Nitric oxide acts as a cotransmitter in a subset of dopaminergic neurons to diversify memory dynamics
Figures
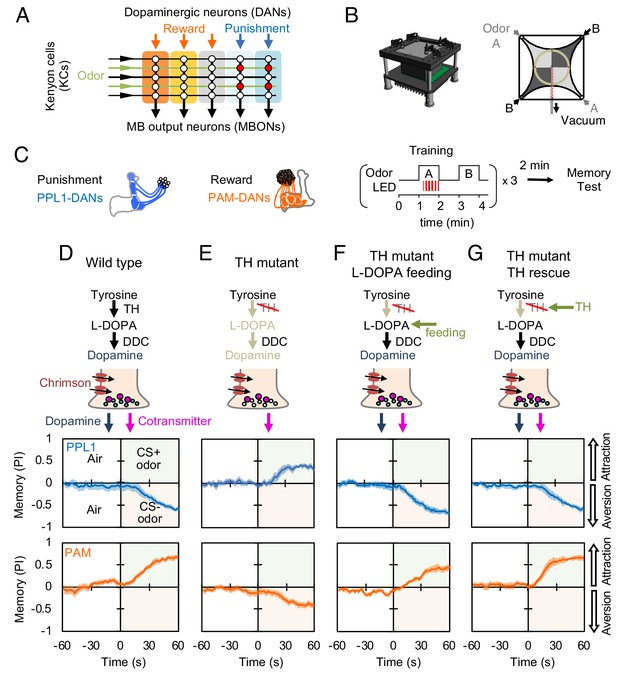
Dopaminergic neurons can induce memories without dopamine, but with opposite valence.
(A) Conceptual diagram of the circuit organization in the MB lobes. Sparse activity in the parallel axonal fibers of the KCs represent odor stimuli. DANs induce plasticity at KC to MBON synapses (represented by circles), when DAN and KC activity are coincident (red circles). The MB compartments (indicated by the colored rectangles) differ in their learning and memory decay rates. The actual MB lobes contains 15 compartments (Tanaka et al., 2008). (B) Design of the optogenetic olfactory arena and a diagram illustrating odor paths in the arena. (C) Schematic representation of the innervation patterns of the PPL1 (blue; R52H03-p65ADZp; TH-ZpGAL4DBD) and PAM cluster (orange; R58E02-p65ADZp; DDC-ZpGAL4DBD) DANs used to train flies. A diagram of the training protocol is also shown. Flies were trained and tested in the olfactory area. A 1 min odor exposure was paired with thirty 1 s pulses of red light (627 nm peak and 34.9 µW/mm2), followed by 1 min without odor or red light, and then presentation of a second odor for 1 min without red light. In one group of flies, odors A and B were 3-octanol and 4-methylcyclohexanol, respectively, while in a second group of flies, the odors were reversed. Memory was tested immediately after three repetitions of training bouts by giving flies a binary choice between the two odors in the olfactory area. (D–G) Odor memories induced by the collective optogenetic activation of PPL1-γ1pedc, PPL1-γ2α'1, PPL1-α'2α2 and PPL1-α3 DANs (upper panels; blue lines) or activation of PAM cluster DANs (lower panels; orange lines) in wild type (D), TH mutant (E), TH mutant feed 1 mg/ml L-DOPA and 0.1 mg/ml carbidopa (F), or TH mutant with cell-type specific expression of a wild-type TH cDNA (G; see the Materials and methods for drug treatment and supplemental information for genotypes). Time courses of the performance index (PI) during the test period are shown as the average of reciprocal experiments. The PI is defined as [(number of flies in the odor A quadrants) - (number of flies in odor B quadrants)]/ (total number of flies). Thick line and shading represent mean ± SEM. N = 12–16. Two split-GAL4 drivers R52H03-p65ADZp in attP40; TH-ZpGAL4DBD in VK00027 and R58E02-p65ADZp in attP40; DDC-ZpGAL4DBD in VK00027 were used for driving 20xUAS-CsChrimon-mVenus in PPL1 or PAM DANs, respectively.
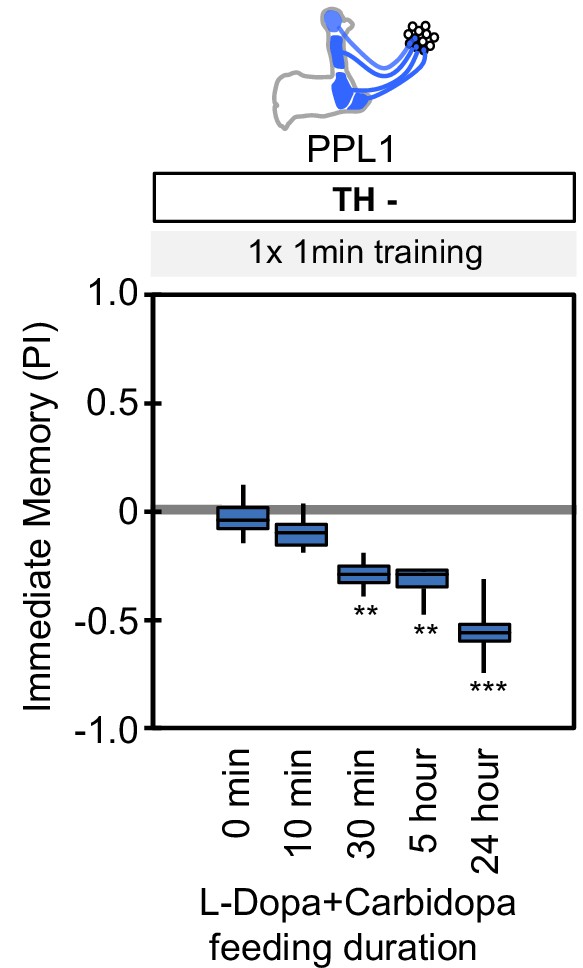
Duration of L-Dopa and Carbidopa feeding.
Flies were starved for 48 hr and then fed with fly food containing 1 mg/ml L-DOPA and 0.1 mg/ml carbidopa for the indicated time period. The flies were then trained by the collective optogenetic activation of PPL1-γ1pedc, PPL1-γ2α'1, PPL1-α'2α2 and PPL1-α3 DANs and tested for immediate memory. Note that NO-dependent memory is not detectable immediately after 1 × 1 min training (Figure 6A). As short as 30 min of feeding restored significant aversive memory. The whiskers represent the minimum and maximum. N = 12. Asterisk indicates significance from the 0 min feeding. **, p<0.01; ***, p<0.001.
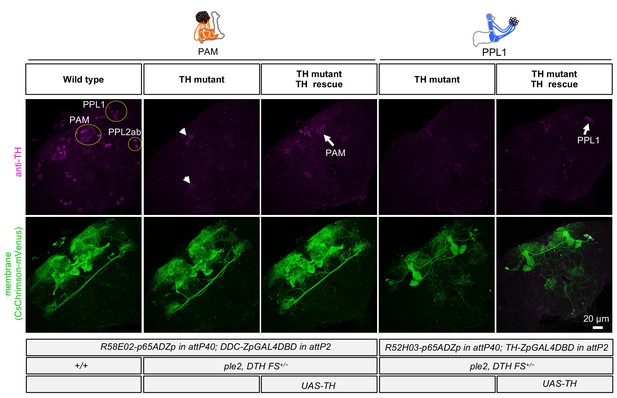
Cell-type-specific rescue of TH.
TH immunoreactivity (from left to right): PAM DANs in wild type; PAM DANs in TH mutant; PAM DANs in TH mutant with cell-type-specific rescue of TH using UAS-TH; PPL1 DANs in TH mutant; and or PPL1 DANs in TH mutant with cell-type-specific rescue of TH using UAS-TH. In the TH mutant background, TH immunoreactivity in the brain was completely abolished, except for a few cells dorsal medial surface of the brain and near the ventral lateral protocerebrum (arrow heads). With UAS-TH, TH immunoreactivity was restored in either PAM or PPL1 DANs depending on the split-GAL4 driver used (arrows). The expression pattern, visualized using UAS-CsChrimson-mVenus, of the PAM and PPL1 drivers used is shown in lower panels.
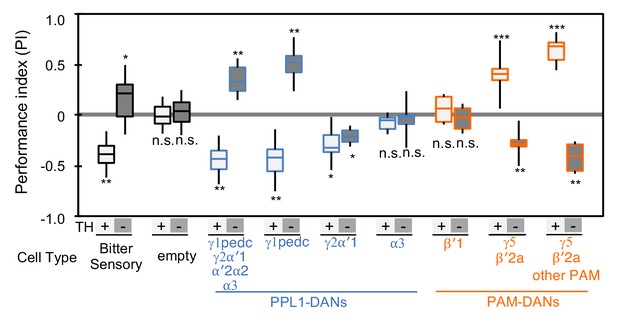
Cotransmitter effects differ among DAN cell types.
