Structural basis for AcrVA4 inhibition of specific CRISPR-Cas12a
Figures
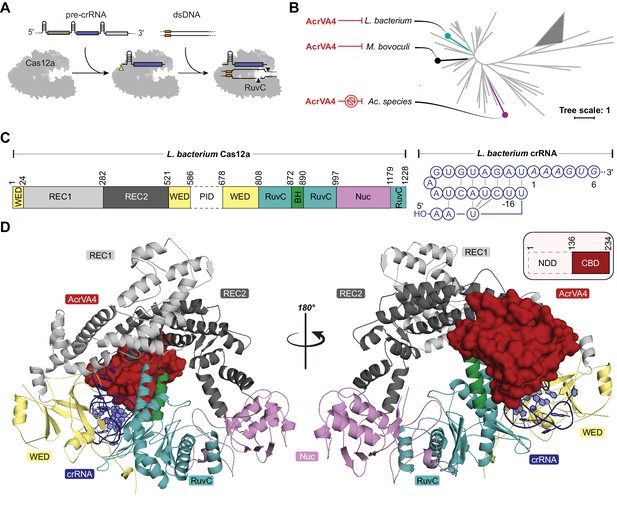
Overall structure of the LbCas12a-crRNA-AcrVA4 complex.
(A) Schematic representation of Cas12a activity. (B) Unrooted maximum likelihood phylogenetic tree of Type V-A CRISPR-Cas12a. Species known to be susceptible or unsusceptible to phage-derived AcrVA4 are highlighted. The triangle denotes collapsed branches of Cas12b-e. (C) Schematic representation of LbCas12a and the mature crRNA modeled within the cryo-EM structures. (D) Two views of the LbCas12a-crRNA complex (cartoon) bound to AcrVA4 (surface) shown related by a 180° rotation. The color scheme for the crRNA, LbCas12a, and AcrVA4 in panels B, C, and D are used throughout the manuscript.
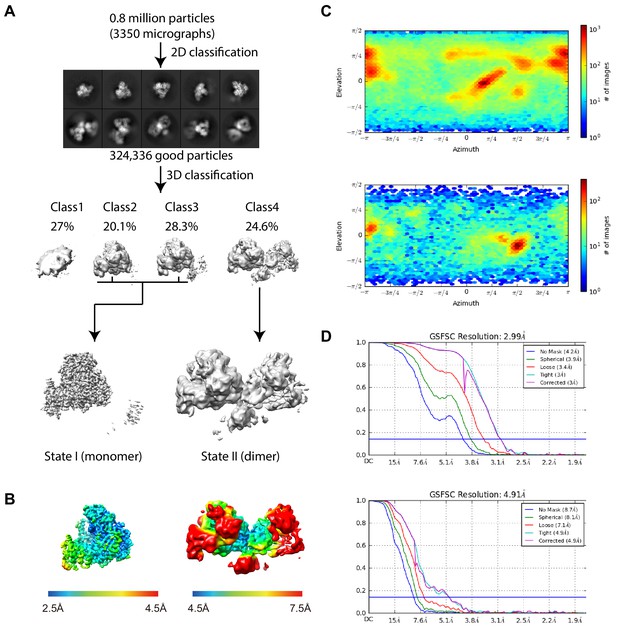
Cryo-EM data collection and 3D reconstruction.
(A) Electron microscopy analysis pipeline. From 3350 drift-corrected micrographs,~800,000 particles were picked and used for 2D classification. By 2D-based manual screening, 324,336 good particles were selected for 3D classification into four classes. From the classes that show a monomeric Cas12a architecture (Class 2 and Class 3), 156,979 particles were further used for refinement to generate reconstruction State I. From the class that shows a dimeric Cas12a architecture (Class 4), 79,786 particles were used for refinement to generate reconstruction State II. State I and State II were then independently refined to 3.0 Å and 5.0 Å, respectively. (B) The density maps for both states, colored by local resolution as calculated in Cryosparc. The resolution ranges from 2.5 Å to 4.5 Å in State I and 4.5 Å to 7.5 Å in State II. (C) Euler angle distribution of the refined particles belonging to State I (top) and State II (bottom). (D) Gold-standard Fourier shell correlation (GSFSC) curve calculated using two independent half-maps for State I (top) and State II (bottom).
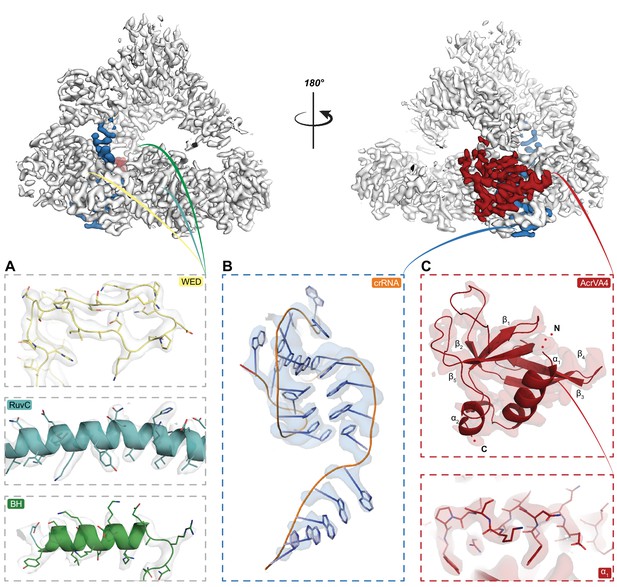
Cryo-EM density for State I.
