0001; dpr11 -/-, DIP-γ -/-151+ /- 18.3 (n = 6), DIP-γ -/-134+ /- 14.3 (n = 6), not significant; dpr11 -/-, DIP-γ -/-151+ /- 18.3 (n = 6), dpr11 -/- 152+ /- 20.4 (n = 6), not significant; DIP-γ -/+ control 270+ /- 13.03 (n = 6), DIP-γ rescue 219.2+ /- 20.7 (n = 6), ***p=0.0004; DIP-γ -/-mutant control 135.2+ /- 6.5 (n = 6), DIP-γ rescue 219.2+ /- 20.7 (n = 6), ****p<0.0001; DIP-γ -/+ control 270+ /- 13.03 (n = 6), yDm8 DIAP rescue 390.8+ /- 8.0 (n = 6), ****p<0.0001; DIP-γ -/-mutant control 135.2+ /- 6.5 (n = 6), yDm8 DIAP rescue 390.8+ /- 8.0 (n = 6), ****p<0.0001. (B) pDm8 numbers are unchanged in mutants and in DIAP rescue. DIP-γ -/+ 179.4+ /- 25.6 (n = 11), DIP-γ -/-204.9+ /- 27.2 (n = 12), *p=0.03; dpr11 -/+ control 176.5+ /- 29.2 (n = 6), dpr11 -/- 190.2+ /- 15.5 (n = 6), not significant (n.s.); dpr11 -/-, DIP-γ -/-180.7+ /- 12.8 (n = 6), DIP-γ -/-204.9+ /- 27.2 (n = 12), not significant; dpr11 -/-, DIP-γ -/-180.7+ /- 12.8 (n = 6), dpr11 -/- 190.2+ /- 15.5, not significant; DIP-γ -/-204.9+ /- 27.2 (n = 12), yDm8 DIAP rescue 191.8+ /- 23.2 (n = 6), not significant (n.s.). (C) Dpr11 in R7 is required for yDm8 survival. yDm8 cell number in wild-type, mutants, dpr11 eye-specific RNAi, and ectopic DIP-γ expression in PRs. DIP-γ -/+ 318+ /- 9.7 (n = 5), DIP-γ -/-110.5+ /- 6.1 (n = 4), ****p<0.0001; DIP-γ -/-110.5+ /- 6.1 (n = 4), dpr11 -/- 129+ /- 15.4 (n = 6), not significant (n.s.); lGMR-Gal4 control at 29°C 190.3+ /- 15.5 (n = 9), lGMR-Gal4 >dpr11 RNAi 110.6+ /- 4.5 (n = 9), ****p<0.0001; lGMR-Gal4 control 271+ /- 18.3 (n = 6), lGMR-Gal4 >UAS-DIP-γ (n = 6) 125.2+ /- 18.5, ****p<0.0001; lGMR-Gal4 control 271+ /- 18.3 (n = 6), lGMR-Gal4 >UAS-DIP-α 283.2+ /- 20.6 (n = 6), not significant (n.s.). (D-H) yDm8 labeling in the neuropil in wild-type, DIP-γ mutant, DIP-γ rescue, DIAP yDm8 rescue, and dpr11 mutant, using DIP-γMi>GFP reporter. Large gaps (asterisks) representing yDm8 cell death are seen in the M6 layer (arrow) in both DIP-γ and dpr11 null mutants (E, H), whereas wild-type, DIP-γ and DIAP rescues showed smaller and fewer gaps (asterisks in D, F-G). Adult optic lobes were labeled with anti-GFP for yDm8 reporter.
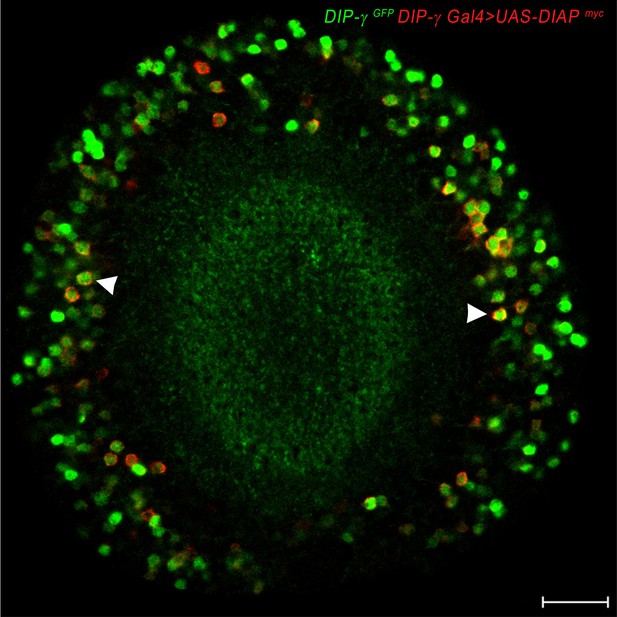
DIAP localizes to DIP-γ expressing cells.
DIAP1 tagged with myc (red) driven with DIP-γ Gal4 shows co-localization of DIAP1 and DIP-γMi>GFP reporter (green). Adult optic lobe labeled with anti-myc (red) and anti-GFP (green). Single section. Scale bar 20 µm.
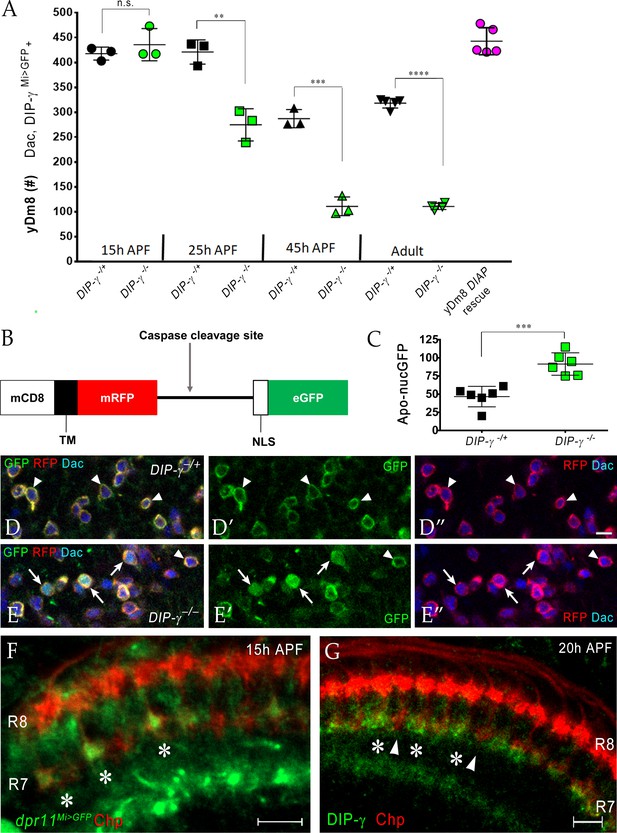
Dpr11-DIP-γ interactions are required early in pupal development to prevent yDm8 cell death.