The PI of olfactory memories measured immediately after 3 × 1 min training in TH mutant/TH+ heterozygotes (+) or TH mutant/TH mutant (-) background. Odor presentation was paired with either activation of bitter taste neurons (Gr66a-GAL4) or the indicated subset of DANs using the optogenetic activation protocol diagramed in Figure 1C. A split-GAL4 driver without enhancer (empty) was used as a control. The bottom and top of each box represents the first and third quartile, and the horizontal line dividing the box is the median; the PI was calculated by averaging the PIs from the final 30 s of each test period (see legend to Figure 1). The whiskers represent the minimum and maximum. N = 8–16. Asterisk indicates significance from the empty-GAL4 control. Comparison with chance level (i.e. PI = 0) resulted in identical statistical significance: *, p<0.05; **, p<0.01; ***, p<0.001; n.s., not significant. See Figure 2—figure supplement 1 and www.janelia.org/split-gal4 for expression patterns of split-GAL4 drivers.
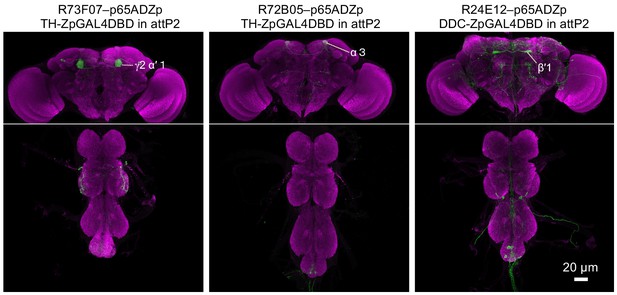
Expression patterns of driver lines.
Expression patterns of CsChrimson-mVenus driven by each of the three new split-GAL4 drivers made as part of this study. All other lines used were described previously. See supplemental table 1 for the list of driver lines and Aso et al., 2014a and www.janelia.org/split-gal4 for primary image data of all split-GAL4 lines.
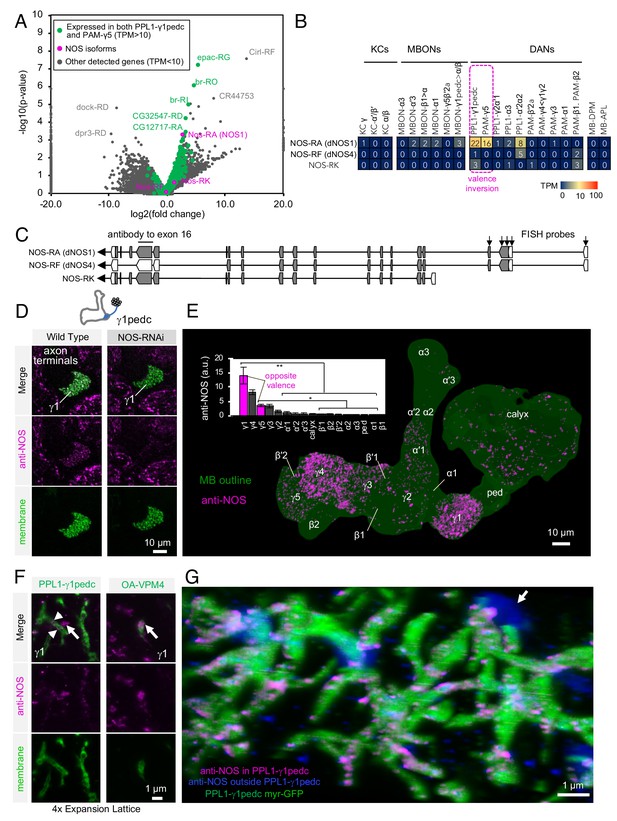
Identification of NOS1 in PPL1-γ1pedc and PAM-γ5 by RNA-Seq.
(A) The RNA-Seq data of two cell types that showed the valence-inversion phenotype (i.e. PPL1-γ1pedc and PAM-γ5) were pooled and compared against the pooled data of all other cell types examined (DANs, MBONs, KCs). The -log10 of p-values for comparing number of counted transcripts between the pooled data of PPL1-γ1pedc and PAM-γ5 relative to that of all other cell types were plotted against the log2 of fold changes observed in the expression levels of transcripts. Different splice isoforms of genes are plotted separately (dots). Green dots represent the 2981 transcripts expressed at levels above 10 transcripts per million (TPM) in both PPL1-γ1pedc and PAM-γ5. Gray dots represent the 31,539 transcripts with expression levels below 10 TPM in one or both of these two DANs. Dots on +20 or −20 x-axis represent splice isoforms that were detected only in the pooled PPL1-γ1pedc and PAM-γ5 or in other cell types. Magenta dots show the three splice isoforms of NOS. (B) Mean TPM of NOS splicing isoforms. The magenta dashed line highlights PPL1-γ1pedc and PAM-γ5, the two cell types that showed the valence-inversion phenotype. (C) Map of the NOS locus. Exons and protein coding sequences are depicted as boxes and gray boxes, respectively. Only the full-length isoform NOS-RA (dNOS1) produces a functional NOS protein (Stasiv et al., 2001). The antibody we used was raised against exon16 (Kuntz et al., 2017; Yakubovich et al., 2010). Arrows indicate the first four exons of NOS1 and NOS4 where 40 FISH probes were designed to recognize these, but not NOS-RK transcripts. See Materials and methods for details of the position and sequence of the probes. (D) NOS immunoreactivity was observed in the γ1pedc compartment of the MB. Immunoreactivity was markedly reduced by expressing NOS-RNAi in PPL1-γ1pedc. (E) Distribution of NOS-immunoreactivity inside the MB is displayed. Voxels above mean +2SD of the entire brain are shown in 12-bit scale in magenta. The insert shows a quantification of NOS-immunoreactivity in each MB compartment. Signal in γ1 was significantly higher than 12 compartments indicated by the bracket (Kruskal-Wallis with Dann’s test for selected pairs). Signal in γ5 was significantly higher than eight compartments indicated by the bracket; *, p<0.05; **, p<0.01; n = 12. (F) Lattice light sheet image of a 4x expanded brain shows colocalization of NOS immunoreactivity and the terminals of PPL1-γ1pedc (left; arrowheads). In addition, large but sparse NOS puncta were observed outside PPL1-γ1pedc (left; arrow); these puncta match the pattern of terminals of OA-VPM3 and/or OA-VPM4 (right; arrow), indicating that these octopaminergic neurons also expresses NOS. (G) 3D reconstruction of lattice light sheet image of the γ1 with pseudo colors for NOS inside (magenta) or outside (blue) the PPL1-γ1pedc DANs (green). The arrow indicates a large NOS puncta outside PPL1-γ1pedc.
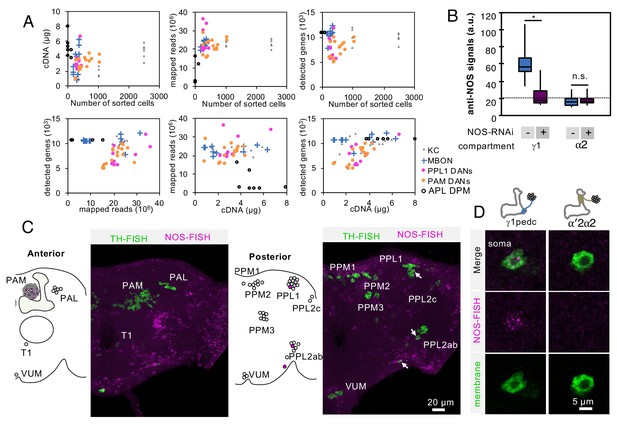
Controls for RNA-Seq reproducibility, anti-NOS antibody specificity and FISH probes.
(A) Relationship between cDNA yields, number of sorted cells, mapped reads, number of detected genes with more than one mapped read. Data for each biological replicate are shown as individual symbols. Original data are presented in Supplementary file 1. (B) Quantification of NOS immunoreactivity (a.u.; arbitrary units) in the γ1 or α2 compartment with or without NOS-RNAi. The whiskers represent the minimum and maximum. N = 12. Asterisk indicates significance from 0: *, p<0.05; n.s., not significant. Horizontal dotted line indicates the mean level of NOS immunoreactivity in the brain. (C) Outside of the MB, only a couple of additional DANs are labeled by the NOS FISH probes. Colabeling by FISH probes against TH (green) and first 4 exons of NOS1 transcripts (magenta) is shown. Diagram and projection of anterior or posterior brains are shown. Circles in diagram represent soma of DANs detected by TH probes; the different clusters of dopaminergic cell bodies (PAM, PAL, T1, PPM1, PPM2, PPM3, PPL1, PPL2c, PPL2ab and VUM) are indicated. Circles filled with magenta represent DANs overlapping with NOS (arrows). In addition to one cell in PPL1 (presumably PPL1-γ1pedc) and about a dozen of PAM cluster cells, we observed that one cell in the PPL2ab cluster and a small TH-positive cell outside, but near, the PPL2ab cluster (arrows) were labeled with NOS-FISH probes. (D) NOS-FISH signal was observed in soma of PPL1-γ1pedc but not in PPL1-α′2α2.