(top) Segmented cryo EM density map for State I illustrating the LbCas12a protein chain (gray), the crRNA (blue) and the CBD of AcrVA4 (red) (map contoured to 10 σ). (A) Detailed views of cryo EM density describing the LbCas12a WED domain (yellow), RuvC domain (blue), and bridge helix (green) (maps contoured to 10 σ). (B) Detailed view of the cryo EM density describing the LbCas12a bound crRNA (map contoured to 15 σ). (C) Detailed view of the cryo EM density describing the overall fold of the AcrVA4 CBD (top) and an example of the high-resolution that allowed for side chain assignment in the map (bottom).
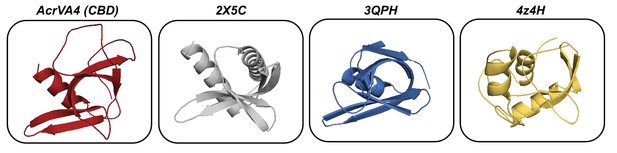
Structurally similar proteins to AcrVA4 CBD as determined by Dali search.
Cartoon representation of (A) AcrVA4 (red), (B) Pyrobaculum spherical virus protein (PDB code: 2X5C, gray), (C) TrmB archaeal transcriptional regulator protein (PDB code: 3QPH, blue), and (D) Argonaute protein PAZ domain (PDB code: 4Z4H, yellow).
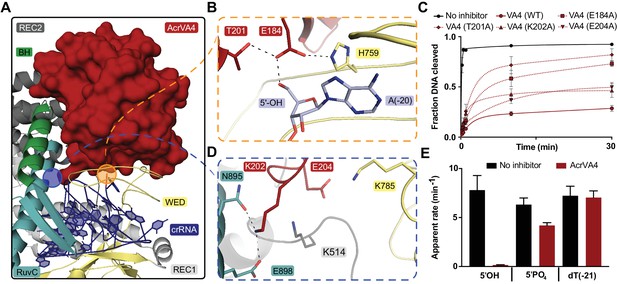
AcrVA4 recognizes the crRNA 5’-OH and Cas12a crRNA processing nuclease.
(A) AcrVA4 (surface) is shown on the LbCas12a WED domain (cartoon, yellow) and crRNA (blue, cartoon) braced against the bridge-helix (green, cartoon), RuvC (teal, cartoon), REC1 (cartoon, gray), and REC2 domains (cartoon, dark gray). (B) Detailed atomistic view of AcrVA4 (red) recognition of the WED domain pre-crRNA processing nuclease (yellow) and crRNA 5’OH (blue). (C) LbCas12a dsDNA cis-cleavage over time measured under single-turnover conditions in the presence or absence of AcrVA4 containing alanine substitutions (mean ∓ s.d., n = 3 independent measurements). Two-phase exponential decay experimental fits are shown as solid or dashed lines. (D) Detailed atomistic view of the AcrVA4 (red) interface with the LbCas12a RuvC (teal), REC2 (dark gray), and WED domains (yellow). (E) Bar-graph illustrating the apparent rate of LbCas12a-mediated dsDNA cis-cleavage under single-turnover conditions guided by a crRNA bearing a 5’-OH, 5’-PO4 or 5’-dT (−21) in the presence or absence of AcrVA4 (mean ∓ s.d., n = 3 independent measurements).
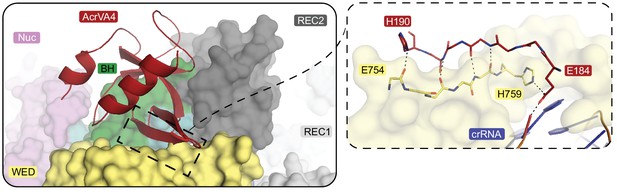
Extended interaction network between the AcrVA4 CBD and the LbCas12a-crRNA complex WED domain.
(left) LbCas12a (surface representation) bound to crRNA (cartoon, blue/orange) and AcrVA4 (cartoon, red). (right) An extended main chain β-sheet interface between AcrVA4 β3-β4 (sticks, red) and the LbCas12a WED domain (surface and sticks, yellow).
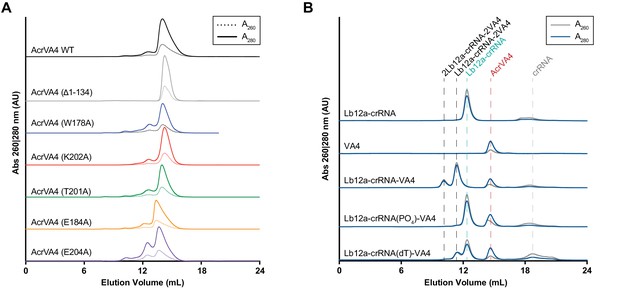
Size exclusion chromatography (SEC) traces for AcrVA4 mutants used in this study and the effect of 5'-crRNA chemistry on AcrVA4 binding to LbCas12a.
(A) SEC traces for AcrVA4 mutants purified and used in this study plotted as absorbance (AU) at 260 (dashed) and 280 (solid) nm against elution volume (mL). (B) SEC traces for LbCas12a and/or AcrVA4 (VA4) in the presence of crRNA bearing modification to the 5’ end chemistry. For each plot, the absorbance (AU) at 260 nm (gray) and 280 nm (blue) are shown against elution volume (mL).
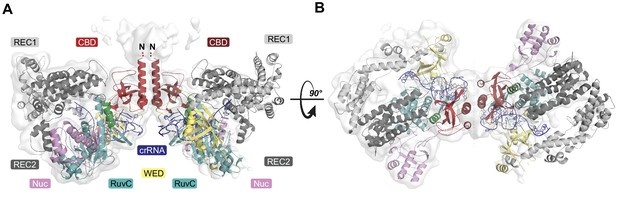
Overall structure of the heterotetrameric LbCas12a-crRNA-AcrVA4 complex.