(A) yDm8 cell death occurs in DIP-γ mutants between 15 hr and 45 hr APF. yDm8 cell death in wild-type occurs between 25 hr and 45 hr APF (p-value below). DIAP expression in the DIP-γ mutant rescues cell number back to the original level at 15 hr APF (p-value below). yDm8 cell number determined with anti-GFP for DIP-γMi>GFP reporter and anti-Dac. DIP-γMi>GFP reporter heterozygote indicated as DIP-γ-/+. Graph shows mean + /- std. deviation and unpaired Student’s t-test p-values. Complete genotypes in Table 2 of Materials and methods (Table 1). 15 hr APF: DIP-γ -/+ 417.7+ /- 13.1 (n = 3), DIP-γ -/-435.7+ /- 32.3 (n = 3), not significant (n.s.); 25 hr APF: DIP-γ -/+ 421+ /- 24.3 (n = 3), DIP-γ -/-274.7+ /- 32.3 (n = 3), **p=0.0033; 45 hr APF: DIP-γ -/+ 287+ /- 18.2 (n = 3), DIP-γ -/-110.7+ /- 18.7 (n = 3), ***p=0.0003; Adult: DIP-γ -/+ 318+ /- 9.7 (n = 5), DIP-γ -/-110.5+ /- 6.1 (n = 4), ****p<0.0001; p-values below are not shown on the graph: 25 hr APF: DIP-γ -/+ 421+ /- 24.3 (n = 3), 45 hr APF: DIP-γ -/+ 287+ /- 18.2 (n = 3), **p=0.0016; yDm8 DIAP rescue 442.6+ /- 27.2 (n = 5), 15 hr APF DIP-γ -/+ 417.7+ /- 13.1, not significant (n.s.) 15 hr APF: DIP-γ -/+ 417.7+ /- 13.1 (n = 3), Adult: DIP-γ -/+ 318+ /- 9.7 (n = 5), ****p<0.0001; 15 hr APF: DIP-γ -/-435.7+ /- 32.3 (n = 3), 25 hr DIP-γ -/-274.7+ /- 32.3 (n = 3), **p=0.004; 25 hr APF: DIP-γ -/-274.7+ /- 32.3 (n = 3), 45 hr DIP-γ -/-110.7+ /- 18.7 (n = 3), **p=0.0016. (B-E) yDm8 cell bodies marked with apoptotic reporter Apoliner are significantly increased in DIP-γ mutant. (B) Schematic of Apoliner (Bardet et al., 2008). In live cells, GFP is tethered to the membrane with RFP. In dying cells, activated caspases cleave Apoliner to release the GFP moiety that localizes to the nucleus. (C) Quantitation of dying yDm8 cell bodies in control and DIP-γ mutant. Graph shows mean + /- std. deviation and unpaired Student’s t-test p-values, n = 6 OLs for each genotype. Complete genotypes in Table 2 of Materials and methods (Table 1). DIP-γ -/+ 46.7+ /- 14.12 (n = 6), DIP-γ -/- 91.5+ /- 15.4 (n = 6), ***p=0.0004. (D-E) Visualization of Apoliner in yDm8 soma in control (D) and DIP-γ mutant (E). UAS-Apoliner was driven with a DIP-γ split Gal4 driver and yDm8 soma were labeled with anti-Dac, anti-GFP and anti-RFP. Dying yDm8 with nuclear GFP are seen in the DIP-γ mutant (arrows in E), while live yDm8 with membrane-localized GFP and RFP are seen in both control and mutant (arrowheads). (F) Dpr11 is expressed in select R7 PRs at 15 hr APF (asterisks). dpr11Mi>GFP reporter labeled with anti-Chp (red) and anti-GFP (green) at 15 hr APF. Note that one of the younger R7 PRs located on the right of the image also expresses Dpr11. Single confocal slice; scale bar 5 µm. (G) DIP-γ is expressed in yDm8 neurons apposed to specific R7 PRs by 20 hr APF (asterisks). pR7 terminals (arrowheads) do not show overlapping DIP-γ labeling (see also Figure 8—figure supplement 1B–B’). Wild-type, labeled at 20 hr APF with anti-DIP-γ (green) and anti-Chp (red). Complete genotypes in Table 2 of Materials and methods. Single confocal slice; scale bar 10 µm.
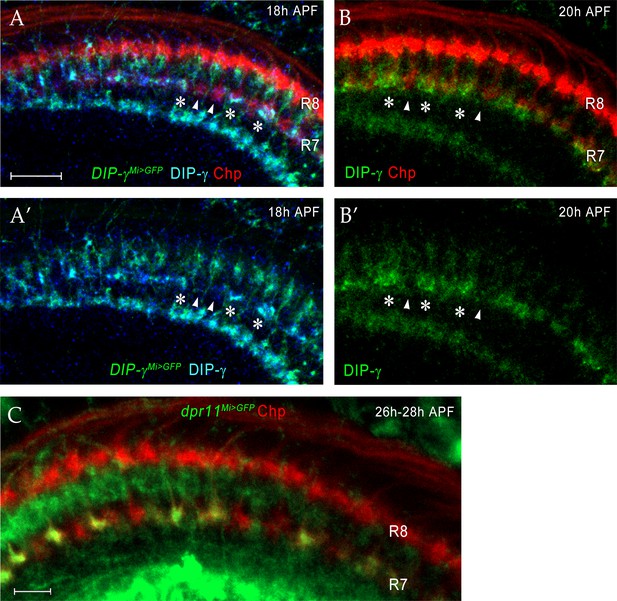
Dpr11 and DIP-γ are expressed in yR7 and yDm8, respectively, around the time yR7 selects yDm8 for survival.
(A–A’) DIP-γ is expressed in yDm8 processes overlapping select R7 terminals by 18 hr APF. DIP-γMi>GFP reporter and DIP-γ antibody show the same pattern in the neuropil. Three R7 PRs with anti-GFP and anti-DIP-γ labeling are indicated by asterisks. There are gaps in DIP-γ labeling in the R7 incipient layer (one such gap covering two R7 terminals indicated by two arrowheads). DIP-γMi>GFP reporter labeled with anti-GFP (green), anti-DIP-γ (blue) and anti-Chp (red). (A’) is without the Chp labeling, for clearer visualization of the gaps. (B–B’) (B) is the same merged image seen in Figure 8F. (B’) shows the DIP-γ channel only for this image, and reveals gaps in DIP-γ labeling, implying that pDm8 are apposed to pR7 within the gaps (arrowheads). DIP-γ labeled yDm8 processes indicated by asterisks. Wild-type labeled at 20 hr APF with anti-DIP-γ (green) and anti-Chp (red). (C) dpr11Mi>GFP expression at 26 hr - 28hr APF labels yR7 but not pR7. dpr11Mi>GFP reporter labeled with anti-Chp (red) and anti-GFP (green). R7 terminals unlabeled by GFP are pR7 (red). Complete genotypes in Table 3 of Materials and methods.