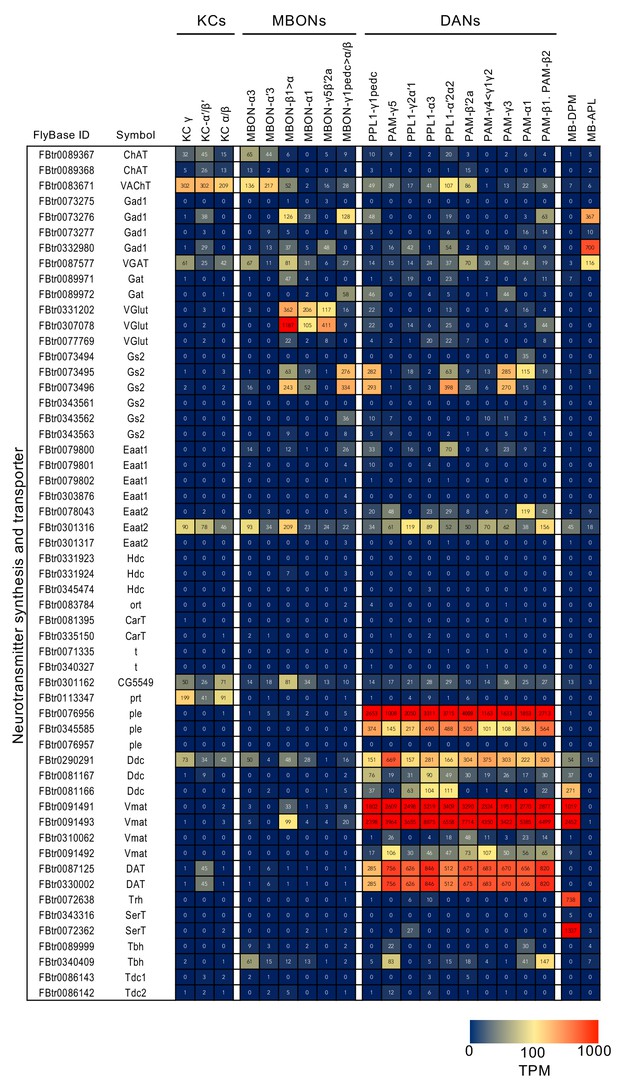
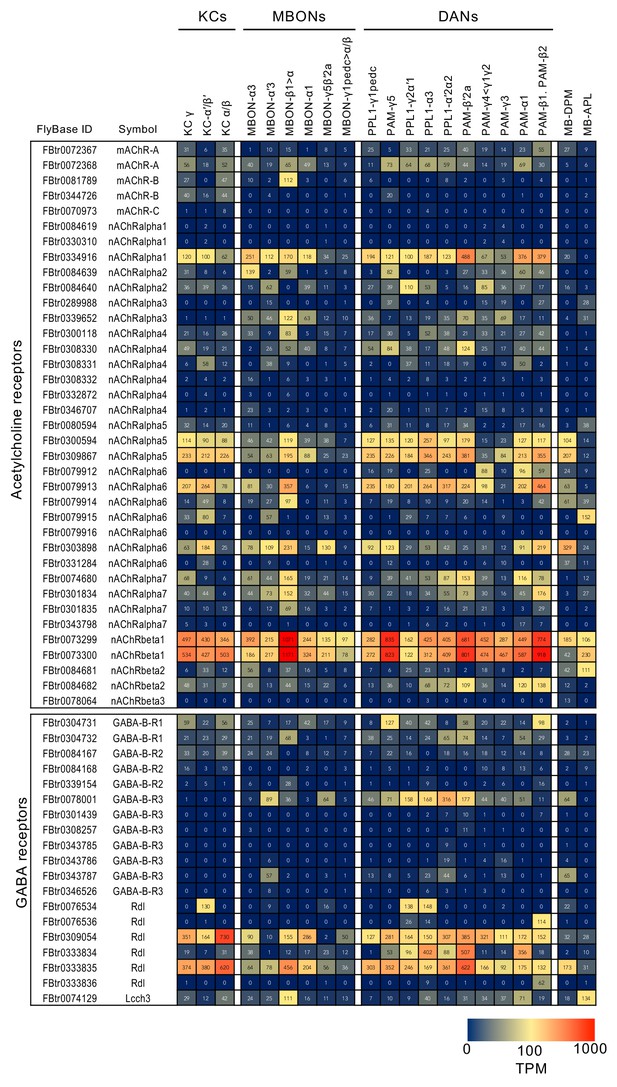
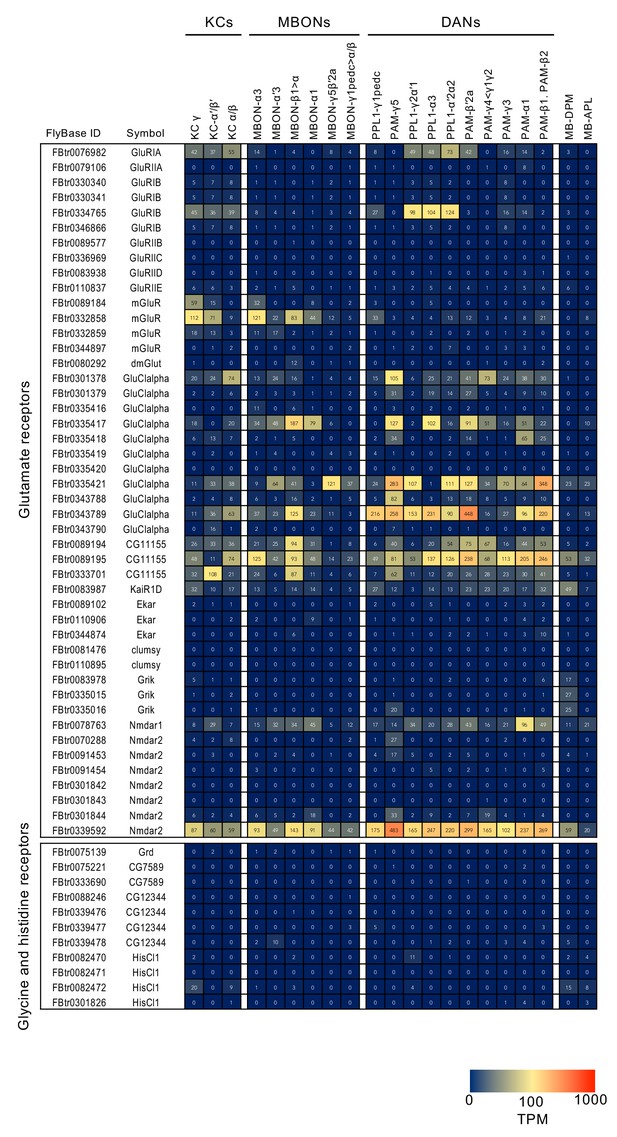
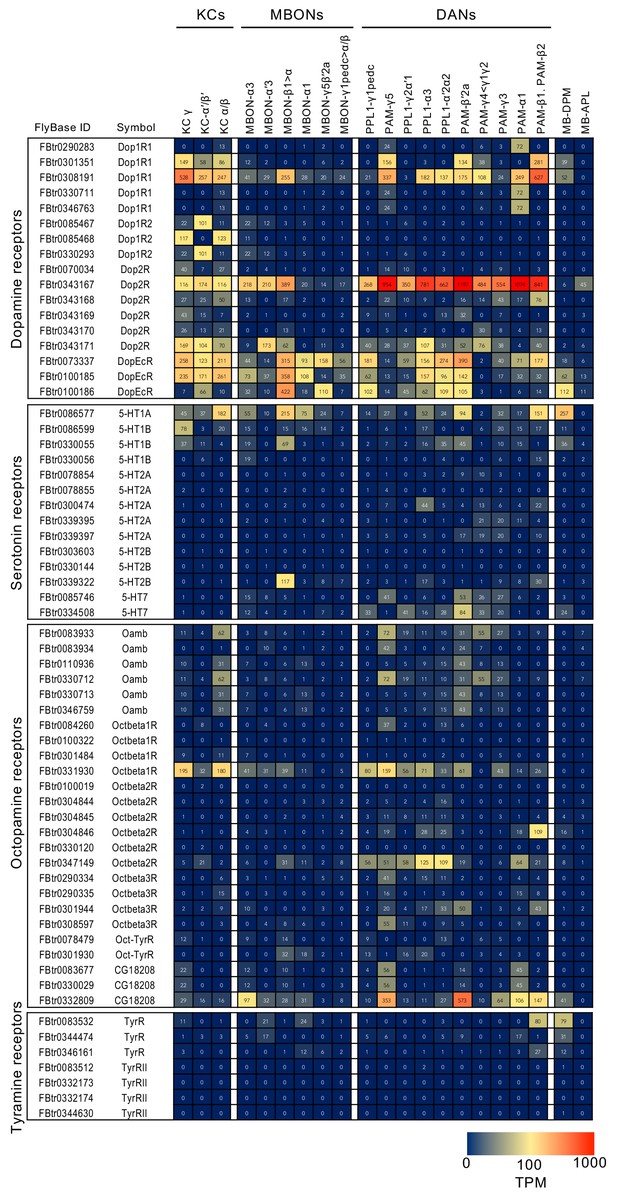
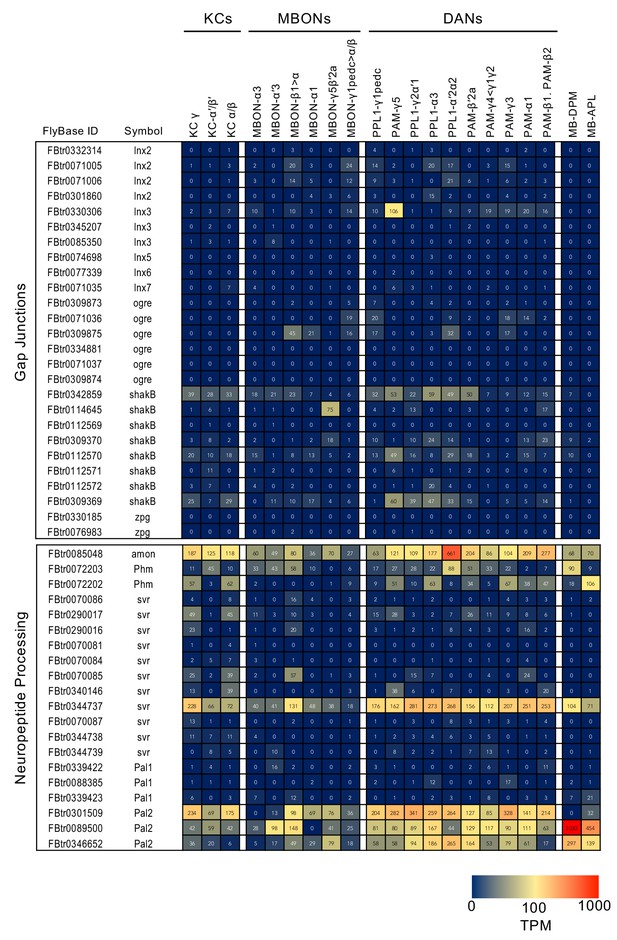
