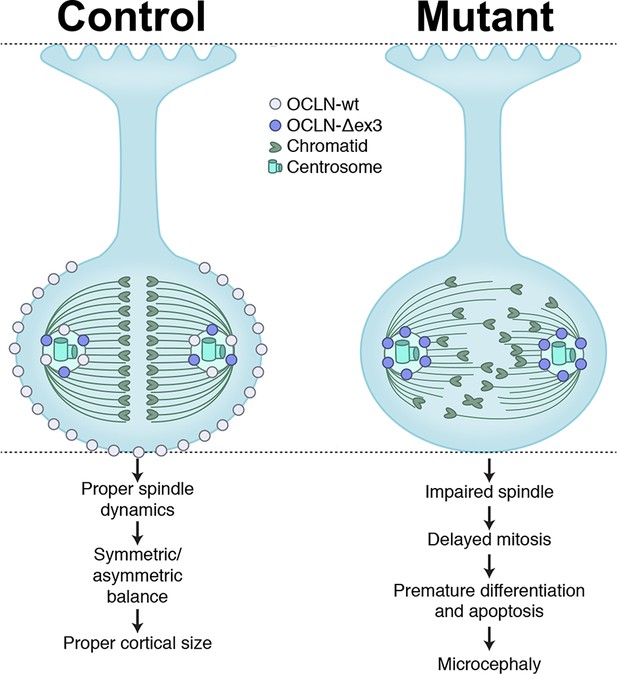
Tight junction protein occludin regulates progenitor Self-Renewal and survival in developing cortex
Figures
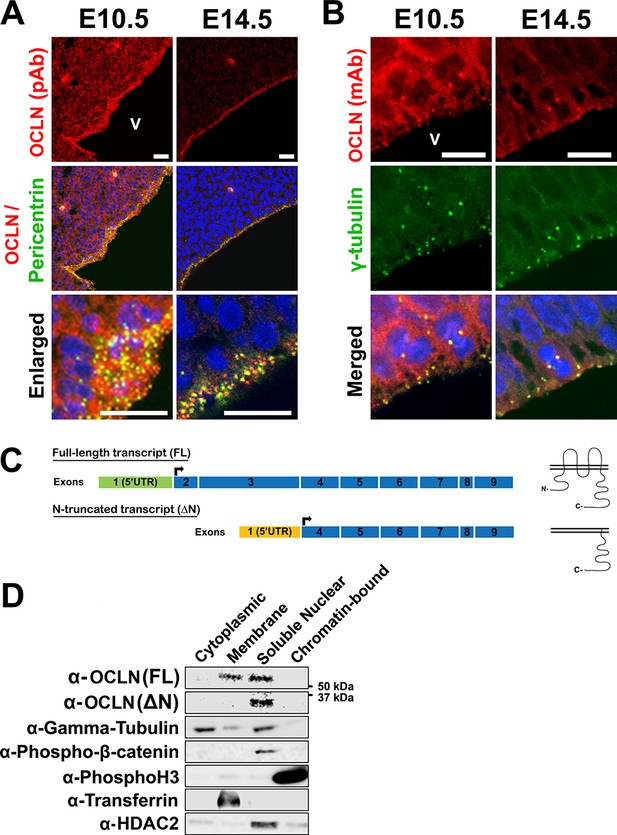
Differential localization of OCLN isoforms in embryonic mouse cortex.
(A,B) Confocal microscope images of coronal sections from E10.5 and E14.5 mouse cortices stained with polyclonal (pAb) (A) and monoclonal (mAb) (B) OCLN antibodies and co-stained with centrosomal markers pericentrin or γ-tubulin. V, ventricle. Scale bar, 25 μm in A (including enlarged images) and 10 μm in B. (C) Schematic of mouse Ocln transcripts and their corresponding presumed protein structure. (D) Subcellular localization of occludin by immunoblot analysis of subcellular fractions (cytoplasmic, membrane, soluble nuclear, and chromatin-bound). Centrosomal markers γ-tubulin and phospho-Beta-catenin colocalize with occludin in the soluble nuclear fraction. Fraction-specific controls used are anti-HDAC2 (soluble nuclear), anti-transferrin (membrane), and anti-Phospho-histone3 (chromatin-bound).
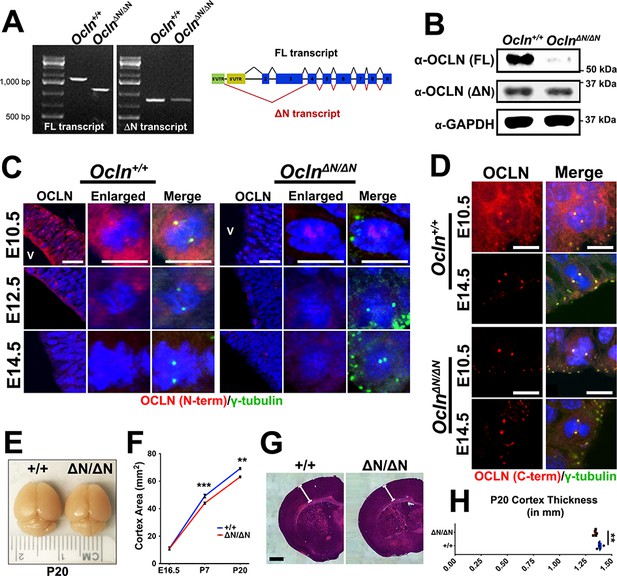
The OclnΔN/ΔN mouse, a hypomorphic mutant line, exhibits microcephaly by P7.