(A–B) Dimeric LbCas12a-crRNA complex (cartoon) bound to homodimeric AcrVA4 (cartoon, two shades of red) shown related by a 90° rotation.
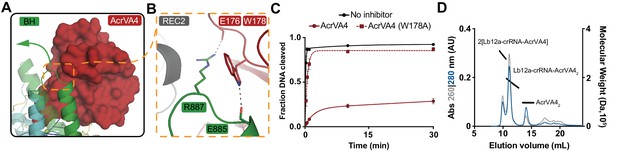
AcrVA4 locks the Cas12a bridge-helix to prevent DNA binding.
(A) AcrVA4 (surface) is shown braced against the bridge-helix (BH, green, cartoon) near the RuvC (teal, cartoon). The conformation of the LbCas12a bridge-helix when bound to DNA is shown semi-transparent with a green arrow denoting the direction of helix motion upon DNA binding. (B) Detailed atomistic view of AcrVA4 (red) recognition of the Cas12a bridge-helix (green). (C) LbCas12a dsDNA cis-cleavage over time measured under single-turnover conditions in the presence or absence of AcrVA4 containing alanine substitutions (mean ∓ s.d., n = 3 independent measurements). Two-phase exponential decay experimental fits are shown as solid or dashed lines. (D) Size-exclusion chromatography coupled di-angle light scattering (SEC-DALS) trace for AcrVA4 W178A in the presence of LbCas12a-crRNA. The absorbance at 280 nm (blue) and 260 nm (gray) are shown (left axis) with the linear region for the mass estimate corresponding to the relevant peaks (black lines, right axis).
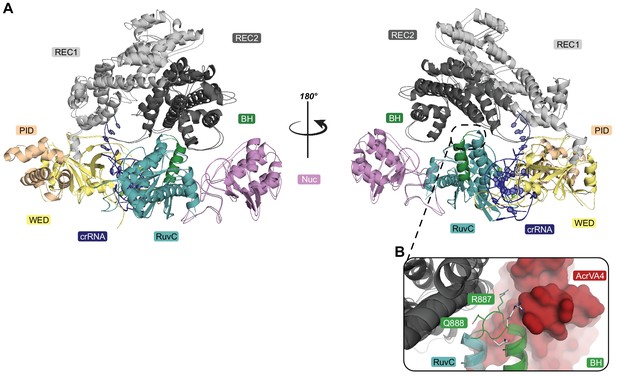
Superposition of LbCas12a-crRNA (PDB code: 5ID6) on our State I LbCas12a-crRNA-AcrVA4 complex.
(A) Both the LbCas12a-crRNA complex (5ID6) and our State I structure are shown as cartoons with the domains colored as in Figure 1. (B) A subtle distortion of the loop between the LbCas12a bridge-helix (cartoon, green) and RuvC (cartoon, teal) is observed in the presence of AcrVA4 (surface, red). R887 and Q888 adopt different conformations between the LbCas12a-crRNA complex (sticks, white) and the LbCas12a-crRNA-AcrVA4 complex (sticks, green).
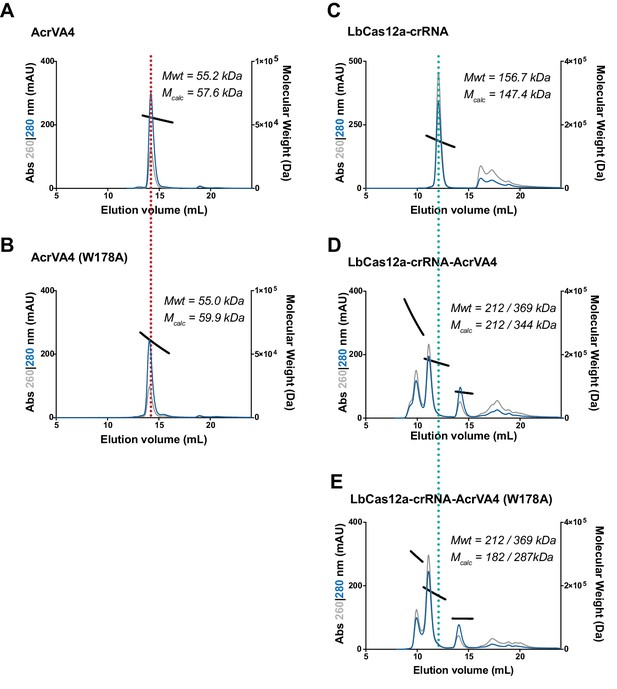
Size exclusion chromatography coupled di-angle light scattering (SEC-DALS) for AcrVA4 and LbCas12a-crRNA.
Size exclusion chromatography coupled light scattering traces for (A) AcrVA4, (B) AcrVA4 (W178A), (C) LbCas12a-crRNA complex, (D) LbCas12a-crRNA-AcrVA4 complex, and (E) LbCas12a-crRNA-AcrVA4 (W178A) complex. For each plot, the absorbance at 260 nm (gray) and 280 nm (blue) are shown (left axis) with the linear region for the mass estimate (black lines) corresponding to the relevant peaks (right axis). The theoretical and calculated molecular weights are annotated on each graph.
LbCas12a undergoes a large conformational change from the crRNA-bound state to a DNA-bound state.