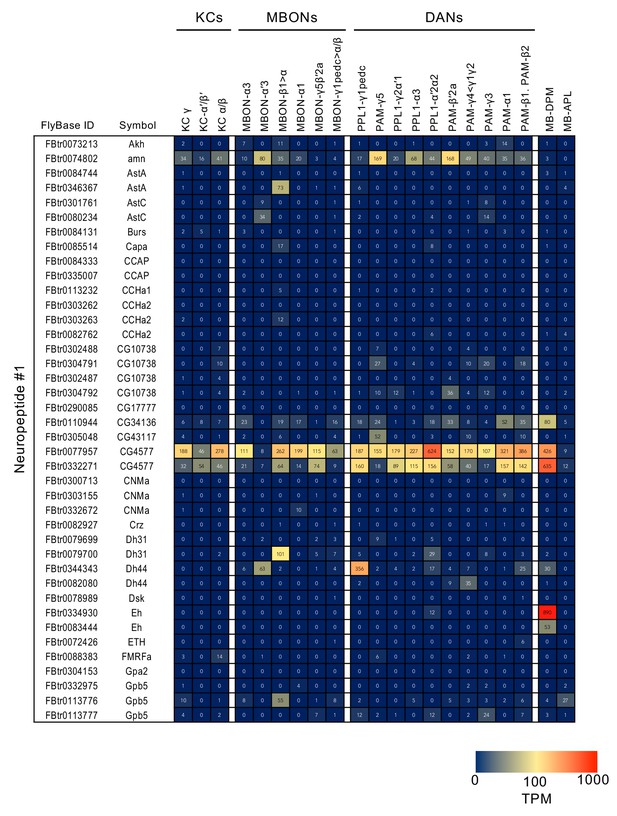
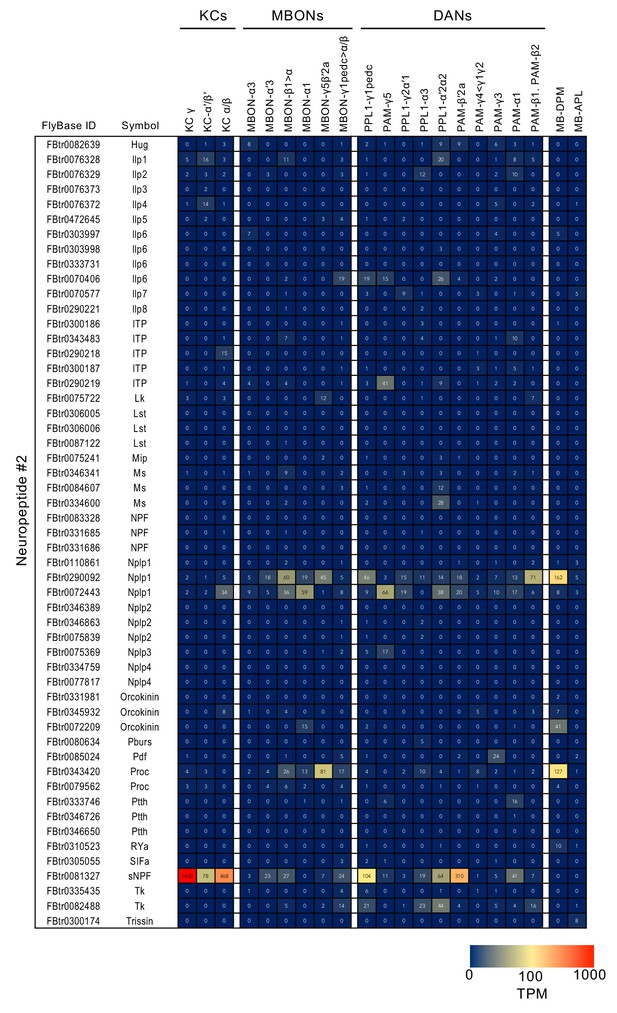
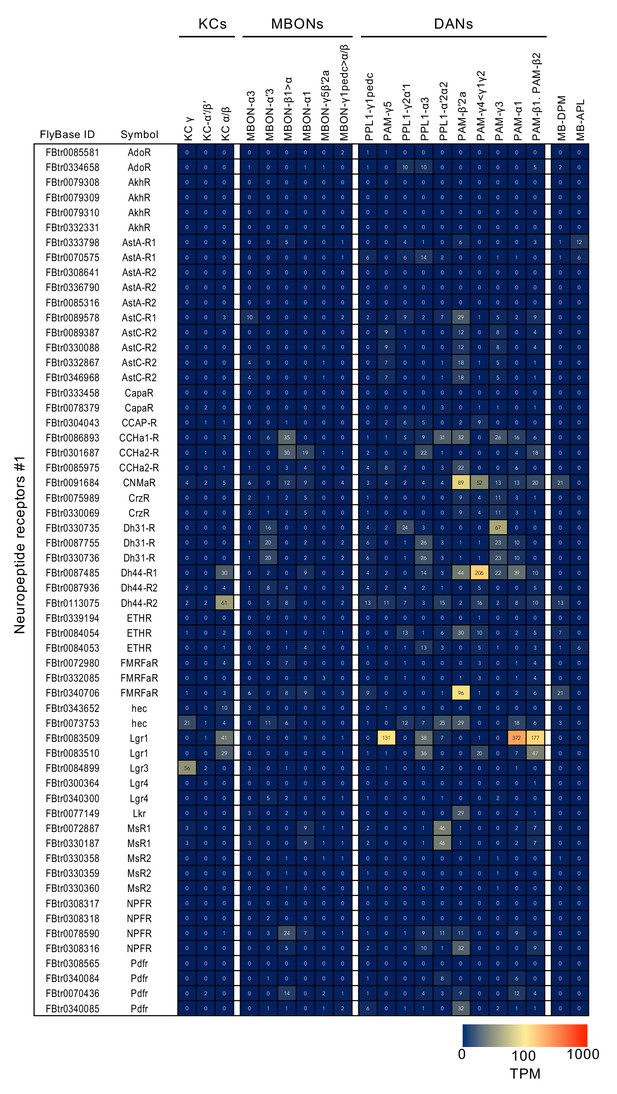
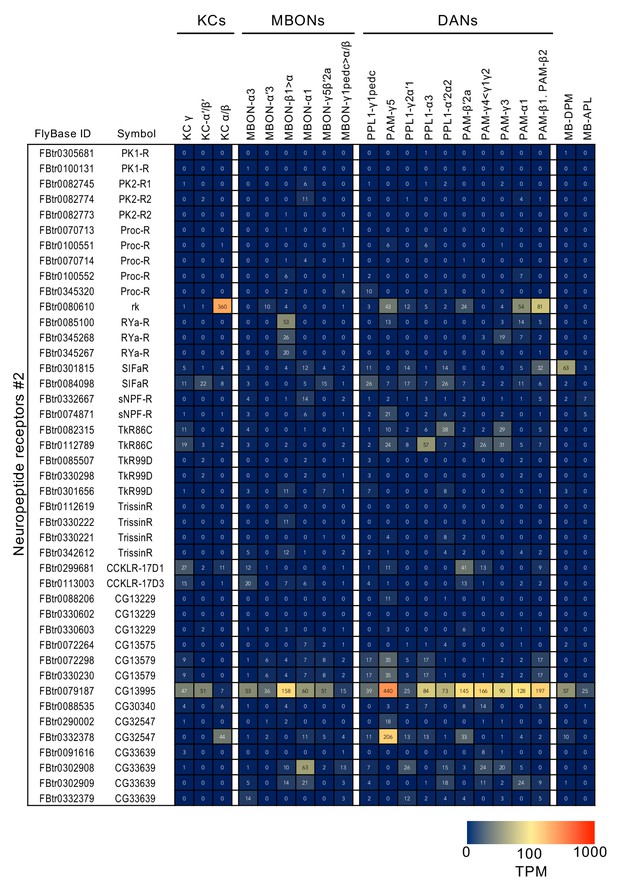
RNA-seq data for genes related to neurotransmitter synthesis and transporters.