(A) Qualitative RT-PCR of Ocln expression in control and mutant E12.5 mouse probing for full-length (FL) and truncated (ΔN) transcripts. Representative image is shown from a sample of three biological replicates (primers in Supplementary file 1). (B) Western blot analysis of E12.5 mouse total brain lysate from Ocln+/+ and OclnΔN/ΔN mice. Representative image from three biological replicates. (C,D) Confocal images of coronal sections from E10.5-E14.5 control and mutant mouse cortices stained with N-terminus (C) and C-terminus-specific (D) occludin antibody and co-stained with centrosomal marker γ-tubulin. V, ventricle. Scale bar, 25 um in C) and 10 um in D. (E) Dorsal view of P20 control and mutant brains. (F) Quantification of cortical area of E16.5, P7, and P20 mouse brains (n = 4 brains for E16.5 control, n = 6 for E16.5 mutant, n = 8 for control and mutant P7, n = 3 for control and mutant P20). Data points represent mean ± SEM. Two-way ANOVA with Sidak’s multiple comparison test; in each age group, +/+ was compared to ΔN/ΔN, **p<0.01,***p<0.001. (G) P20 brain section of wild-type and mutant mice cortices stained with H and E (Hematoxylin and Eosin). Scale bar, 1 mm (H) Quantification of P20 cortical thickness (n = 5 for each genotype). Data points represent mean ± SEM. Student’s t-test, **p<0.01. Detailed tabulation of means, SEMs, sample sizes, and exact p-values can be found in Figure 2—source data 1.
-
Figure 2—source data 1
Mean, SEM, sample size (n), and exact p-values for Figure 2 quantifications.
- https://cdn.elifesciences.org/articles/49376/elife-49376-fig2-data1-v1.xlsx

mOCLN-FL co-localizes with vascular endothelial marker, CD31, in Ocln+/+ but not Ocln∆N/∆N embryonic cortex.
(A) Confocal images of mouse coronal sections at E12.5 stained with OCLN C-term antibody (red) and vascular endothelial cell marker CD-31 (green). The OCLN antibody detects both full-length and truncated OCLN isoforms. Insets show OCLN labeling of the mitotic spindle apparatus of dividing progenitors at the ventricular surface in both WT and ∆N/∆N genotypes. Scale bar 30 μm (B) Confocal images of mouse coronal sections at E14.5 stained with C-term OCLN and CD-31 antibodies. Full-length OCLN continues to be robustly expressed in vascular endothelial cells despite developmental down regulation of the full length OCLN protein in neural cells of Ocln+/+ cortex. However, mOCLN-FL is absent in vessels in the mutant brain. Scale bar, 30 μm.
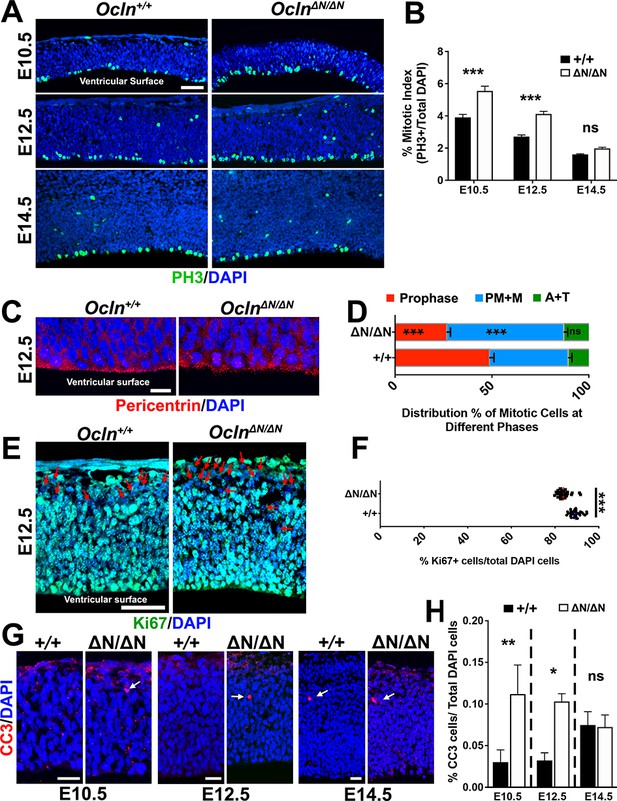
Progenitor proliferation abnormalities and apoptosis in OclnΔN/ΔN embryonic cortex.