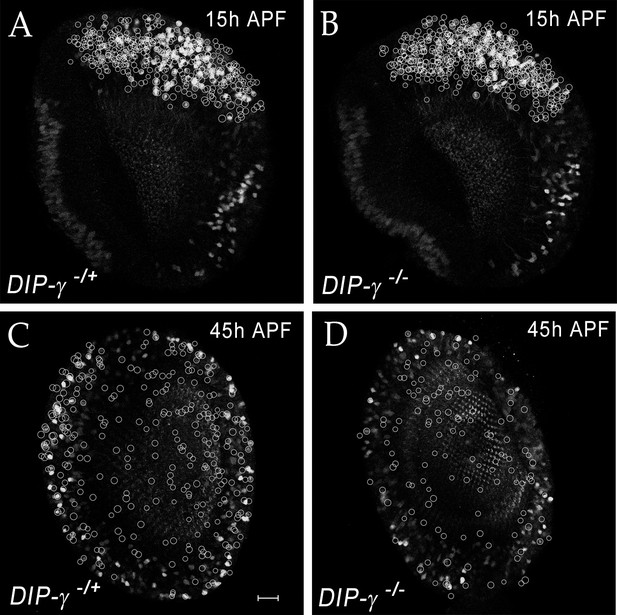
yDm8 cell death in DIP-γ mutant during migration of the cell bodies.
(A–D) yDm8 cell death in DIP-γ mutant occurs as yDm8 soma are migrating during early pupal development. White circles indicate yDm8 cell bodies that were labeled with anti-Dac (not shown here) and anti-GFP for DIP-γMi>GFP reporter to indicate position in the cortex in wild-type (A, C) and in DIP-γ mutant (B, D) at 15 hr (A–B) and 45 hr APF (C–D). There is no difference in their relative positions in DIP-γ mutant as compared to that in wild-type. Complete genotypes in Table 3 of Materials and methods.
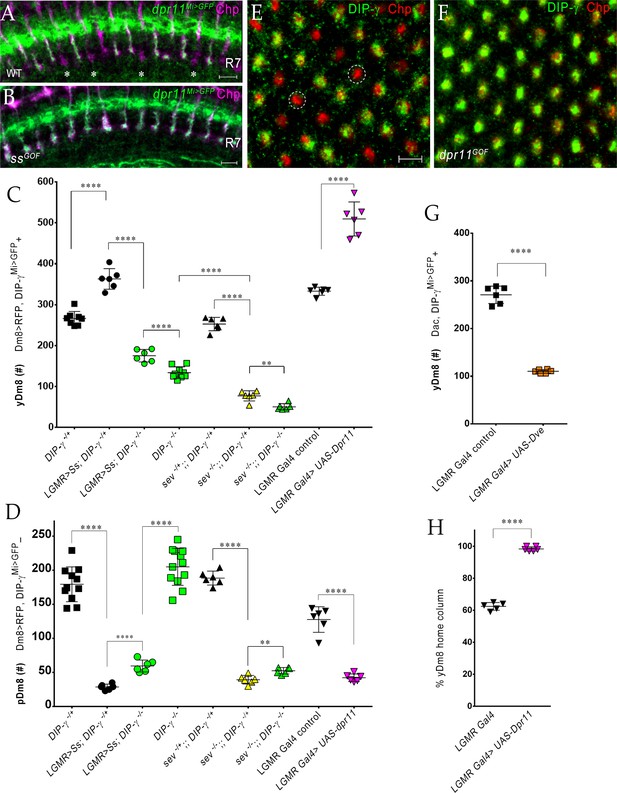
The representation of Dm8 subtypes in the medulla is altered by manipulation of R7 fates in the retina.
(A–B) Conversion of all R7 PRs to the yR7 fate in ssGOF results in Dpr11 expression in every R7. Dpr11 is expressed in yR7 PRs in (A) wild-type and (B) ssGOF. pR7 PRs are present in wild-type (asterisk) and absent in ssGOF. Mid-pupal optic lobes labeled with anti-GFP (green) for dpr11Mi>GFP reporter and anti-Chp (magenta) for all PRs. Maximum intensity projection; scale bar 5 µm. (C–D) yDm8 and pDm8 cell number determined using pan-Dm8 driver >RFP and DIP-γMi>GFP reporter. yDm8 expresses both RFP and GFP and pDm8 expresses only RFP. Graphs show mean + /- std. deviation and unpaired Student’s t-test p-values. Complete genotypes in Table 2 of Materials and methods (Table 1). DIP-γMi>GFP reporter heterozygote indicated as DIP-γ-/+. (C) More yDm8 neurons survive when all R7 PRs are converted to the yR7 fate (ssGOF) or when they all express Dpr11 (dpr11GOF). yDm8 neurons are also lost when R7 PRs are absent (sev) and/or DIP-γ is mutant. yDm8 cell number in wild-type, ssGOF, sev mutant, dpr11GOF: DIP-γ -/+ 266.4+ /- 17.2 (n = 8), ssGOF, DIP-γ -/+ 363+ /- 25.3 (n = 6), ****p<0.0001; ssGOF, DIP-γ -/+ 363+ /- 25.3 (n = 6), ssGOF, DIP-γ -/-175.3+ /- 15.2 (n = 6), ****p<0.0001; DIP-γ -/-134+ /- 14.3 (n = 9), ssGOF, DIP-γ -/-175.3+ /- 15.2 (n = 6), ***p=0.0001; sev -/+; ; DIP-γ -/+ 252.8+ /- 16.3 (n = 6), sev -/-; ; DIP-γ -/+ 77.3+ /- 12.5 (n = 6), ****p<0.0001; sev -/-; ; DIP-γ -/+ 77.3+ /- 12.5 (n = 6), sev -/-; ; DIP-γ -/- 50.5+ /- 7.8 (n = 6), **p=0.0012; DIP-γ -/- 134+ /- 14.3 (n = 9), sev -/-; ; DIP-γ -/- 50.5+ /- 7.8 (n = 6), ****p<0.0001; lGMR Gal4 control 333+ /- 9.8 (n = 6), lGMR Gal4 >UAS-dpr11 509.7+ /- 41.5 (n = 6), ****p<0.0001. (D) pDm8 cell numbers decrease dramatically when R7 PRs are absent, when they are converted to the yR7 fate (ssGOF), or when they all express Dpr11 (dpr11GOF). pDm8 cell number in wild-type, ssGOF, sev mutant, Dpr11 overexpression: DIP-γ -/+ 179.4+ /- 25.6 (n = 11), ssGOF, DIP-γ -/+ 28.7+ /- 4.3 (n = 6), ****p<0.0001; ssGOF, DIP-γ -/+ 28.7+ /- 4.3 (n = 6), ssGOF, DIP-γ -/-59.5+ /- 8.8 (n = 6), ****p<0.0001; DIP-γ -/-204.9+ /- 27.2 (n = 12), ssGOF, DIP-γ -/-59.5+ /- 8.8 (n = 6), ****p<0.0001; lGMR Gal4 control 127.7+ /- 18.7 (n = 6), lGMR Gal4 >UAS-dpr11 42.2+ /- 5.6 (n = 6), ****p<0.0001; sev -/+; ; DIP-γ -/+ 188.5+ /- 10.2 (n = 6), sev -/-; ; DIP-γ -/+ 39.3+ /- 6.3 (n = 6), ****p<0.0001; sev -/-; ; DIP-γ -/+ 39.3+ /- 6.3 (n = 6), sev -/-; ; DIP-γ -/- 52.5+ /- 4.8 (n = 6), **p=0.002. (E-F, H) Dpr11 overexpression in the retina converts all medulla columns to y by selecting for yDm8. Pupal medullary neuropil (~45 hr - 48hr APF) of (E) lGMR-Gal4 control and (F) lGMR-Gal4 >UAS-dpr11 labeled with anti-Chp (red) and anti-DIP-γ (green). Cross-section views of the medulla shown (E-F). Two p columns (red only) are circled in (E). Quantitation in (H). Maximum intensity projection; scale bar 5 µm. (G) Conversion of all R7 PRs to pR7 fate results in loss of yDm8 neurons. yDm8 cell number in dveGOF counted with anti-Dac and anti-GFP for DIP-γMi>GFP reporter. lGMR Gal4 control 271+ /- 18.3 (n = 6), lGMR Gal4 >UAS Dve 110.3+ /- 3.8 (n = 6), ****p<0.0001. Graph shows mean + /- std. deviation and unpaired Student’s t-test p-values. (H) Percentage of Dm8 home columns that are yDm8 (quantitated from images like those in E and F). lGMR Gal4/+ 0.62+ /- 0.02 (n = 199, 5 OLs), lGMR Gal4 >UAS-dpr11 0.98+ /- 0.01 (n = 234, 6 OLs), ****p<0.0001; n represents total number of columns analyzed. Graph shows mean + /- std. deviation and unpaired Student’s t-test p-values.