Mean TPM values for each gene isoform are listed with the corresponding FlyBase ID. See Supplementary file 1 for the full data set and NCBI Gene Expression Omnibus (accession number GSE139889) for the raw data.
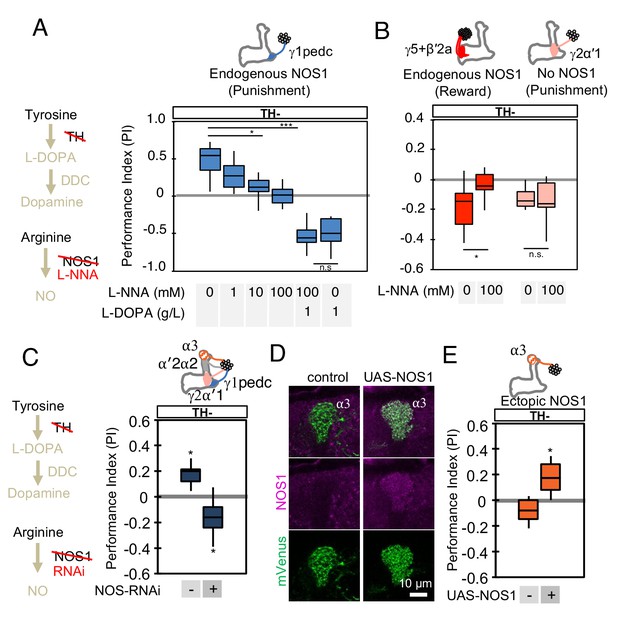
NOS in DANs contributes to memory formation.
(A) Increasing the dose of L-NNA reduced the positive-valence memory induced by activation of PPL1-γ1pedc in a TH mutant background. The ability to form an negative-valence memory was restored by feeding of L-DOPA plus carbidopa and this memory formation was not affected by L-NNA. N = 8–12. (B) Feeding of L-NNA in a TH mutant background reduced the negative-valence memory induced by activation of the combination of PAM- γ5 and PAM-β′2a, but not of PPL1-γ2α′1. N = 12–16 (C) Activation of PPL1 DANs (PPL1-γ1pedc, PPL1-γ2α′1, PPL1-α′2α2 and PPL1-α3) induced significant positive-valence memory in a TH mutant background. The valence of the induced memory was negative when NOS-RNAi was expressed in the same DANs. We postulate that the negative-valence memory observed when NOS-RNAi is expressed results from an as yet unidentified cotransmitter released by PPL1-γ2α′1 (see also panel B and Figure 2). N = 8 (D) NOS immunoreactivity in the α3 compartment in wild type (left) and after ectopic expression of NOS (right). (E) Activation of PPL1-α3 in which NOS was ectopically expressed induced significant positive-valence memory after 3 × 1 min training protocol in a TH mutant background (Figure 1C). Note that activation of PPL1-α3 can induce negative-valence memory in a wild-type background, but only after 10x spaced training (Aso and Rubin, 2016). N = 12 In A-C and E, memories assessed immediately after 3 × 1 min training are shown. The bottom and top of each box represents the first and third quartile, and the horizontal line dividing the box is the median. The whiskers represent the minimum and maximum. N = 8–16. Asterisk indicates significance of designated pair in A and B, or from 0 in C and D: *, p<0.05; ***, p<0.001; n.s., not significant.
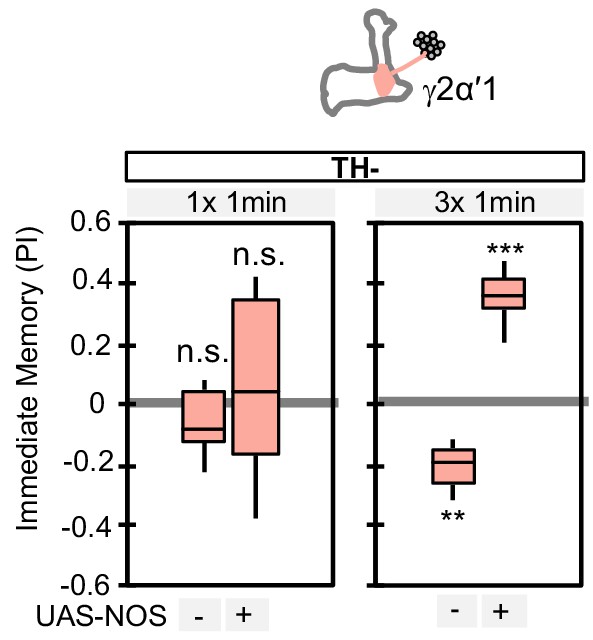
Ectopic expression of NOS in PPL1-γ2α′1 can change the valence of the memory formed by this cell type.
PPL1-γ2α′1, a cell type that lacks endogenous NOS expression, induced significant negative-valence memory after a 3 × 1 min training protocol (but not with a 1 × 1 min training protocol) in a TH mutant background. When NOS was ectopically expressed in PPL1-γ2α′1, the valence of the memory formed by the 3 × 1 min training protocol switched to positive. N = 12–16. Asterisk indicates significance from 0: **, p<0.01; ***, p<0.001; n.s., not significant.
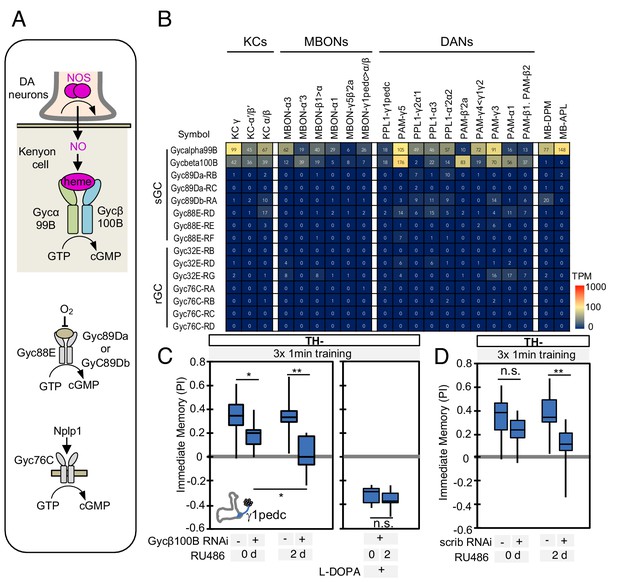
Soluble guanylate cyclase in the KCs is required to form NO-dependent memory.