(A) Confocal microscope images of coronal section from E10.5-E14.5 control and mutant mouse cortices stained with M-phase marker phospho-histone3 (PH3; green) and counterstained with DAPI (blue). Scale bar, 50 μm (B) Mitotic index quantification, defined as the percentage of DAPI cells labeled with PH3, in Ocln+/+ and OclnΔN/ΔN mice. All data points represent mean ± SEM, n = 30–39 brain sections for each genotype, from three independent experiments. Two-way ANOVA with Sidak’s multiple comparison test; in each age group, +/+ compared to ΔN/ΔN, ***p<0.001, ns, not significant. (C) Confocal microscope images of coronal section from E12.5 Ocln+/+ and OclnΔN/ΔN mouse cortices stained with centrosomal marker pericentrin (red) and counterstained with DAPI (blue). Phases of mitosis are identified by their unique DAPI and centrosomal staining patterns. Scale bar, 10 um. (D) Distribution percentages of M-phase stages in E12.5 control and mutant mice cortices at the ventricular surface. All data points represent mean ± SEM, n = 28 Ocln+/+ sections and n = 29 OclnΔN/ΔN sections, compiled from three independent experiments. One-way ANOVA with Sidak’s multiple comparison test comparing control and mutant data sets in each stage of mitosis, ***p<0.001, (E) Confocal images of E12.5 control and mutant mouse cortices in coronal sections stained with proliferation marker Ki67 (green) and counterstained with DAPI (blue). Red arrows denote Ki67-negative cells. Scale bar, 50 μm (F) Percentage of total DAPI cells labeled with KI67 in control and mutant mouse cortices. All data points represent mean, n = 30 brain sections for each genotype, from three independent experiments. Student’s t-test, ***p<0.001. (G) Confocal images of coronal section from E10.5-E14.5 control and mutant mouse cortices stained with apoptosis marker activated caspase-3 (CC3; red) and counterstained with DAPI (blue). Scale bar, 20 um for all groups. (H) Percentage of DAPI cells that are also caspase3+ from E10.5-E14.5. All data points represent mean ± SEM, n = 35–46 brain sections for each genotype, from three independent experiments. Two-way ANOVA with Sidak’s multiple comparison test; within each age group, +/+ was compared to ΔN/ΔN; *p<0.05, **p<0.01, ns, not significant. Detailed tabulation of means, SEMs, sample sizes, and exact p-values can be found in Figure 3—source data 1.
-
Figure 3—source data 1
Mean, SEM, sample size (n), and exact p-values for Figure 3 quantifications.
- https://cdn.elifesciences.org/articles/49376/elife-49376-fig3-data1-v1.xlsx
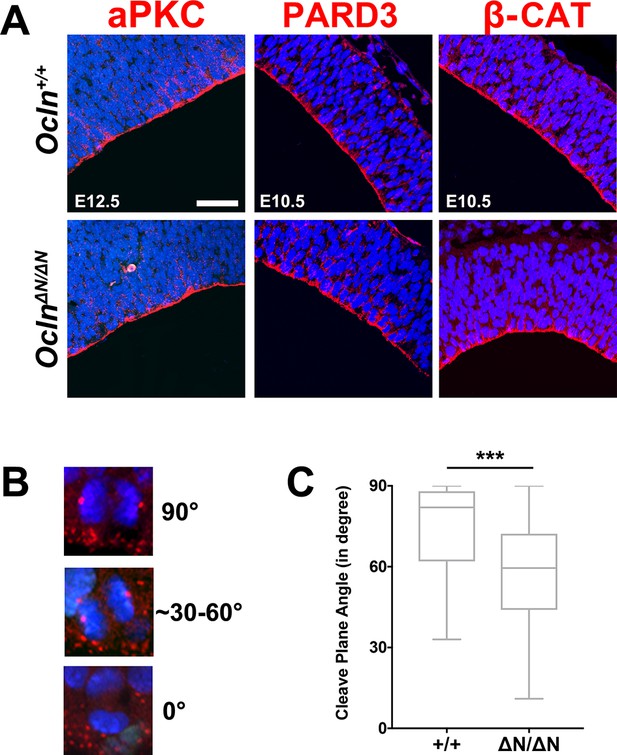
Intact organization of apical complex proteins and skewed cleavage plane orientation in Ocln-mutant mouse.
(A) Confocal microscope images of coronal section from E12.5 control and mutant mouse cortices stained with apical complex markers atypical PKC (aPKC), Pard3, and B-catenin (all red) and counterstained with DAPI (blue). Scale bar, 50 μm. (B) Representative images for quantification of cleavage plane orientation angle. (C) Quantification of cleavage plane angle in control and mutant mice. All data points represent mean ± SEM, n = 52 Ocln+/+ cells and n = 70 OclnΔN/ΔN cells, from three independent experiments. Student’s t-test, ***p<0.001. Detailed tabulation of means, SEMs, sample sizes, and exact p-values can be found in Figure 3—figure supplement 1—source data 1.
-
Figure 3—figure supplement 1—source data 1
Mean, SEM, sample size (n), and exact p-values for Figure 3—figure supplement 1 quantifications.