Structural morph illustrating the conformational change between the LbCas12a-crRNA complex (PDB code: 5ID6) and the LbCas12a-crRNA-DNA complex (PDB code: 5XUS). Protein domains are colored as in Figure 1. The crRNA phosphate backbone is shown in orange with nucleobases in the cartoon ring representation.
The LbCas12a bridge-helix undergoes a large conformational change upon DNA binding.
Structural morph illustrating the conformational change between the LbCas12a-crRNA complex (PDB code: 5ID6) and the LbCas12a-crRNA-DNA complex (PDB code: 5XUS). Protein domains are colored as in Figure 1. The bases of the crRNA and the target DNA strand are shown in orange and white, respectively. Bridge-helix Arg 887 is shown with a stick representation.
AcrVA4 binding prevents LbCas12a bridge-helix conformational dynamics upon DNA binding.
Structural morph illustrating the conformational change between the LbCas12a-crRNA-AcrVA4 complex (PDB code: 6P7M) and the LbCas12a-crRNA-DNA complex (PDB code: 5XUS). Protein domains are colored as in Figure 1. Protein domains are colored as in Figure 1. The phosphate backbone of the crRNA and the target DNA strand are shown in orange and white, respectively. Bridge-helix Arg 887 is shown with a stick representation. AcrVA4 is shown as a semi-transparent surface.
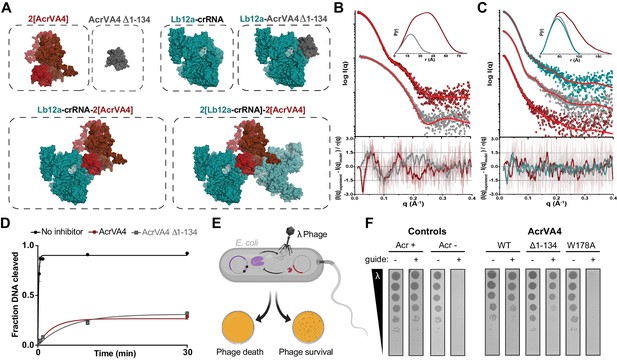
The C-terminal binding domain is sufficient for Cas12a inhibition.
(A) Atomistic models for the AcrVA4, AcrVA4 Δ1–134, LbCas12a-crRNA, LbCas12a-crRNA-AcrVA4 Δ1–134 and LbCas12a-crRNA-AcrVA4 that were used to match experimental small-angle X-ray scattering (SAXS) data shown in panel B and panel C. (B) Experimental data for AcrVA4, AcrVA4 Δ1–134 (red and gray) and theoretical (red line) SAXS profiles for the solution state models shown in the panel A. SAXS fits are shown together with the fit-residuals. (C) Experimental data for LbCas12a-crRNA, LbCas12a-crRNA-AcrVA4 Δ1–134 and LbCas12a-crRNA-AcrVA4 (teal, gray and red) and theoretical (red line) SAXS profiles for the solution state models shown in the panel A. SAXS fits are shown together with the fit-residuals. (B/C-insets) Normalized P(r) determined from the experimental SAXS curves. The area of each P(r) is normalized relative to the SAXS calculated molecular weights (Table 2). (D) LbCas12a dsDNA cis-cleavage over time measured under single-turnover conditions in the presence or absence of AcrVA4 or AcrVA4 Δ1–134 (mean ∓ s.d., n = 3 independent measurements). Two-phase exponential decay experimental fits are shown as solid lines. (E) Schematic representation of phage lambda plaque assay in E. coli. All strains harbor both a CRISPR-Cas plasmid (purple) and anti-CRISPR plasmid (red). Cas12a confers immunity to phage lambda, while anti-CRISPR inhibition of Cas12a restores plaquing. (F) Phage plaque assay to compare inhibition of LbCas12a by wild-type AcrVA4 and mutants relative to positive (AcrVA1: Acr +) and negative (AcrIIA4: Acr -) control anti-CRISPRs in E. coli. Ten-fold serial dilutions of heat-inducible phage lambda spotted on lawns of E. coli strains expressing the specified anti-CRISPR protein and a non-targeting guide (-) or lambda-targeting guide (+). Images shown are representative of the effect seen in replicates (n = 3 independent measurements).
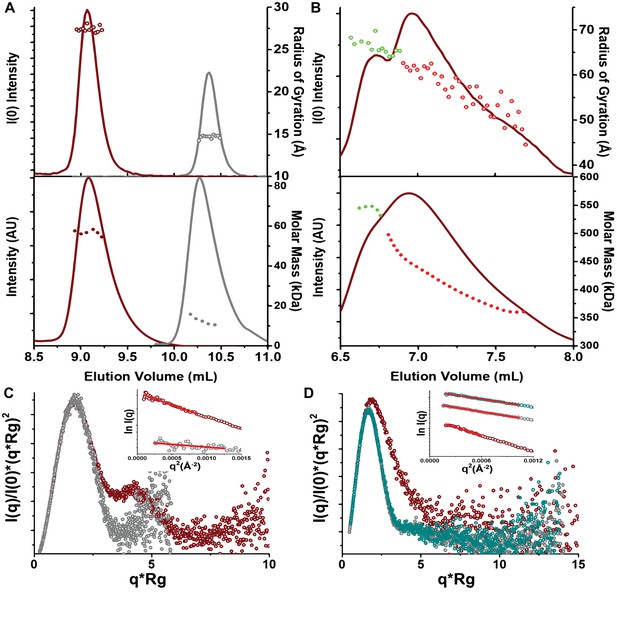
Small-angle X-ray scattering (SAXS) and multi-angle light scattering (MALS) data.