(A) Diagram of soluble or receptor guanylyl cyclases in Drosophila. (B) RNA-seq data indicate coexpression of Gycα99B and Gycβ100B in KCs, MBONs and DANs. For comparison, the expression levels of other guanylate cyclase genes are also shown. Note that RNA-Seq detected transcripts of neuropeptide gene Nplp1 in both PPL1-γ1pedc and PAM-γ5 (Figure 3—figure supplement 8), but expression of its receptor Gyc76C was barely detectable compared to Gycα99B and Gycβ100B. (C) Induction of Gycβ100B-shRNA in Kenyon cells by activating MB247-switch driver (Mao et al., 2004) with RU-486 feeding reduced the positive-valence memory induced by PPL1-γ1pedc. We also observed a partial effect in the flies without RU-486, presumably due to leaky expression (Figure 5—figure supplement 1E and F). Negative-valence memory with additional feeding of L-DOPA and carbidopa was not affected by Gycβ100B-shRNA induction in KCs. Memories immediately after 3 × 1 min training are shown. The bottom and top of each box represents the first and third quartile, and the horizontal line dividing the box is the median. (D) Induction of scrib-shRNA in KCs also reduced the positive-valence memory induced by activation of PPL1-γ1pedc in a TH mutant background. The whiskers represent the minimum and maximum. N = 12–16. Asterisk indicates significance of designated pair: *, p<0.05; **,p<0.01; n.s., not significant.
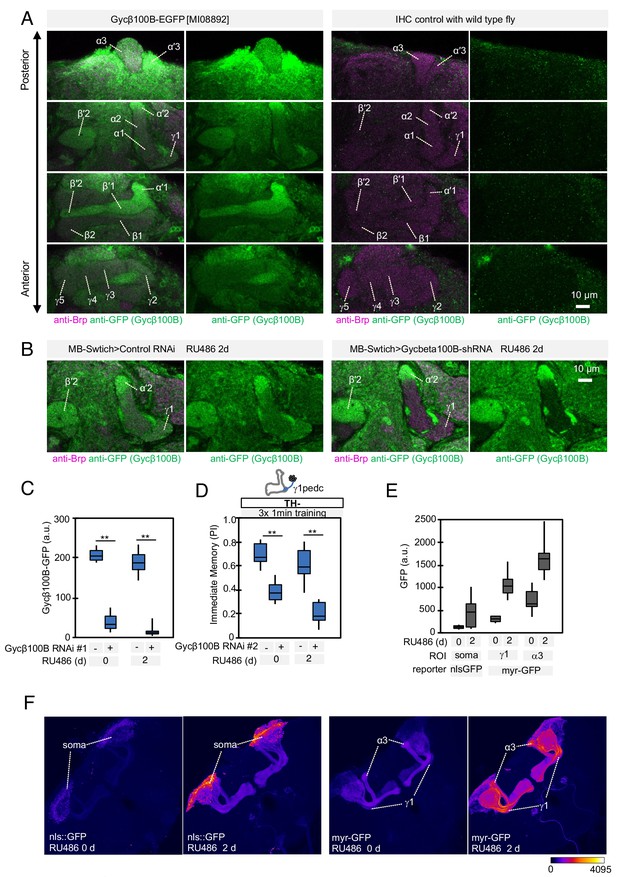
Expression of Gycbeta100B in the mushroom body lobes (A) Distribution of Gycbeta100B-EGFP in flies carrying the Gycbeta100B[MI08892-GFSTF.2] construct in the MB lobes is shown in a series of anterior to posterior confocal sections.
Right panels show images of wild-type fly, which did not contain the Gycbeta100B-EGFP insertion, prepared with the identical immunolabelling procedure. (B–D) Gycbeta100B-EGFP signals in the γ1 (B) were markedly reduced with induction of Gycβ100B-shRNA HMJ22589 (C) or another RNAi line KK100706 (Dietzl et al., 2007) (D) in Kenyon cells. (E–F) We detected enhanced induction of reporters by MB-switch after feeding RU486 with low but significant basal expression without RU486 feeding.
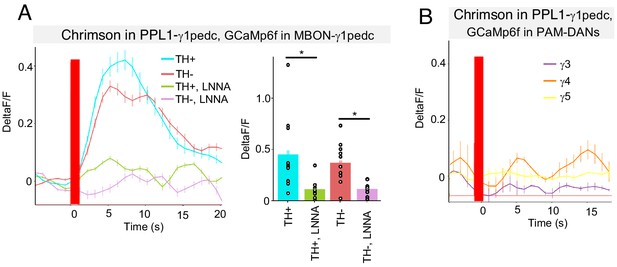
NO from PPL1-γ1pedc activates MBON-γ1pedc but not PAM-DANs in γ3, γ4 and γ5.
(A) In an ex vivo preparation, optogenetic activation of PPL1-γ1pedc evoked slow and sustained calcium response in MBON-γ1pedc. This response was observed in both TH mutant and wild-type background, but diminished in flies fed with L-NNA. N = 6–10. Error bars indicate SEM. Asterisk indicates significance of designated pair: *, p<0.05. (B) Optogenetic activation of PPL1-γ1pedc evoked no obvious calcium response in terminals of PAM-DANs in γ3, γ4 and γ5 compartments.
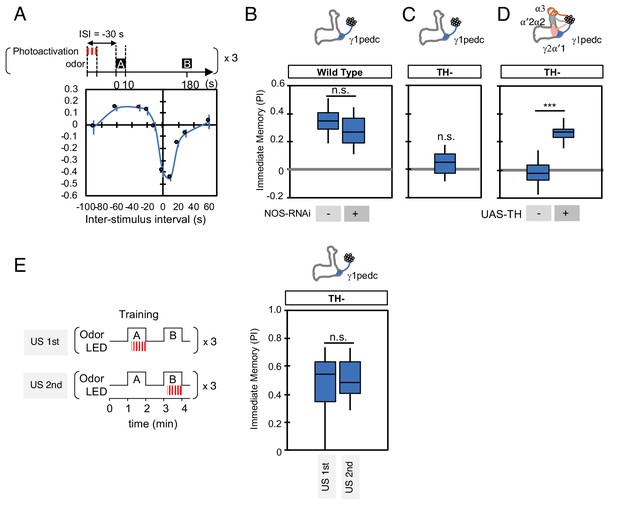
NO is not involved in timing-dependent inversion of valence.
(A) Activation of PPL1-γ1pedc 30 s prior to odor onset induced appetitive memory (known as ‘relief learning’) in a TH wild type background as shown in this panel reproduced from Aso and Rubin (2016). These data were obtained with 1x training whereas a 3x training protocol was used for the experiments shown in (B). N = 10–12. (B) Relief learning was not affected by NOS-knockdown in PPL1-γ1pedc. N = 10. (C–D) Relief learning was completely abolished in a TH mutant background and could be restored with TH expression in PPL1-DANs. N = 12. (E) Paring optogenetic activation of PPL1-γ1pedc with either the first or second presented odor gave similar levels of valence-inverted memory. N = 12. Error bars indicate SEM. Asterisk indicates significance of designated pair in B, D, E or zero in C: *, p<0.001; n.s., not significant.
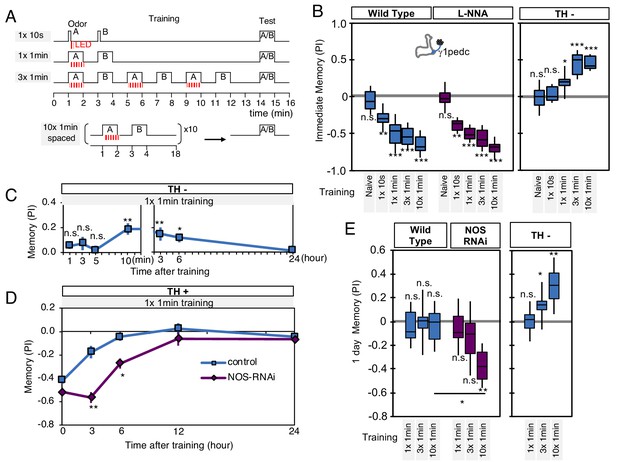
NO-dependent effect develops slowly, requires longer training than dopamine-dependent memory, and shortens memory retention.