- https://cdn.elifesciences.org/articles/49376/elife-49376-fig3-figsupp1-data1-v1.xlsx
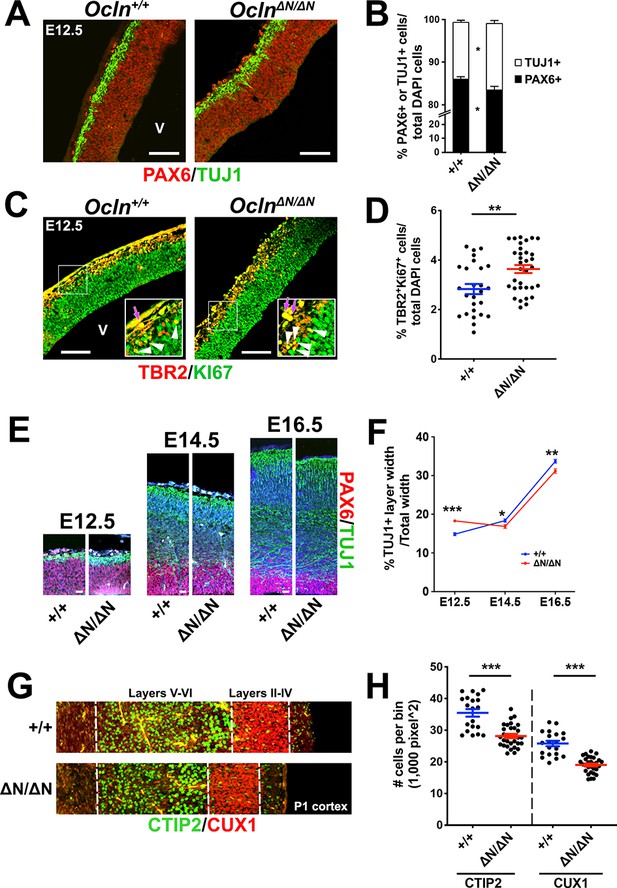
Progenitor depletion, precocious neuronal differentiation, and microcephaly in Ocln∆N/∆N cortex.
(A) Confocal images of coronal sections from E12.5 control and mutant mouse cortices stained with RGC marker PAX6 (red) and neuronal marker TUJ1 (green). V, ventricle. Scale bar, 50 um. (B) Quantification of TUJ1+ or PAX6+ cell percentages in control and mutant mice. All data points represent mean ± SEM, n = 26 brain sections from three independent experiments for each genotype. Student’s t-test, *p<0.05. (C) Confocal images of coronal section from E12.5 control and mutant mouse cortices stained with intermediate progenitor marker TBR2 (red) and proliferation marker KI67 (green). Inset enlargements show regions outlined by white box. Dual-stained TBR2+/Ki67+ yellow IPCs (white arrowhead) were quantified while auto-fluorescent red blood cells (purple arrows) were disregarded in quantification. V, ventricle. Scale bar, 50 μm. (D) Quantification of dual-stained TBR2+ and KI67+ cell percentages in control and mutant mice. All data points represent mean ± SEM, n = 26 sections for Ocln+/+, n = 33 sections for OclnΔN/ΔN, from three independent experiments for each genotype. Student’s t-test, **p<0.01. (E) Confocal microscope images of E12.5-E16.5 wild-type and mutant mouse cortices stained with PAX6 (red) and TUJ1 (green) and counterstained with DAPI (blue). Scale bar, 20 um (F) Time-course quantification of TUJ1+ layer width of E12.5 to E16.5 mouse cortices. All data points represent mean ± SEM, n = 34 brain sections for E12.5 Ocln+/+, n = 48 for E12.5 OclnΔN/ΔN, n = 41 for E14.5 Ocln+/+, n = 51 for E14.5 OclnΔN/ΔN, n = 28 for E16.5 Ocln+/+, and n = 30 for E16.5 OclnΔN/ΔN, from three independent experiments for each age and each genotype. Two-way ANOVA with Sidak’s multiple comparison test was performed comparing control and mutant in each age group, *p<0.05; **p<0.01; ***p<0.001. (G) Confocal images of P1 wild type and mutant mice cortices stained for CTIP2 and CUX1 to label deep layers (V and VI) and upper layers (II-IV), respectively. (H) Quantification of Ctip2+ or Cux1+ cell numbers per bin. All data points represent mean ± SEM, n = 20 for Ocln+/+ and n = 30 for OclnΔN/ΔN from three independent experiments for each genotype. Student’s t-test, ***p<0.001. Detailed tabulation of all means, SEMs, sample sizes, and exact p-values can be found in Figure 4—source data 1.
-
Figure 4—source data 1
Mean, SEM, sample size (n), and exact p-values for Figure 4 quantifications.
- https://cdn.elifesciences.org/articles/49376/elife-49376-fig4-data1-v1.xlsx
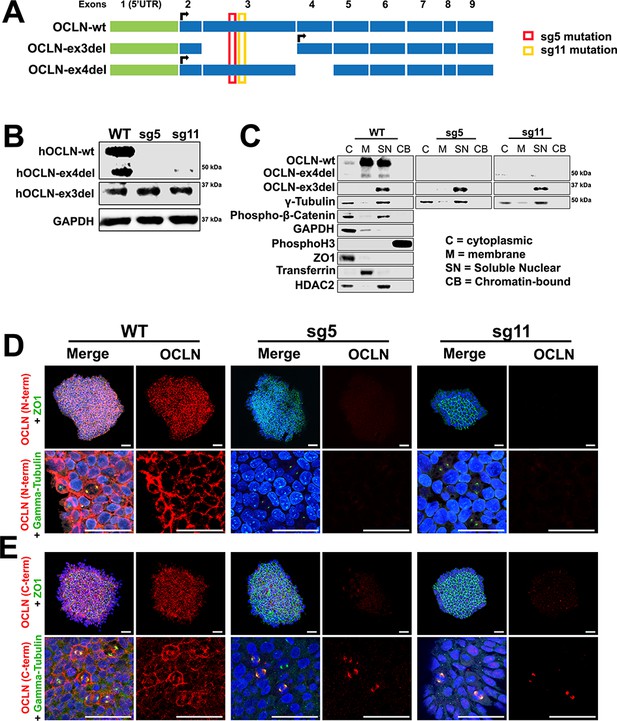
Human full-length and truncated OCLN isoform expression in WT and genome-edited embryonic stem cells.