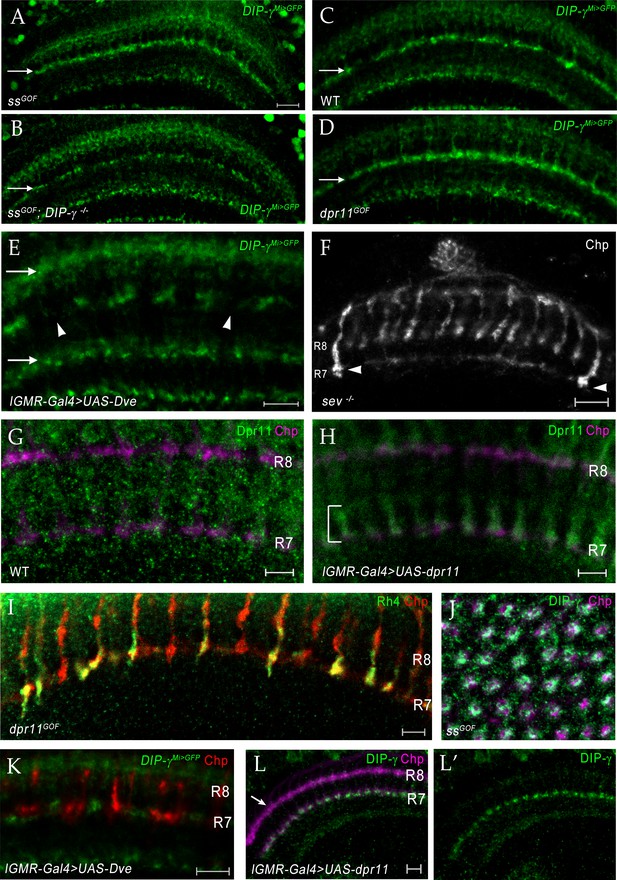
Changing R7 fate or expressing Dpr11 affects yDm8 and pDm8 survival.
(A–D) Conversion of all R7 PRs to yR7 fate in ssGOF results in loss of pDm8 arbors. (A) ssGOF; DIP-γ -/+ (B) ssGOF; DIP-γ -/- (C) Wild-type control (D) lGMR >UAS-dpr11 (dpr11GOF). DIP-γMi>GFP reporter in M6 layer (arrow) is shown for all panels. Gaps representing pDm8 arbors are present in wild-type control (C) but absent in ssGOF (A), where pR7 PRs have been replaced by yR7 PRs. Ectopic expression of Dpr11 in all PRs mimics ssGOF (compare panels A and D). (E) yDm8 arbors are lost when yR7 are converted to pR7 by expressing Dve in all PR. There are large gaps (arrowheads) in yDm8 labeling in layer M6 of the neuropil indicating extensive yDm8 cell death (compare to control (C), which has only small gaps). yDm8 arbors in M6 were examined with DIP-γMi>GFP reporter in lGMR-Gal4 >UAS Dve. The other layers in the distal medulla (M3, top arrow) and in the proximal medulla (bottom arrow) that label with the reporter, are unaffected in dveGOF (arrows in (E); compare to those layers in (C)). (A)-(E) single confocal slices; all scale bars are 10 µm. (F) Some R7 PRs remain in a sev ‘null’ mutant. A single confocal slice of a sev14/sev14 (putative amorphic mutant; see Flybase) adult, labeled with anti-Chp. There are two R7 axons that project to M6 visible in this slice (arrowheads). Scale bar, 10 µm. (G–H) Overexpressed Dpr11 can be detected on R7 terminals in lGMR-Gal4 >UAS-dpr11. (G) lGMR-Gal4 control (H) lGMR-Gal4 >UAS-dpr11. Pupal optic lobes (~43 hr APF) were labeled with anti-Dpr11 (green) and anti-Chp (magenta). Note distinct Dpr11 labeling (bracket) of R7 axons/terminals in (H), and its absence in (G). This antibody is weak and shows no labeling of specific neurons in wild-type. Maximum intensity projection; scale bar 5 µm. (I) Expression of Dpr11 in pR7 PRs does not convert them to the y fate. R7 terminals in dpr11GOF adult labeled with Rh4-lacZ and Chp. Note that 4 of the R7 terminals are labeled only by Chp and are therefore pR7. If Dpr11 expression in all PRs produced the same effect as ss expression, all R7 terminals would express both Rh4-lacZ and Chp, because they would all be y. Scale bar, 5 µm. (J) Ss overexpression (ssGOF) converts all R7 PRs to yR7, and converts almost all columns to yellow due to selection of yDm8, which ensures their survival. Cross-section view of medulla neuropil at 54 hr APF labeled with anti-Chp (magenta) and anti-DIP-γ (green). Compare to Figure 9F. Maximum intensity projection. (K) R7 and R8 PR labeling in DveGOF. lGMR-Gal4 >UAS Dve labeled for Chp (red) and GFP (green) in DIP-γ Mi>GFP background. Single confocal slice. (L) yDm8 do not mistarget to the M3 layer in dpr11GOF when Dpr11 is expressed in all PRs with lGMR-Gal4. Horizontal view of 20 hr APF in lGMR-Gal4 >UAS-dpr11 labeled with anti-Chp (magenta) and anti-DIP-γ (green) at 20 hr APF.