(A) Schematic diagram of training protocols. (B) Learning rate by activation of PPL1-γ1pedc in wild type (left; blue), wild type with L-NNA feeding (center; purple) or TH mutant backgrounds (right; blue). Memory scores were not significantly affected by L-NNA feeding in any of the training protocols in wild-type flies. A single 10 s training was insufficient to induce any detectable memory in a TH mutant background, but induced significant negative-valence memory in wild-type background. (C) Time course of NO-dependent, positive-valence memory induced by PPL1-γ1pedc in a TH mutant background after 1 × 1 min training. Data point and error bars indicate mean and SEM. N = 8–10. Note that the plot is split in the time axis to better display the kinetics. (D) After 1 × 1 min training, a cell-type-specific knock down of NOS in PPL1-γ1pedc prolonged the retention of negative-valence memory induced by PPL1-γ1pedc in wild-type background measured at 3 and 6 hr. Note that expression of NOS-RNAi did not affect the score of immediate memory. N = 12. (E) Effect of repetitive trainings on 1-day memory. Repetitive training with activation of PPL1-γ1pedc did not induced significant 1-day memory in wild-type background irrespective of training protocols (blue; left). Flies expressing NOS-shRNA showed significant 1 day memory after 10x spaced training (purple; center). In a TH mutant background (right), appetitive memory was induced by 3X and 10X repetitive training. For activation of PPL1-γ1pedc, VT045661-LexA was used as the driver for experiments in B, C and in the TH mutant background data in E, and MB320C split-GAL4 was used for wild-type and NOS-RNAi data in D. We made consistent observations with both global L-NNA inhibition of NOS and cell-type-specific NOS-RNAi (see Figure 6—figure supplement 1). The bottom and top of each box represents the first and third quartile, and the horizontal line dividing the box is the median. The whiskers represent the minimum and maximum. N = 12–16. Asterisk indicates significance between control and NOS-RNAi in D, between designated pair in E, or from 0 in all others: *, p<0.05; **, p<0.01; ***, p<0.001; n.s., not significant.
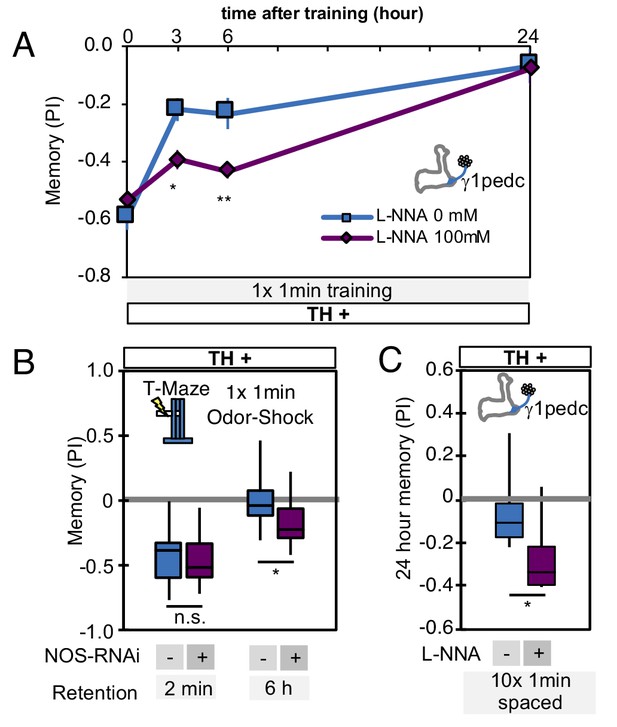
Comparison of the effects of NOS-RNAi and L-NNA.
(A) L-NNA feeding prolonged retention of memory induced by 1 × 1 min training with PPL1-γ1pedc activation at 3- and 6 hr time points, as observed with NOS-RNAi (Figure 6C). VT045661-LexA was used as a driver to express CsChrimson-mVenus. Data point and error bars indicate mean and SEM. *, p<0.05; **, p<0.01 (B) Knockdown of NOS in PPL1-γ1pedc with NOS-RNAi prolonged memory retention after 1 × 1 min training with 60V electric shock. N = 12. (C) Inhibition of NOS by L-NNA prolonged the retention of memory that was induced by 10x spaced training with activation of PPL1-γ1pedc using the VT045661-LexA driver. N = 12. The bottom and top of each box in (B) and (C) represents the first and third quartile, and the horizontal line dividing the box is the median. The whiskers represent the minimum and maximum. *, p<0.05.
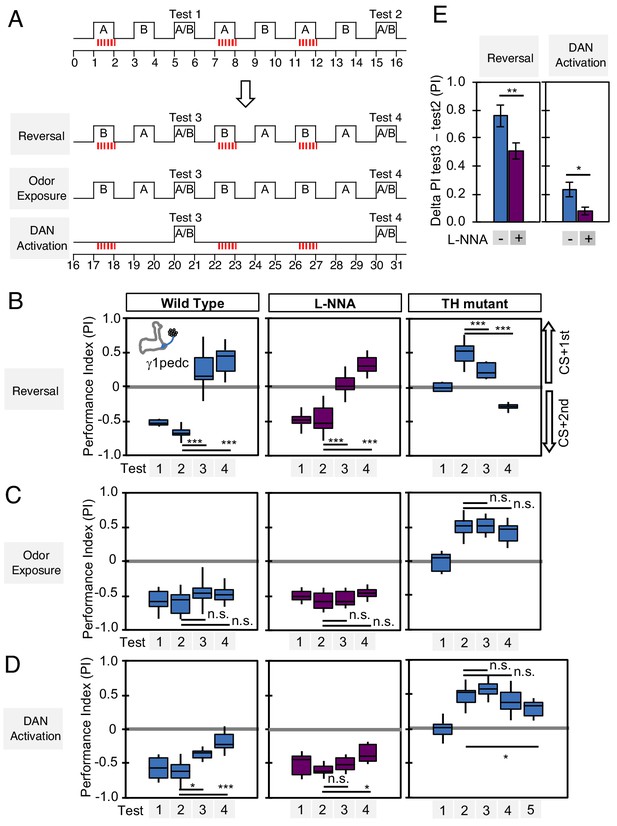
Nitric oxide enhances fast update of memory.
(A) Experimental design to measure dynamics of memory when flies encounter new experiences after establishing an initial odor memory by the 3 × 1 min spaced training pairing odor presentation and optogenetic PPL1-γ1pedc activation. (B) In reversal learning, the control odor in the first three trainings was now paired with activation of PPL1-γ1pedc. In all three cases, the first reversal learning was sufficient to modify the odor preference. However, only in wild-type flies was this change large enough that the flies preferred the new odor. In L-NNA fed or TH mutant (dopamine deficient) flies changing the odor preference required multiple training sessions with the new odor. (C) Three exposures to each of the two odors did not significantly change the odor preference in any of the three sets of flies. (D) One activation of PPL1-γ1ped without odor quickly reduced the conditioned response in wild type. L-NNA fed flies or dopamine deficient flies required three or five times, respectively, repetitions of PPL1-γ1ped activation to significantly reduce the conditioned response. The bottom and top of each box in (B–D) represents the first and third quartile, and the horizontal line dividing the box is the median. The whiskers represent the minimum and maximum. (E) Changes in PI induced by 3x training (as measured in Test 2) resulting from the first reversal training (left) or from DAN activation without odor presentation (right) (as measured in Test 3). The observed changes were significantly larger in wild-type flies compared to L-NNA fed flies. *, p<0.05; **, p<0.01.
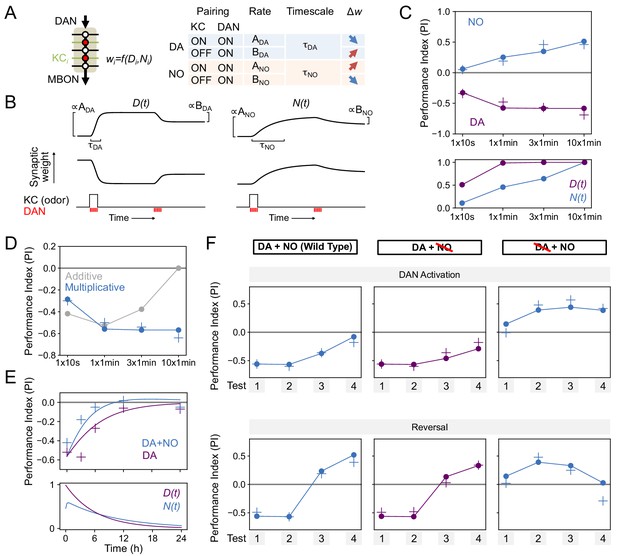
Modeling dopamine and NO mediated plasticity (A) Summary of plasticity model for independent dopaminergic (DA) and NO pathways.
Synaptic weight wi from KCi to an MBON is increased or decreased depending on the pairing protocol. A and B determine the magnitude of the depression or potentiation induced by pairing, and τ determines the timescale over which weight changes occur. (B) Illustration of the effects of the model in (A), for only DA (left) or only NO (right) dependent plasticity. In each plot, KC and DANs are first co-activated, followed by a later DA activation without KC activation. (C) Top: Model performance index (PI) for different pairing protocols. In (C)-(F), crosses represent the means of data from Figure 7. Bottom: Dynamics of D(t) and N(t) in the model. (D) Modeling effects of combined DA and NO dependent plasticity. Gray curve: synaptic weights w(t) are modeled as an additive function of DA and NO dependent effects D(t) and N(t), w(t) ∝ N(t) – D(t). Blue curve: a multiplicative interaction with w(t) ∝ (1 + N(t)) (1 – D(t)). (E) Modeling 24 hr memory decay following 1 × 1 min odor pairing. We assume a low level of spontaneous DAN activity and choose BDA and BNO to fit the data. Top: Performance index in model and data. Blue curve: control. Purple curve: only DA-dependent plasticity (compared to data from NOS-RNAi experiment). Bottom: Dynamics of D(t) and N(t) in the model. (F) Modeling effects of DAN activation and reversal learning. BDA and BNO are chosen to fit the effects of DAN activation (top). The model qualitatively reproduces the effects of reversal learning (bottom) with no free parameters.