(A) Human OCLN transcripts explored in this study. Shown are three OCLN isoforms expressed the human brain and that exhibited the lowest intersample variation amongst human liver samples (Kohaar et al., 2010). The location of Cas9/CRISPR targeting is denoted in red and yellow for the sg5 and sg11 mutants, respectively. (B) Western blot analysis of hESC WT, sg5, and sg11 lysates. Representative image is from four independent replicates. GAPDH is used as loading control. (C) Subcellular localization of OCLN by immunoblot analysis of subcellular fractions (cytoplasmic, membrane, soluble nuclear, and chromatin-bound) in WT, sg5, and sg11 hESC lines. Centrosomal markers γ-tubulin and phospho-β-catenin colocalize with OCLN in the soluble nuclear fraction. Fraction-specific controls used are GAPDH and ZO1 (cytoplasmic), HDAC2 (soluble nuclear), Transferrin (membrane), Phospho-histone3 (chromatin-bound). (D and E) Confocal microscope images of hESC colonies from WT, sg5, and sg11 cultures stained with N-terminus-specific (D) and C-terminus-specific (E) OCLN antibodies and either tight junction marker ZO1 or centrosomal marker γ-tubulin. Nuclei are stained with DAPI. Scale bar, 50 um.
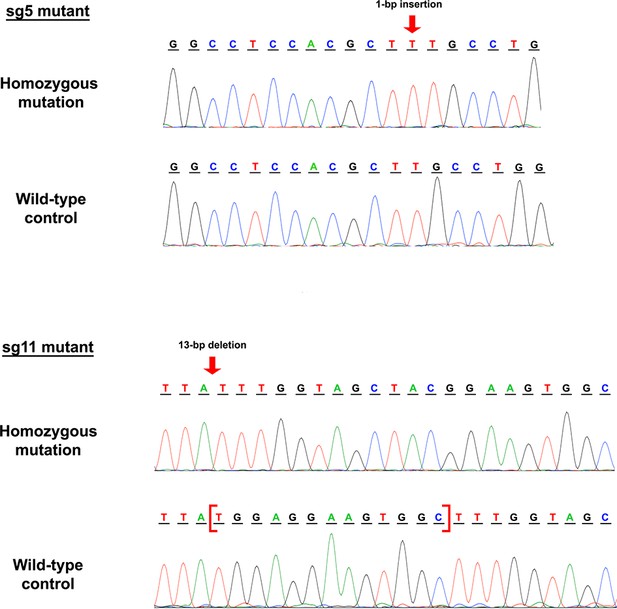
CRISPR mutagenesis targets OCLN at two separate loci on exon 3.
Schematics of OCLN sg5 and sg11 mutant DNA sequence and corresponding chromatograms. Both mutants contain homozygous mutations at their respective targeted loci and their wild-type controls are shown as comparison.

Off-target analysis of sg5 and sg11 OCLN mutant lines.
Schematics of potential off-target loci for OCLN sg5 and sg11 mutant lines, obtained at http://chopchop.cbu.uib.no/.
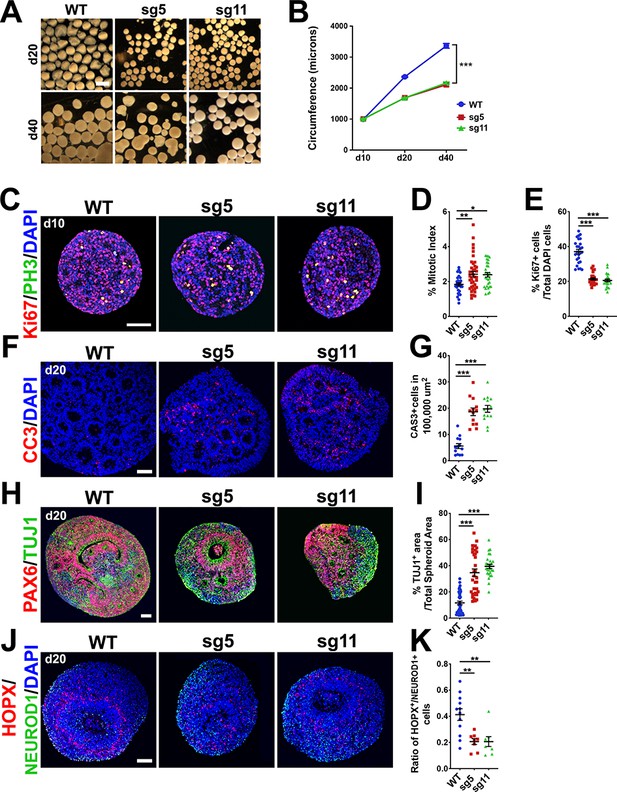
Progenitor proliferation defects, precocious neuronal differentiation and apoptosis in 3-D cortical spheroids from OCLN sg5 and sg11 hESC mutants.