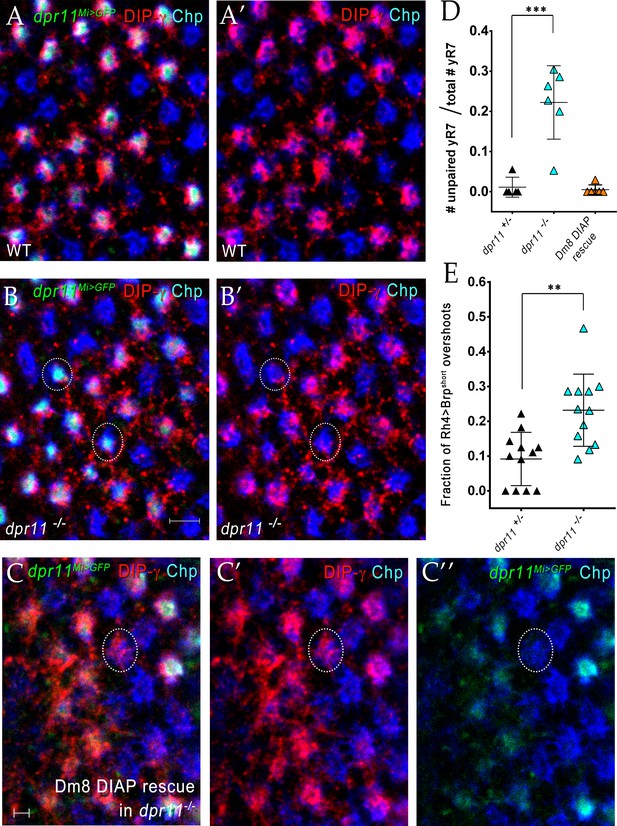
yDm8 arbors do not innervate pR7 home columns in dpr11 mutants.
(A)-(B) Surviving yDm8 do not mistarget to pR7 home columns in a dpr11 mutant. Mid-pupal optic lobes of dpr11Mi>GFP heterozygote reporter line (WT) and dpr11Mi>GFP /- (dpr11 -/- were labeled with anti-GFP for yR7 (green), anti-DIP-γ for yDm8 (red) and Chp for all PRs (blue). (A) and (B) show all three channels, and (A’) and (B’) show only red and blue. A yR7 column labeled by DIP-γ is defined as one in which there are red pixels directly on top of the blue and green Chp and dpr11Mi>GFP labeling. Note that all yR7 columns with green labeling in (A) (these appear white) are labeled by both red and blue in (A’). However, in dpr11 mutants, some yR7 columns (circled in (B) and (B’)) with green labeling have no red DIP-γ labeling on top of the column, indicating that these are vacant yellow columns that have no yDm8. These are quantitated in (D). All dpr11 mutant columns are either blue (pR7-Chp only), or blue+red+green (Chp+DIP-γ+ dpr11Mi>GFP), showing that no yDm8 mistarget to pR7 (i.e. there are no red+blue columns in (B)). (C) Rare mistargeting of yDm8 to pR7 in DIAP rescue of yDm8 in dpr11 mutant. Pupal optic lobes of Traffic Jam Gal4 >UAS DIAP in a dpr11-/- mutant were labeled as in A-B: anti-GFP for yR7, anti-DIP-γ for yDm8, and Chp for all PRs. 2/119 pR7 (one is circled) showed yDm8 mistargeting (n = 6 OLs). (D) Some yR7 home columns are uninnervated in dpr11 mutants due to yDm8 cell death, and this loss of innervation is rescued when cell death is prevented. Pupal optic lobes of dpr11Mi>GFP reporter line (dpr11-/+), dpr11-/- and DIAP rescue in dpr11 mutant were labeled with anti-GFP for yR7, anti-DIP-γ for yDm8 and Chp for all PRs. Number of yR7 columns in the medulla were quantitated in 6 × 6 grids drawn on images of cross-section views. Number of yR7 columns without yDm8 partners was determined by examining how many dpr11Mi>GFP labeled yR7 did not have yDm8 labeling adjacent to them. Graph shows mean + /- std. deviation and unpaired Student’s t-test p-values. dpr11-/+ 0.011+ /- 0.02, dpr11-/- 0.22+ /- 0.09, ***p=0.0008 (total number of yR7 columns examined: dpr11-/+: 87, dpr11-/-: 122) DIAP rescue in dpr11 mutant 0.005+ /- 0.01 (total number of yR7 columns examined: 181). (E) yR7 overshoots detected with a truncated Brp marker are increased in the dpr11 null mutant. We repeated our previously published analysis of yR7 overshoots in dpr11 mutants using the CRISPR-generated dpr11 null allele instead of dpr11Mi>GFP/Df (Carrillo et al., 2015; Xu et al., 2018). We used the same reporter as before, Rh4 driving a truncated version of Brp (Brp-short; Berger-Müller et al., 2013) and determined the number of overshoots in which Brp-short labeling was observed beyond (proximal to) M6. The quantitation was done blind by a person not involved in the experiment. Graph shows mean + /- std. deviation and unpaired Student’s t-test p-values. dpr11-/+ 0.09+ /- 0.08, dpr11-/- 0.23+ /- 0.1, **p=0.0011.
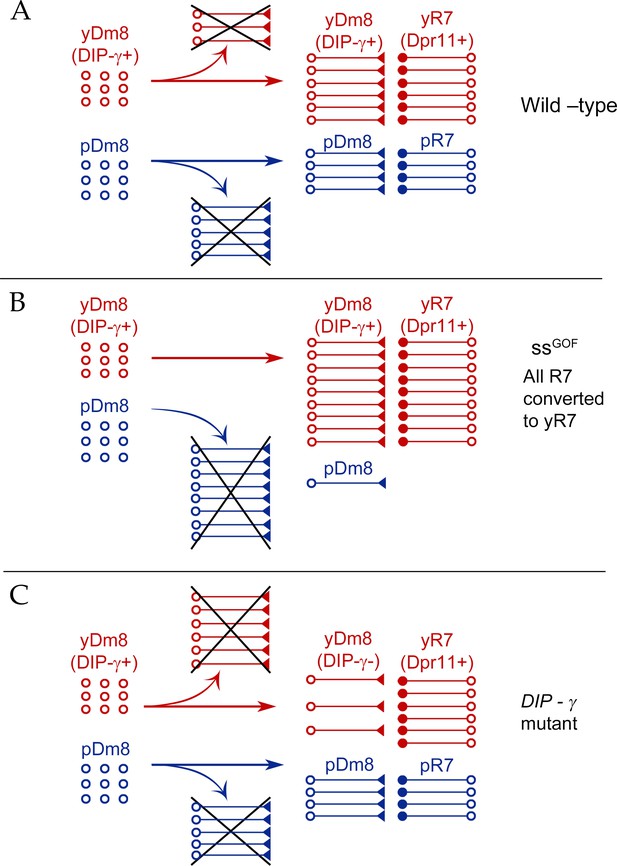
Model for Dm8 selection and survival.