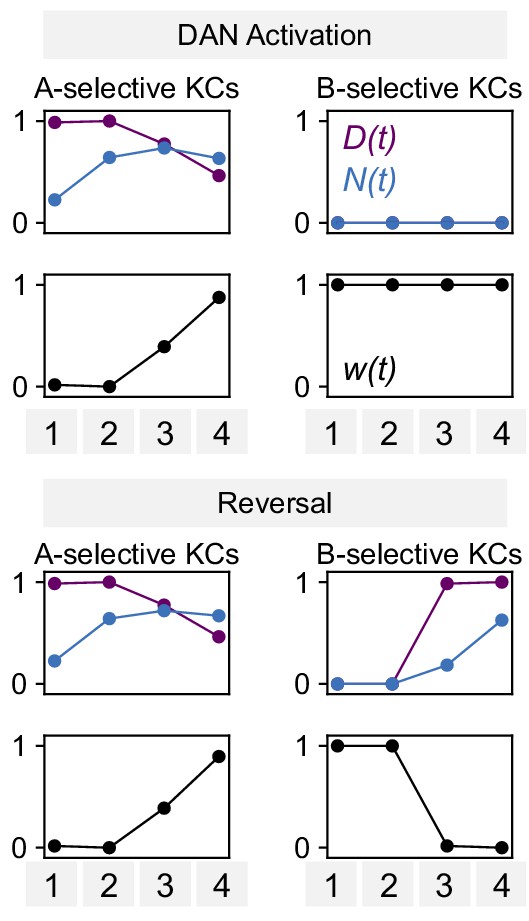
Parameters for modeling dopamine and NO mediated plasticity.
Values of N(t), D(t), and synaptic weight w(t) for the model of DAN-activation and reversal protocols in Figure 8F.
Plots of ‘A-selective KCs’ show N(t) and D(t)for KCs that represent the odor paired with DAN activation in the first two pairings, whereas ‘B-selective KCs’ show N(t) and D(t) for KCs that represent the corresponding odor for the second two pairings (Figure 7A).
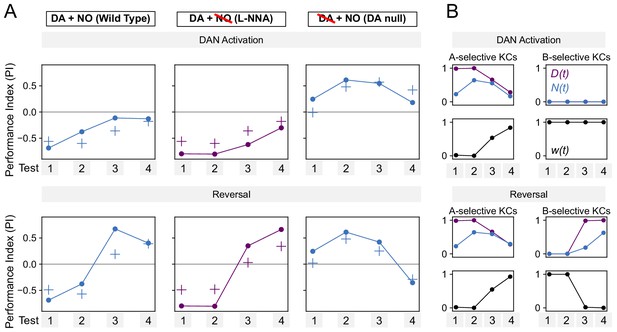
Performance of additive model on DAN activation and reversal paradigms.
(A) Same as Figure 7F, but for a model in which DA and NO effects are additive, w(t) ∝ N(t) – D(t).
The model produces a worse fit than the multiplicative model (Figure 7F). (B) Same as Figure 8—figure supplement 1, but for the additive model.
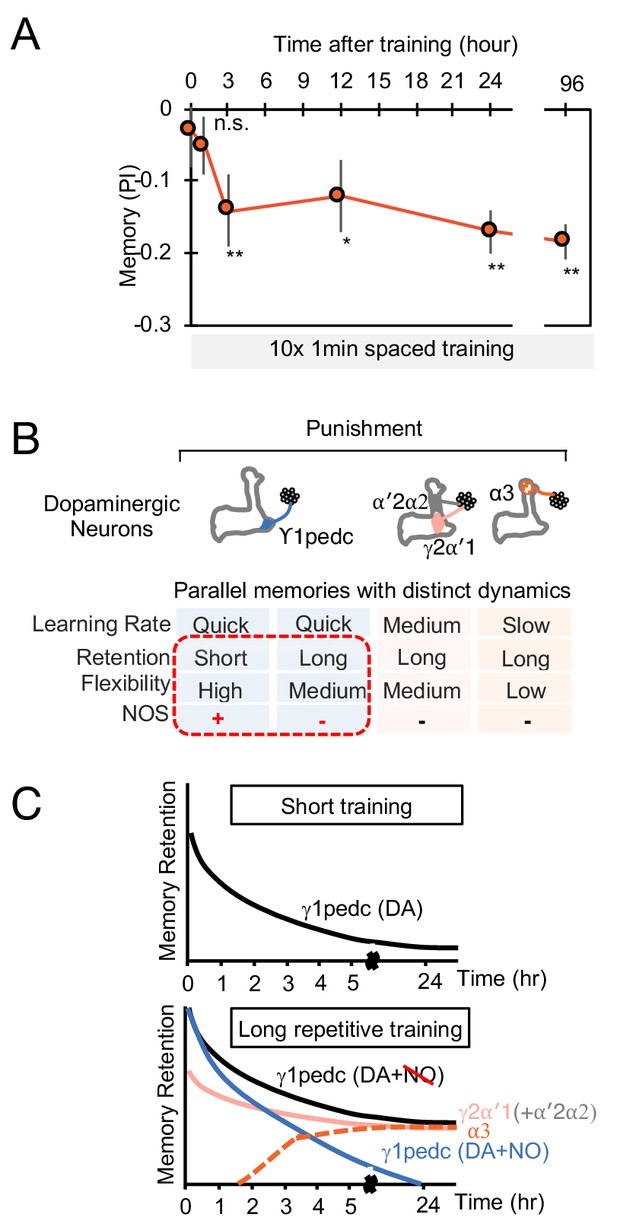
The MB stores parallel memories with distinct dynamics.
(A) Flies were trained with 10 × 1 min spaced protocol with activation of PPL1-α3 (Figure 6A) and tested at different retention times. Memory was not detectable immediately after training and at 1 hr, but became significant after 3 hr. Data point and error bars indicate mean and SEM. N = 12–16. *, p<0.05; **, p<0.01 (B) Summary of this study. NOS diversify memory dynamics of MB compartments by reducing memory retention but enhancing flexibility. (C) A conceptual model of how naturalistic punishment (e.g. heat) induce memories that have distinct dynamics as flies experience training events. Punishment activate all PPL1-α3, γ2α′1 (MB-MV1) and γ1pedc (MB-MP1) to similar level (Kirkhart and Scott, 2015; Mao and Davis, 2009). PPL1-α′2/α′2 (MB-V1) does not respond to heat and shock (Kirkhart and Scott, 2015; Mao and Davis, 2009) and cannot induce aversive memory by itself, but can have a synergistic effect on memory retention when coactivated with PPL1-γ2α′1 (Aso and Rubin, 2016). Top: A one-time short training event first induces memory only in the γ1pedc compartment because of the fast learning rate by PPL1-γ1pedc (Aso and Rubin, 2016; Hige et al., 2015). This short-training is insufficient to induce NO-dependent memory (Figure 6B). Bottom: When flies experience similar training repeatedly, stable memories will form in the compartments with slower learning rates (i.e. γ2α′1 and α3) (Aso and Rubin, 2016). Repetitive training also promotes NO-dependent processes to reduce memory retention and enhance flexibility in the γ1pedc. After repetitive training, PPL1-γ1pedc also regulates stability of memories (Awata et al., 2019; Plaçais et al., 2012), but NO’s role in that process is unknown.
Videos
Anti-NOS signals inside or outside the PPL1-γ1pedc.
Rotation movie of the 3D reconstruction of lattice light sheet image of DANs in γ1 (green) with pseudo colors for NOS inside (magenta) or outside (blue).
Additional files
-
Supplementary file 1
RNA-seq data.
Transcripts Per Kilobase Million (TPM) TPM values for each splicing isoform are listed with the corresponding FlyBase ID. The source data is available at NCBI Gene Expression Omnibus (accession number GSE139889).
- https://cdn.elifesciences.org/articles/49257/elife-49257-supp1-v2.xlsx
-
Supplementary file 2
Key resource table.
The list of key reagents used in this study.
- https://cdn.elifesciences.org/articles/49257/elife-49257-supp2-v2.docx
-
Supplementary file 3
Drosophila genotypes.
The list of Drosophila genotypes and drug treatment used in each experiment.
- https://cdn.elifesciences.org/articles/49257/elife-49257-supp3-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/49257/elife-49257-transrepform-v2.docx