(A) Brightfield images of d20 and d40 organoids derived from WT, sg5, and sg11 hESC cultures. Scale bar, 1 mm. (B) Time-course of d10, d20, and d40 spheroid circumference (in microns). All data points represent mean ± SEM, n = 61 d10 WT organoids, n = 67 d10 sg5 organoids, n = 40 d10 sg11 organoids, n = 246 d20 WT, n = 170 d20 sg5 organoids, n = 208 d20 sg11 organoids, n = 91 d40 WT organoids, n = 98 d40 sg5 organoids, and n = 50 d40 sg11 organoids, collected from five independent experiments for each genotype and each age.. Two-way ANOVA was performed with Tukey’s multiple comparison test, comparing all three lines to each other within each age group, ***p<0.001. (C) Confocal microscope images of 10-micron sections from d10 WT, sg5, and sg11 spheroids stained with proliferation marker KI67 (red), mitotic marker PH3 (green) and counterstained with DAPI (blue). Scale bar, 50 um. (D) Mitotic index, defined as the percentage of proliferating, KI67+ cells co-labeled with PH3 in WT, sg5, and sg11 individual organoids. All data points represent mean ± SEM, n = 32 WT, 39 sg5, and 30 sg11 full spheroids, from three independent experiments. One-way ANOVA with Tukey’s multiple comparison test, **p<0.01, *p<0.05. (E) Percentage of DAPI labeled cells that are KI67+ in WT, sg5, and sg11 individual organoids. All data points represent mean ± SEM, n = 26 WT, n = 23 sg5, and n = 23 sg11 organoids, from three independent experiments. One-way ANOVA with Tukey’s multiple comparison test, ***p<0.001. (F) Confocal images of 10-micron sections from d20 WT, sg5, and sg11 organoids stained with apoptosis marker activated caspase-3 (red) and counterstained with DAPI (blue). Scale bar, 50 μm. (G) Percentage of DAPI labeled cells that are caspase3+. All data points represent mean ± SEM, n = 13–14 organoids for each genotype, from two independent experiments. One-way ANOVA with Tukey’s multiple comparison test, ***p<0.001. (H) Confocal images of 10-micron sections from d20 WT, sg5, and sg11 organoids stained with RGC marker PAX6 (red), neuronal marker TUJ1 (green) and counterstained with DAPI (blue). Scale bar, 50 μm. (I) Quantification of TUJ1+ area per total organoid area or PAX6+ cell percentages in WT, sg5, and sg11 organoids. All data points represent mean ± SEM, n = 38 organoids for WT, n = 34 for sg5, and n = 27 for sg11, from four independent experiments for each group. One-way ANOVA with Tukey’s multiple comparison test, ***p<0.001. (J) Confocal images of 10-micron sections from d20 WT, sg5, and sg11 organoids stained with oRG marker HOPX (red), neuronal marker NeuroD1 (green) and counterstained with DAPI (blue). Scale bar, 50 μm. (K) Ratio of HOPX+ cells to NeuroD1+ cells in WT, sg5, and sg11 individual organoids. All data points represent mean ± SEM, n = 12 WT organoids, n = 8 sg5 organoids, and n = 8 sg11 organoids, from two independent experiments for each group. One-way ANOVA with Tukey’s multiple comparison test, **p<0.01. Detailed tabulation of all means, SEMs, sample sizes, and exact p-values can be found in Figure 6—source data 1.
-
Figure 6—source data 1
Mean, SEM, sample size (n), and exact p-values for Figure 6 quantifications.
- https://cdn.elifesciences.org/articles/49376/elife-49376-fig6-data1-v1.xlsx
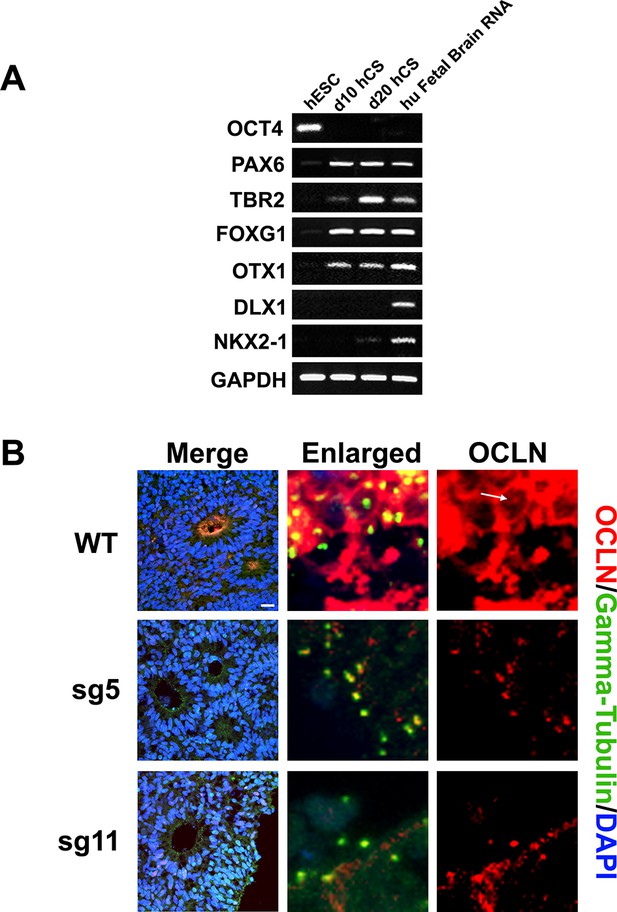
Human Cortical Spheroid (hCS) characterization and OCLN localization.