(A) In wild-type, yDm8 neurons (red) are produced in excess and are selected by yR7 PRs for connectivity and survival. pDm8 neurons (blue) are also shown as being generated in excess. We have indicated their cell numbers prior to selection as being the same as yDm8, but this is arbitrary, since we do not know how many pDm8 are born. Unselected yDm8 and pDm8 neurons die due to the absence of survival signals (indicated by Xs).~30% of the yDm8 neurons present at 15 hr APF die in wild-type (Figure 8A). (B) When all R7 PRs are converted to yR7 in ssGOF (or when they all express Dpr11), more yDm8 neurons survive and almost all pDm8 neurons die (Figure 9). (C) In DIP-γ mutants, yDm8 neurons are not selected by yR7 PRs and more of them die. ~75% of the yDm8 neurons present at 15 hr APF die in DIP-γ mutants (Figure 8A). Similar results are observed for dpr11 mutants. This means that some yR7 PRs remain uninnervated (Figure 9—figure supplement 2).
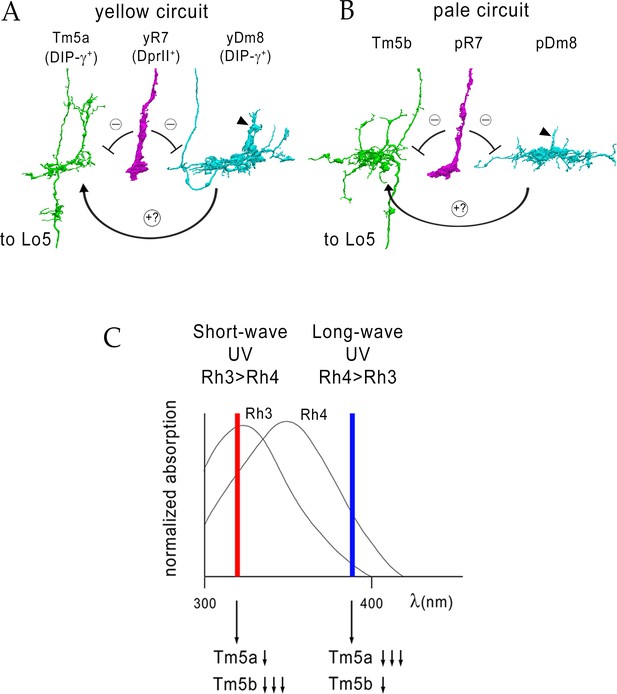
Model for UV wavelength discrimination by yellow and pale circuits.
(A) Cells in a yellow circuit, from column E. yR7-E, yDm8-E, and Tm5a-E are shown. yR7 inhibits both yDm8 and Tm5a (repression bars). yDm8 makes glutamatergic synapses (probably excitatory) onto Tm5a (arrow). Tm5a and Tm5b axons project to the 5th layer of the lobula. (B) Cells in a pale circuit, from column D. pR7-D, pDm8-D, and Tm5b-D are shown. The connections among the cells are the same as for the yellow circuit. The implication of these connection patterns is that R7 stimulation might inhibit Tm5a/b by both direct (histaminergic) and indirect (histaminergic inhibition of excitatory Dm8 glutamatergic output) pathways. yDm8-E (in A) and pDm8-D (in B) sprigs indicated by arrowheads. (C) Short-wave UV (red bar) would stimulate Rh3+ pR7 more than Rh4+ yR7, and might therefore produce more inhibition of Tm5b than of Tm5a. Long-wave UV (blue bar) would produce more inhibition of Tm5a than of Tm5b. These signals could be read out by Lo neurons that receive Tm5a and Tm5b inputs.
Tables
Lines and sources.
| Genotype | Source |
|---|---|
| yw hsFlp; UAS > CD2, y+>mCD8::GFP/Cyo; TM2/TM6B | Gift from C-H Lee |
| Rh4-lacZ | Bloomington |
| R24F06-p65.AD | Bloomington |
| DIP-γ Gal4-DBD | Gift from C Desplan |
| OK371-VP16AD | Gift from C-H Lee |
| R24F06-Gal4 | Bloomington, (Nern et al., 2015) |
| DIP-γMI03222(DIP-γMi>GFP), DIP-γMI03222 Gal4 | HJ Bellen |
| DIP-γnull | Gift from L Zipursky (Xu et al., 2018) |
| dpr11MI02231-GFP (dpr11Mi>GFP) | HJ Bellen |
| dpr11null | Gift from L Zipursky (Xu et al., 2018) |
| dpr11 RNAi GD23243 (III) | VDRC |
| UAS-DIP-γsh (II) | This study |
| UASp-DIAP I (II) | Bloomington |
| UAS-DIAP1-myc/TM6b | Gift from L Zipursky (Xu et al., 2018) |
| UAS-dveA-9B2/TM3 | Gift from Hideki Nakagoshi (Nakagawa et al., 2011) |
| lGMR-Ss (II) | Gift from C Desplan |
| lGMR-Gal4 | Bloomington |
| sev14 | Bloomington |
| UAS-dpr11sh (II) | This study |
| Rh4-lexA::p65, lexAop2-brp-shortcherry | Gift from T Suzuki |
| UAS-DIP-α-V5 | Gift from L Zipursky (Xu et al., 2018) |
List of genotypes in figures and graphs.