(A) Top, SEC-SAXS profiles for full-length AcrVA4 (red) and the truncated AcrVA4-Δ1–134 (gray). I(0) (lines) and Rg (symbols, Å) are shown for each collected frame across the relevant SEC peak. Bottom, SEC-MALS profiles for full-length AcrVA4 (red) and the truncated AcrVA4-Δ1–134 (gray). Intensity (AU) and calculated Molar Mass (kDa) are shown for each collected frame across the relevant SEC peak. (B) Top, SEC-SAXS profiles for LbCas12a-crRNA in the presence of full-length AcrVA4 (red). I(0) (lines) and Rg (symbols, Å) are shown for each collected frame across the relevant SEC peak. Bottom, SEC-MALS profiles for LbCas12a-crRNA in the presence of full-length AcrVA4 (red). Intensity (AU) and calculated Molar Mass (kDa) are shown for each collected frame across the relevant SEC peak. (C) Normalized Kratky plots for full-length AcrVA4 (red), truncated AcrVA4 Δ1–134 (gray) adjacent to (D) normalized Kratky plots for LbCas12a-crRNA (teal), LbCas12a-crRNA in the presence of full-length AcrVA4 (red), and LbCas12a-crRNA in the presence of truncated AcrVA4 Δ1–134 (gray).
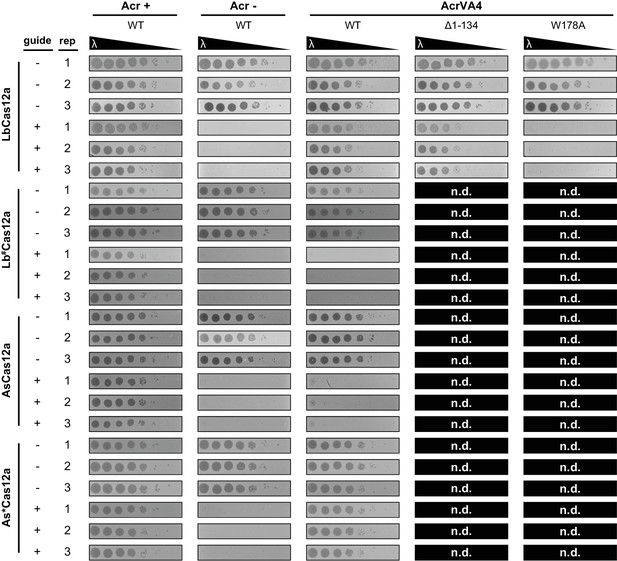
Phage lambda plaque assays.
Comparing inhibition of LbCas12a, Lb#Cas12a, AsCas12a, and chimeric As*Cas12a by wild-type AcrVA4 relative to positive (AcrVA1) and negative (AcrIIA4) control anti-CRISPRs in E. coli. Inhibition of Cas12-mediated phage immunity by AcrVA4 mutants was tested for LbCas12a and was not determined (n.d) for Lb#Cas12a, AsCas12a, and As*Cas12a. Ten-fold serial dilutions of heat-inducible phage lambda spotted on lawns of E. coli strains expressing the specified anti-CRISPR protein and a non-targeting guide (-) or lambda-targeting guide (+). Biological triplicates are shown.
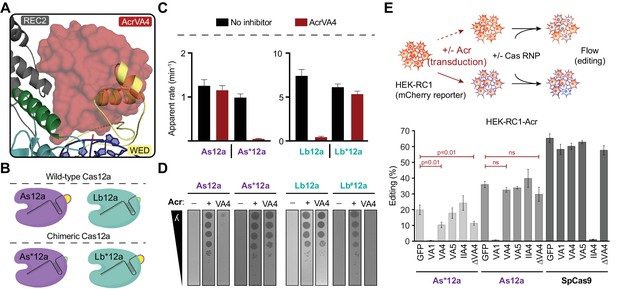
AsCas12a evades AcrVA4 by concealing its pre-crRNA processing nuclease.
(A) The C-terminal binding domain of AcrVA4 (surface) is shown superposed on the AsCas12a structure where it clashes with the WED domain (yellow, cartoon). (B) Schematic representation of the wild-type (Lb and As) and engineered chimeric (Lb* and As*) Cas12a constructs. (C) Bar-graph illustrating the apparent rate of wild-type or chimeric Cas12a-mediated dsDNA cis-cleavage under single-turnover conditions in the presence or absence of AcrVA4 (mean ∓ s.d., n = 3 independent measurements). Single-phase exponential fits to the cleavage kinetics from which the rate was derived can be found in Figure 6—figure supplement 2C. (D) Phage plaque assays to determine susceptibility of chimeric Cas12a to AcrVA4 in E. coli. Ten-fold serial dilutions of heat-inducible phage lambda spotted on lawns of E. coli strains expressing lambda-targeting guide, wild-type or chimeric (*/#) Cas12a, and AcrVA4 or the indicated control anti-CRISPR protein. Images shown are representative of the effect seen in replicates (n = 3 independent measurements). (E) CRISPR-Cas12a inhibition specificity in human cells. Schematic (top panel) showing human cells stably expressing a fluorescence reporter and doxycycline-inducible anti-CRISPR (Acr) constructs. Acr expression blocks genome editing upon transfection of susceptible Cas ribonucleoprotein (RNP) complexes, quantifiable by flow cytometry of mCherry fluorescence. Assessment of editing efficiency in HEK-RC1 reporter cells (bottom panel) expressing GFP or GFP-Acr polycistronic constructs (AcrVA1, AcrVA4, AcrVA5, AcrIIA4, AcrVA4 Δ1–134) and transfected with various Cas9 and Cas12a RNPs targeting the reporter. Note, in contrast to wild-type AsCas12a (As12a), editing by the AsCas12a-chimera (As*12a) was moderately susceptible to AcrVA4 and AcrVA4 Δ1–134 inhibition.