(A) RT-PCR analysis of undifferentiated hESC, d10, and d20 cortical spheroids for pluripotency marker OCT4, forebrain-specific markers PAX6, TBR2, FOXG1, and OTX1, and ventral forebrain markers DLX1 and NKX2.1. GAPDH is used as loading control. Human Fetal brain RNA is used a positive control. (B) OCLN immunoreactivity in d20 organoids. WT organoids are positive for OCLN at the apical surface as well as at the centrosome (white arrow) while sg5 and sg11 mutants only express OCLN at the centrosome. Scale bar, 20 μm.
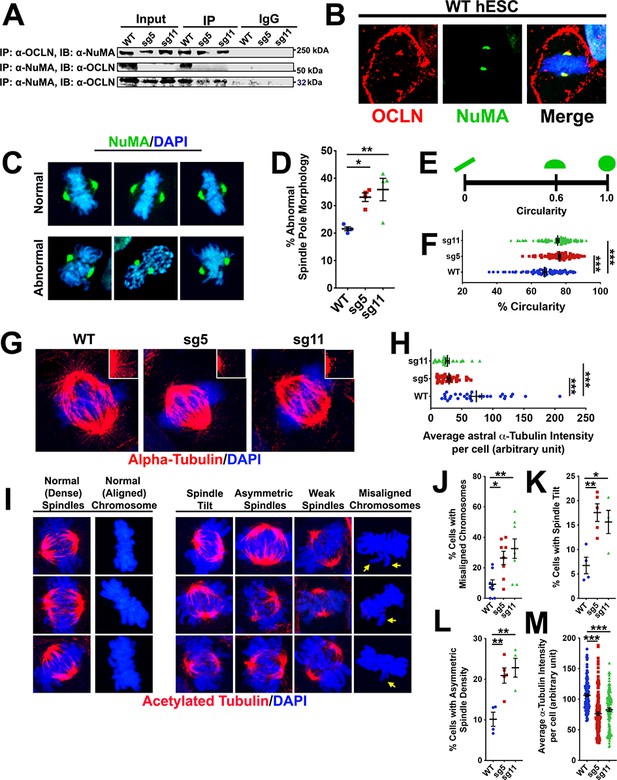
Full-length OCLN deficit impairs hESC mitotic spindle and astral microtubule organization and integrity.
(A) Immunoprecipitation (IP) of endogenous OCLN protein with anti-OCLN antibody and of endogenous NuMA protein with anti-NuMA from hESCs lysates. IP = antibody used to pulldown protein of interest and its adjacent interaction partners; immunoblot (IB) = antibody used to probe for presence of interaction partners. The input represents about 3% of total lysate. Non-specific IgG used as negative control. (B) Confocal images of WT hESCs stained with anti-NuMA (green) and anti-OCLN (red) antibodies. DAPI (blue) stains nuclei. (C) Representative confocal images of normal and abnormal NuMA-labeled spindle pole morphology in green and counterstained with DAPI (blue). (D) Percentage of cells with abnormal spindle pole morphology in WT, sg5, and sg11 culture. All data points represent mean ± SEM, n = 4 independent experiments for each group, with each experiment analyzing 15 cells from that group. One-way ANOVA with Tukey’s multiple comparison test, *p<0.05, **p<0.01. (E,F) Schematic of circularity metric in E) used to analyze the percent circularity of each spindle pole in WT, sg5, sg11. Quantification of % circularity in F) of WT, sg5, sg11 culture. All data points ± SEM, n = 92–115 for all groups, from four independent experiments. One-way ANOVA with Tukey’s multiple comparison test, ***p<0.001. (G) Confocal images of astral spindle as stained with alpha-tubulin (red) and counterstained with DAPI (blue). (H) Quantification of average alpha-tubulin fluorescent intensity specifically of astral spindles in WT, sg5, and sg11 culture. All data points represent mean ± SEM, n = 30 cells for all groups, from three independent experiments for each group. One-way ANOVA with Tukey’s multiple comparison test, ***p<0.001. (I) Representative confocal images of hESCs stained with acetylated alpha-tubulin (red) and nuclear stain DAPI (blue), grouped by their mitotic spindle structure, density, and orientation. (J–L) Percentage of cells exhibiting misaligned chromosomes (J), spindle tilts (K), or asymmetric spindle density (L) in WT, sg5, and sg11 culture. All data points represent mean ± SEM. For spindle tilt and asymmetric spindle densities, n = 4–5 independent experiments, with each experiment analyzing 15 cells each, for all groups. For chromosome misalignment, n = 8 independent experiments for all groups, with each experiment analyzing 15 cells. One-way ANOVA with Tukey’s multiple comparison test, *p<0.05, **p<0.01. (M) Quantification of relative alpha-tubulin fluorescent intensity in WT, sg5, and sg11 culture. All data points represent mean ± SEM, n = 128 cells for WT, n = 137 cells for sg5, and n = 116 cells for sg11, from four independent experiments for each group. One-way ANOVA with Tukey’s multiple comparison test, ***p<0.001. Detailed tabulation of all means, SEMs, sample sizes, and exact p-values can be found in Figure 7—source data 1.