| Figures | Short genotype | Complete genotype |
|---|---|---|
| 1E | dpr11Mi>GFP/+ | dpr11MI02231-GFP/+ |
| 1 F-F’ | dpr11MI02231-GFP/Rh4-LacZ | |
| 1 G-G’, H | 10xUAS-mCD8::RFP/+; DIP-γMI03222/R24F06-Gal4 | |
| 1I | dpr11MI02231-GFP/Rh4-LacZ and 10xUAS-mCD8::RFP/+; DIP-γMI03222/R24F06-Gal4 | |
| 2B, D | yw hsflp/+; UAS > CD2, y+>mCD8::GFP/R24F06-p65.AD; Rh4-lacZ/DIP-γMI03222Gal4.DBD | |
| 2C, E | yw hsflp/+; UAS > CD2, y+>mCD8::GFP/+; Rh4LacZ/R24F06-Gal4 | |
| 2F | dpr11Mi>GFP/+ | dpr11MI02231-GFP/+ |
| 2G | DIP-γ Gal4 > Flp, Pan-Dm8 LexA > FSF GFP | 20xUAS-flp/+; R24F06-LexA/+; DIP-γMI03222Gal4, LexAop FRT > stop > FRT mCD8::GFP/+ |
| 2 H-H’ | DIP-γ Mi>GFP, Pan-Dm8 > RFP | 10xUAS mCD8::RFP/+; DIP-γMI03222- GFP/R24F06-Gal4 |
| 2I-J | yDm8 | yw-hsflp/+; UAS > CD2,y+>mCD8GFP/R24F06-p65.AD; Rh4LacZ/DIP-γ Gal4 DBD |
| 2I-J | pDm8 | yw-hsflp/+; UAS > CD2,y+>mCD8 GFP/Sp; R24F06Gal4, DIP-γ Gal80/Rh4LacZ |
| 2I-J | pDm8 | yw-hsflp/+; UAS > CD2,y+>mCD8GFP/+; Rh4LacZ/R24 F06Gal4 |
| 5A-B, I, K | DIP-γ -/+ | yw hsflp/+; UAS > CD2, y+>mCD8::GFP/R24F06-p65.AD; Rh4-lacZ/DIP-γMI03222Gal4.DBD |
| 5C-D, I, K | DIP-γ -/- | yw hsflp/+; UAS > CD2, y+>mCD8::GFP/R24F06-p65.AD; DIP-γnull/DIP-γMI03222Gal4.DBD |
| 5E-F, I, K | dpr11 -/- | yw hsflp/+; UAS > CD2, y+>mCD8::GFP/Rh4LacZ; R24F06-Gal4, dpr11null/dpr11null |
| 5G, H | DIP-γ -/+ | yw-hsflp/+; UAS > CD2,y+>mCD8GFP/R24F06p65AD; Rh4LacZ/DIP-γ Gal4DBD |
| 5G | DIP-γ -/- | yw hsflp/+; UAS > CD2, y+>mCD8::GFP/R24F06-p65.AD; DIP-γnull/DIP-γMI03222Gal4.DBD |
| 5J, L | DIP-γ -/+ | yw-hsflp/+; UAS > CD2,y+>mCD8GFP/R24F06p65AD; Rh4LacZ/DIP-γ Gal4 DBD |
| 5J, L | DIP-γ -/- | yw-hsflp/+; UAS > CD2,y+>mcd8GFP/R24F06p65AD; DIP-γ Gal4 DBD, Rh4LacZ/DIP-γnull |
| 5J, L | dpr11 -/- | yw-hsflp/+; UAS > CD2,y+>mcd8GFP/Rh4LacZ; R24F06Gal4, dpr11null/dpr11null |
| 6A-C and Figure 6—videos 1–2 | WT | yw hsflp/+; UAS > CD2, y+>mCD8::GFP/R24F06-p65.AD; Rh4-lacZ/DIP-γMI03222Gal4.DBD |
| 6D and Figure 6—videos 3–4 | DIP-γ -/- | yw hsflp/+; UAS > CD2, y+>mCD8::GFP/R24F06-p65.AD; DIP-γnull/DIP-γMI03222Gal4.DBD |
| 7A-B, D | DIP-γ -/+ | 10xUAS-mCD8::RFP/+; R24F06-Gal4/DIP-γMI03222-GFP |
| 7A-B | DIP-γ -/- | 10xUAS-mCD8::RFP/+; R24F06-Gal4, DIP-γ null/DIP-γMI03222-GFP |
| 7A-B | dpr11 -/+ control | 10xUAS-mCD8::RFP/+; R24F06-Gal4, dpr11null/DIP-γMI03222-GFP |
| 7A-B, H | dpr11 -/- | 10xUAS-mCD8::RFP/+; R24F06-Gal4, dpr11null/dpr11null, DIP-γMI03222-GFP |
| 7A-B | dpr11 -/-, DIP-γ -/- | 10xUAS-mCD8::RFP/+; R24F06-Gal4, dpr11null, DIP-γnull/dpr11null, DIP-γMI03222-GFP |
| 7A | DIP-γ -/+ control | R24F06-LexA/+; 13xLexAop-tdTomato::myr, DIP-γMI03222-GFP /+ |
| 7A, E | DIP-γ -/-control | R24F06-LexA/+; DIP-γMI03222 Gal4/ 13xLexAop-tdTomato::myr, DIP-γMI03222-GFP |
| 7A, F | DIP-γ rescue | UAS-DIP-γsh/R24F06-LexA; DIP-γMI03222 Gal4/ 13xLexAop-tdTomato::myr, DIP-γMI03222-GFP |
| 7A-B, G | yDm8 DIAP rescue | UASp-DIAP/R24F06-LexA; DIP-γMI03222 Gal4/ 13xLexAop-tdTomato::myr, DIP-γMI03222-GFP |
| 7C | DIP-γ -/+ | DIP-γMI03222-GFP/+ |
| 7C | DIP-γ -/- | DIP-γMI03222-GFP/DIP-γnull |
| 7C | dpr11 -/- | dpr11null/dpr11null, DIP-γMI03222-GFP |
| 7C | lGMR Gal4 control | lGMR-Gal4/+; dpr11null, DIP-γMI03222-GFP/+ @29 deg |
| 7C | lGMR Gal4/dpr11 RNAi | lGMR-Gal4/+; dpr11null, DIP-γMI03222-GFP/dpr11 RNAi GD23243 (III) @29 deg |
| 7C | lGMR Gal4/+ | lGMR-Gal4/+; DIP-γMI03222-GFP/+ |
| 7C | lGMR Gal4/UAS-DIP-γ | lGMR-Gal4/UAS-DIP-γ; DIP-γMI03222-GFP/+ |
| 7C | lGMR Gal4/UAS-DIP-α | lGMR-Gal4/+; DIP-γMI03222-GFP/UAS-DIP-α-V5 |
| 8A, B-D | DIP-γ -/+ | DIP-γMI03222-GFP/+ |
| 8A | DIP-γ -/- | DIP-γMI03222-GFP/DIP-γnull |
| 8A | yDm8 DIAP rescue | UASp-DIAP/+; DIP-γMI03222Gal4/DIP-γMI03222-GFP |
| 8C-E | DIP-γ -/+ | OK371 VP16/UAS-Apo; DIP-γ Gal4 DBD/+ |
| 8C-E | DIP-γ -/- | OK371 VP16/UAS-Apo; DIP-γ Gal4 DBD/DIP-γnull |
| 8E | dpr11Mi>GFP | dpr11MI02231-GFP |
| 8F | yw | WT control |
| 9A | WT | dpr11MI02231-GFP/+ |
| 9B | ssGOF | lGMR-ss/+; dpr11MI02231-GFP/+ |
| 9C-D | DIP-γ -/+ | 10xUAS-mCD8::RFP/+; R24F06-Gal4/DIP-γMI03222-GFP |
| 9C-D | LGMR > ss; DIP-γ -/+ | 10xUAS-mCD8::RFP/lGMR ss; DIP-γMI03222-GFP/R24 F06-Gal4 |
| 9C-D | LGMR > ss; DIP-γ -/- | 10xUAS-mCD8::RFP/lGMR-ss; DIP-γMI03222-GFP/R24F06-Gal4, DIP-γnull |
| 9C-D | DIP-γ -/- | 10xUAS-mCD8::RFP/+; DIP-γMI03222-GFP/R24F06-Gal4, DIP-γnull |
| 9C-D | sev -/+;; DIP-γ -/+ | sev14/+;; R24F06-Gal4/UAS-mCD8::RFP, DIP-γMI03222-GFP |
| 9C-D | sev -/-;; DIP-γ -/+ | sev14/sev14;; R24F06-Gal4/UAS-mCD8::RFP, DIP-γMI03222-GFP |
| 9C-D | sev -/-;; DIP-γ -/- | sev14/sev14;; R24F06-Gal4, DIP-γnull/UAS-mCD8::RFP, DIP-γMI03222-GFP |
| 9C-D | lGMR Gal4 control | R24F06-LexA/+; 13xLexAop-tdTomato::myr, DIP-γMI03222-GFP/lGMR Gal4 |
| 9C-D | lGMR Gal4> UAS-dpr11 | UAS-Dpr11-sh/R24 F06-LexA; 13xLexAop-tdTomato::myr, DIP-γMI03222-GFP/lGMR Gal4 |
| 9E | WT | lGMR- Gal4/+ |
| 9F | dpr11GOF | lGMR-Gal4/UAS-dpr11sh |
| 9G | lGMR Gal4 control | lGMR-Gal4/+; DIP-γMI03222-GFP/+ |
| 9G | lGMR Gal4> UAS-Dve | lGMR-Gal4/+; DIP-γMI03222-GFP/UAS-dveA-9B2 |
| 9H | lGMR Gal4 | lGMR- Gal4/+ |
| 9H | lGMR Gal4> UAS-dpr11 | lGMR-Gal4/UAS-dpr11sh |
List of genotypes in Supplementary figures and graphs.