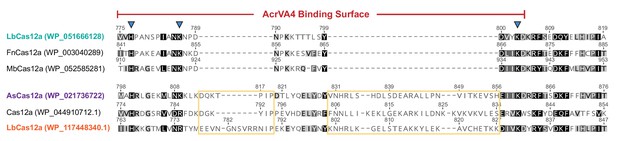
Multiple sequence alignment of Cas12a orthologs.
(A) Multiple sequence alignment of LbCas12a (teal), AsCas12a (purple), and other representative orthologs. The sequence the LbCas12a WED domain where AcrVA4 binds is denoted (red) along with the conserved catalytic triad of the pre-crRNA processing nuclease (blue arrows). The site of the additional sequence that occludes AcrVA4 binding is boxed in yellow.
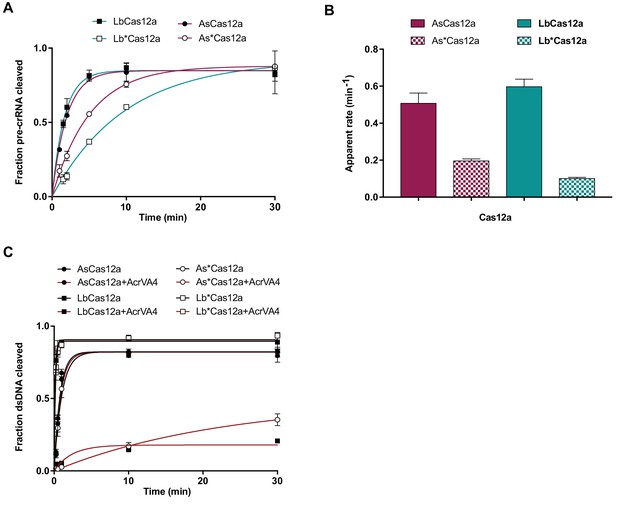
Wild-type and chimeric Cas12a pre-crRNA processing and dsDNA cleavage.
(A) Quantified fraction of 5’-radiolabeled pre-crRNA cleaved by wild-type or chimeric (denoted by *) Cas12a over time under single turnover conditions (mean ∓ sd, n = 3, independent experiments). Experimental fits are shown as solid lines and the calculated pseudo-first-order rate constant (kobs) (mean ∓ sd, n = 3) are plotted in (B). (B) Bar-graph illustrating the apparent rate of wild-type or chimeric (denoted by *) Cas12a-mediated pre-crRNA processing under single-turnover conditions as derived from panel (A). (C) Quantified fraction of 5’-radiolabeled dsRNA cleaved by wild-type or chimeric (denoted by *) Cas12a over time under single turnover conditions in the presence or absence of AcrVA4 (mean ∓ sd, n = 3, independent experiments). Experimental fits are shown as solid lines and the calculated pseudo-first-order rate constant (kobs) (mean ∓ sd, n = 3) are plotted in Figure 6C.
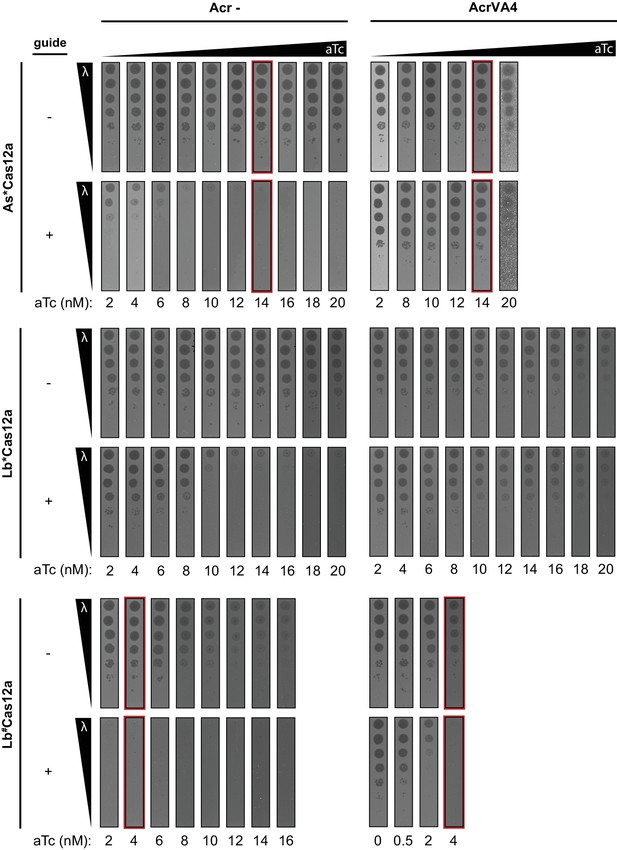
Optimizing induction of Cas12a chimeras in phage lambda plaque assay.
Determining aTc inducer concentration required to observe full As*Cas12a-, Lb*Cas12a-, and Lb#Cas12a-mediated immunity to phage lambda using E. coli plaque assay in the absence of a type V-A anti-CRISPR. Ten-fold serial dilutions of heat-inducible phage lambda spotted on lawns of E. coli strains expressing the specified anti-CRISPR protein (negative control AcrIIA4 and wild-type AcrVA4) and a non-targeting guide (-) or lambda-targeting guide (+). Inducer concentrations achieving full immunity without growth deficiency are highlighted (red) and indicate optimum conditions shown in Figure 6D and Figure 5—figure supplement 2. Images are representative of biological triplicates.