| Supplementary figures | Short genotype | Complete genotype |
|---|---|---|
| Figure 2—figure supplement 1A | yDm8 split-Gal4 driver | yw hsflp/+; UAS > CD2, y+>mCD8::GFP/R24 F06-p65.AD; Rh4-lacZ/DIP-γMI03222Gal4.DBD |
| Figure 2—figure supplement 1A | Pan-Dm8 driver | yw hsflp/+; UAS > CD2, y+>mCD8::GFP/+; Rh4LacZ/R24F06-Gal4 |
| Figure 2—figure supplement 1C–C’ | yw hsflp/+; UAS > CD2, y+>mCD8::GFP/OK371 dVP16.AD; Rh4 LacZ/DIP-γMI03222Gal4.DBD | |
| Figure 5—source data 1 | WT: Pan-Dm8 driver | yw hsflp/+; UAS > CD2, y+>mCD8::GFP/+; Rh4LacZ/R24 F06-Gal4 |
| Figure 5—source data 1 | WT: yDm8 split-Gal4 driver | yw hsflp/+; UAS > CD2, y+>mCD8::GFP/R24F06-p65.AD; Rh4-lacZ/DIP-γMI03222Gal4.DBD |
| Figure 5—source data 1 | DIP-γ -/-: yDm8 split -Gal4 driver | yw hsflp/+; UAS > CD2, y+>mCD8::GFP/R24F06-p65.AD; DIP-γnull/DIP-γMI03222Gal4.DBD |
| Figure 5—source data 1 | dpr11 -/-: Pan-Dm8 driver | yw hsflp/+; UAS > CD2, y+>mCD8::GFP/Rh4LacZ; R24F06-Gal4, dpr11null/dpr11null |
| Figure 7—figure supplement 1 | DIP-γ>>GFP, DIP-γ- Gal4 > UAS-DIAPmyc | UAS-diap1.myc, DIP-γMI03222-GFP/ DIP-γMI03222 Gal4 |
| Figure 8—figure supplement 1A–A’ | DIP-γMI03222-GFP/+ | |
| Figure 8—figure supplement 1B–B’ | WT control | |
| Figure 8—figure supplement 1C | dpr11MI02231-GFP/+ | |
| Figure 8—figure supplement 2A,C | DIP-γ -/+ | DIP-γMI03222-GFP/+ |
| Figure 8—figure supplement 2B,D | DIP-γ -/- | DIP-γMI03222-GFP/DIP-γnull |
| Figure 9—figure supplement 1A | ssGOF | lGMR-ss/+; DIP-γMI03222-GFP |
| Figure 9—figure supplement 1B | ssGOF; DIP-γ -/- | lGMR-ss/+; DIP-γMI03222-GFP/DIP-γnull |
| Figure 9—figure supplement 1C | WT | lGMR-Gal4/+; DIP-γMI03222-GFP/+ |
| Figure 9—figure supplement 1D | dpr11GOF | lGMR-Gal4/UAS-dpr11sh; DIP-γMI03222-GFP/+ |
| Figure 9—figure supplement 1E | lGMR-Gal4 > UAS-Dve | lGMR-Gal4/+; DIP-γMI03222-GFP/UAS-dveA-9B2 |
| Figure 9—figure supplement 1F | sev -/- | sev14/sev14;; R24F06Gal4/DIP-γMI03222-GFP, UAS-mCD8RFP |
| Figure 9—figure supplement 1G | WT | lGMR Gal4/+ |
| Figure 9—figure supplement 1H | lGMR-Gal4 > UAS-dpr11 | lGMR Gal4/+; UAS-dpr11sh |
| Figure 9—figure supplement 1I | dpr11GOF | lGMR Gal4/Rh4-lacZ; UAS-dpr11sh |
| Figure 9—figure supplement 1J | ssGOF | lGMR-ss/+ |
| Figure 9—figure supplement 1K | lGMR-Gal4 > UAS-Dve | lGMR-Gal4/+; DIP-γMI03222-GFP/UAS-dveA-9B2 |
| Figure 9—figure supplement 1L–L’ | lGMR-Gal4 > UAS-dpr11 | lGMR Gal4/+; UAS-dpr11sh |
| Figure 9—figure supplement 2A–A’ | WT | dpr11MI02231-GFP/+ |
| Figure 9—figure supplement 2B–B’ | dpr11 -/- | dpr11MI02231-GFP/dpr11null |
| Figure 9—figure supplement 2C–C” | dpr11 -/- | Traffic jam Gal4/UASp DIAP; dpr11MI02231-GFP/dpr11null |
| Figure 9—figure supplement 2D | dpr11 +/- | dpr11MI02231-GFP/+ |
| Figure 9—figure supplement 2D | dpr11 -/- | dpr11MI02231-GFP/dpr11null |
| Figure 9—figure supplement 2D | Dm8 DIAP rescue | Traffic jam Gal4/UASp DIAP; dpr11MI02231-GFP/dpr11null |
| Figure 9—figure supplement 2E | dpr11 +/- | Rh4-lexA::p65, lexAop2-brp-shortcherry/+; dpr11null/+ |
| Figure 9—figure supplement 2E | dpr11 -/- | Rh4-lexA::p65, lexAop2-brp-shortcherry/+; dpr11null/dpr11null |