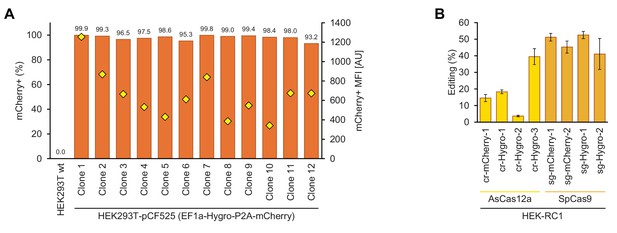
Human CRISPR-Cas and anti-CRISPR (Acr) genome editing assay.
(A) Monoclonal human genome editing reporter cell lines. Flow cytometry analysis of mCherry fluorescence percentage and median fluorescence intensity (MFI) in various monoclonal cell lines derived from HEK293T stably transduced with the lentiviral vector pCF525-EF1a-Hygro-P2A-mCherry and selected on hygromycin B. Clone 1, referred to as HEK-RC1, was selected for further work and used for CRISPR-Cas and Acr assays. (B) Quantification of guide RNA efficiencies for AsCas12a and SpCas9 RNP-mediated genome editing. HEK-RC1 reporter cells were transiently transfected with various AsCas12a or SpCas9 RNP complexes targeting the Hygro-P2A-mCherry polycistronic construct. Editing efficiency was measured as the loss of mCherry fluorescence per sample, compared to non-transfected control. Error bars indicate the standard deviation of triplicates. The cr-Hygro-3 (AsCas12a) and sg-Hygro-1 (Cas9, dual-guide system) were chosen for further CRISPR-Cas and Acr assays.
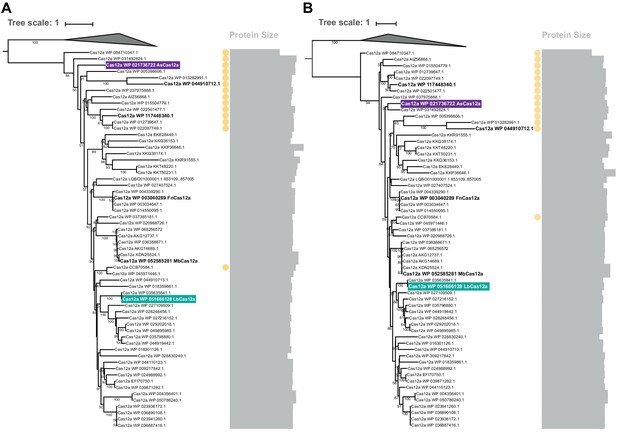
Phylogenetic reconstruction for Cas12a orthologs.
(A) Phylogenetic reconstruction of Cas12a orthologs relative to TnpB using the full-length Cas12a sequence or (B) the Cas12a RuvC alone. LbCas12a (teal) and AsCas12a (purple) are indicated along with other representative orthologs (bold). The presence of the additional sequence is denoted by a yellow circle to the right of the tree. The size of the Cas12a protein is shown as a gray bar adjacent to the relevant Cas12a ortholog.
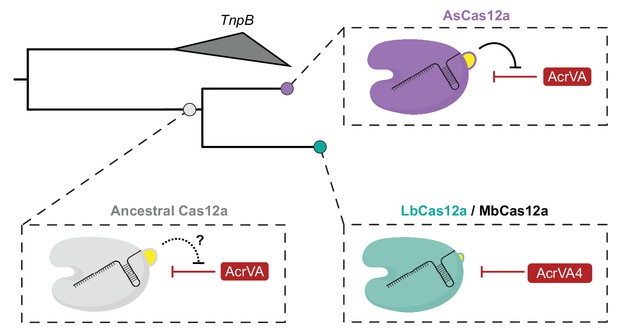
Model for the co-evolution of AcrVA4 susceptibility.
The ancestral Cas12a likely possessed a helical bundle, hindering any exploitation of the processing nuclease and bridge-helix (bottom left). While AsCas12a maintained the ancestral helical bundle and resistance to Acr activity at that site (top right), LbCas12a and related orthologs lost the helical bundle, providing a co-evolutionary opportunity for AcrVA4 allosteric inhibition.
Tables
Cryo-EM data collection, reconstruction, and model statistics.
https://doi.org/10.7554/eLife.49110.006| State I | State II | |
|---|---|---|
| PDB: 6P7M | EMDB: 20266 | PDB: 6P7N | EMDB: 20267 | |
| Data Collection | ||
| Microscope | FEI Titan Krios | FEI Titan Krios |
| Voltage (kV) | 300 | 300 |
| Camera | Gatan K2 Summit | Gatan K2 Summit |
| Defocus range (µm) | 0.7 ~ 2.2 | 0.7 ~ 2.2 |
| Pixel Size (Å) | 0.9 | 0.9 |
| Magnification | 135000 | 135000 |
| Electron Dose (e/Å2) | 47 | 47 |
| Total Particles | 324336 | 324336 |
| Reconstruction | ||
| Software | CryoSparc | CryoSparc |
| Symmetry Imposed | C1 | C1 |
| Final number of refined particles | 156979 | 79787 |
| Resolution of polished unmasked map (Å) | 4.2 | 8.7 |
| Resolution of polished masked map (Å) | 3 | 5 |
| Map Sharpening B-factor (Å2) | −101.4 | −123.5 |
| Refinement | ||
| Model Resolution cutoff (Å) | 3 | 5 |
| FSC threshold | 0.143 | 0.143 |
| Map CC (box) | 0.74 | 0.78 |
| Map CC (mask) | 0.79 | 0.51 |
| R..m.s Deviations | ||
| Bond lengths (Å) | 0.006 | 0.002 |
| Bond angles (°) | 0.607 | 0.524 |
| Molprobity score | 1.77 | 1.84 |
| Clashscore | 7.87 | 6.63 |
| Rotamer Outliers (%) | 0 | 1.33 |
| Ramachandran plot | ||
| Favored (%) | 94.43 | 94.43 |
| Allowed(%) | 5.57 | 5.57 |
| Outliers (%) | 0 | 0 |
SEC-SAXS-MALS-UV-vis data.
https://doi.org/10.7554/eLife.49110.020| Sample | Theoretical Mwt (kDa) | SAXS Mwt (kDa) | MALS Mwt (kDa) | Rg (Å) | Dmax (Å) | Px | Fit χ2 |
|---|---|---|---|---|---|---|---|
| AcrVA4 | 55.2 | ~55 | 55.9 (±0.092%) | 27.71 (±0.27) | 85 | 4 | 1.68 |
| AcrVA4 (Δ1–134) | 12.1 | ~12 | 12.6 (±0.628%) | 14.72 (±0.16) | 42 | 4 | 1.71 |
| Lb12a-crRNA | 156 | ~140 | 160.2 (±0.041%) | 36.15 (±0.46) | 103 | 4 | 1.39 |
| Lb12a-crRNA-AcrVA4 | 212 (369) | ~265 | 415.1 (±0.012%) | 52.76 (±2.55) | 183 | 3.9 | 1.75 |
| Lb12a-crRNA-AcrVA4 (Δ1–134) | 169 | ~155 | 176.5 (±0.044%) | 36.90 (±0.42) | 105 | 4 | 1.31 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Peptide, recombinant protein | AcrVA4 | Watters et al., 2018 | Addgene #115656 | |
| Peptide, recombinant protein | LbCas12a | Watters et al., 2018 | Addgene #115656 | |
| Peptide, recombinant protein | AsCas12a | Watters et al., 2018 | Addgene #113430 | |
| Strain, strain background (E. coli) | NEB 10-beta | New England Biolabs | ||
| Strain, strain background (E. coli) | MG1655 λ+(cI857 bor::kanR) | doi: 10.1073/pnas.0808831105 | ||
| Strain, strain background (E. coli) | Rosetta 2 (DE3) | Novagen | ||
| Strain, strain background (lambda) | λ phage | This paper | cI857 bor::kanR | |
| Sequence-based reagent | LbCas12a pre-crRNA | IDT | lab archive:rGJK_006 | rArGrArUrUrArArArUrArArUrUrUrCrUrArCrUrArArGrUrGrUrArGrArUrArArArGrUrGrCrUrCrArUrCrArUrUrGrGrArArArArCrGrU |
| Sequence-based reagent | AsCas12a pre-crRNA | IDT | lab archive:rGJK_008 | rGrArCrCrUrUrUrUrUrArArUrUrUrCrUrArCrUrCrUrUrGrUrArGrArUrArArArGrUrGrCrUrCrArUrCrArUrUrGrGrArArArArCrGrU |
| Sequence-based reagent | LbCas12a crRNA | IDT | lab archive:rGJK_017 | rArArUrUrUrCrUrArCrUrArArGrUrGrUrArGrArUrGrArUrCrGrUrUrArCrGrCrUrArArCrUrArUrGrA |
| Sequence-based reagent | AsCas12a crRNA | IDT | lab archive:rGJK_018 | rArArUrUrUrCrUrArCrUrCrUrUrGrUrArGrArUrGrArUrCrGrUrUrArCrGrCrUrArArCrUrArUrGrA |
| Sequence-based reagent | Non-target DNA strand | IDT | lab archive:dGJK_006 | GAC GAC AAA ACT TTA GAT CGT TAC GCT AAC TAT GAG GGC TGT CTG TGG AAT GCT A |
| Sequence-based reagent | Target DNA strand | IDT | lab archive:dGJK_007 | TAG CAT TCC ACA GAC AGC CCT CAT AGT TAG CGT AAC GAT CTA AAG TTT TGT CGT C |
| Sequence-based reagent | LbCas12a crRNA (dT −21) | IDT | lab archive:rGJK_051 | rArGrArUrUrArArATrArArUrUrUrCrUrArCrUrArArGrUrGrUrArGrArUrArArArGrUrGrCrUrCrArUrCrArUrUrGrGrArArArArCrGrU |
| Sequence-based reagent | LbCas12a crRNA (5'-PO4) | IDT | lab archive:rGJK_053 | /5Phos/rArArUrUrUrCrUrArCrUrArArGrUrGrUrArGrArUrArArArGrUrGrCrUrCrArUrCrArUrUrGrGrArArArArCrGrU |
| Software, algorithm | Prism7 | GraphPad | ||
| Software, algorithm | ImageQuantTL | GE Healthcare | ||
| Software,algorithm | ScÅtter 3.0 | BioIsis |
Additional files
-
Supplementary file 1
SDS-PAGE of purified proteins used in this study.
Purified proteins used in this study.
- https://doi.org/10.7554/eLife.49110.028
-
Supplementary file 2
Cas12a multiple sequence alignment.
Multiple sequence alignment of Cas12a proteins as described in Materials and methods.
- https://doi.org/10.7554/eLife.49110.029
-
Transparent reporting form
- https://doi.org/10.7554/eLife.49110.030