-
Figure 7—source data 1
Mean, SEM, sample size (n), and exact p-values for Figure 7 quantifications.
- https://cdn.elifesciences.org/articles/49376/elife-49376-fig7-data1-v1.xlsx
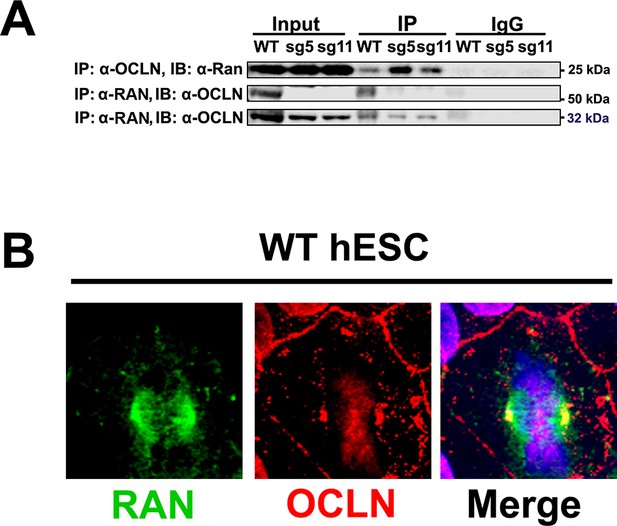
OCLN interacts with mitotic spindle protein RAN.
(A) Immunoprecipitation of endogenous OCLN protein with anti-OCLN antibody and of endogenous Ran protein with anti-Ran from hESCs. Immunoprecipitated proteins (IP) were analyzed by immunoblotted (IB) with indicated antibodies. The input represents about 3% of total lysate. Non-specific IgG used as negative control. (B) Confocal microscope images of WT hESCs stained with Ran (green) and OCLN (red) antibodies. DAPI (blue) was used for nuclear stain.
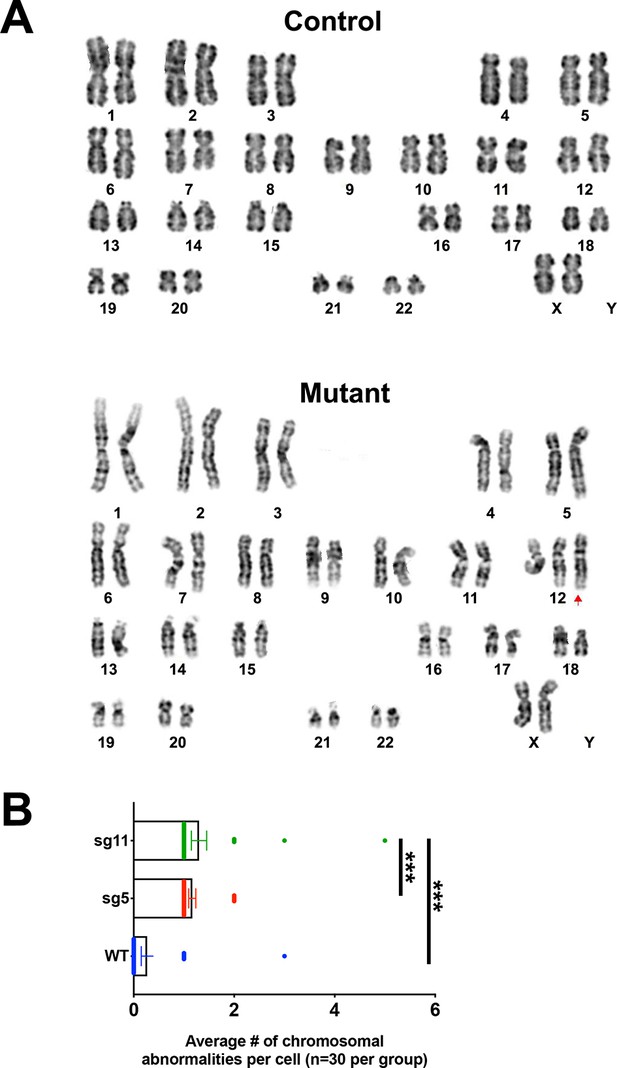
Mutant hESC-derived cortical organoid neural progenitors exhibit increased aneuploidy.
(A) Representative image of G-banded karyotype analysis of WT, sg5, and sg11 metaphase cells isolated from d20 cortical organoids. (B) Quantification of average number of chromosomal abnormalities found per analyzed cell. n = 30 cells per group. One-way ANOVA with Tukey’s multiple comparison test, ***p<0.001.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information | RRID |
|---|---|---|---|---|---|
| Cell line (Homo-sapiens) | Human embryonic stem cell | WiCell | WAe009-A | NIH registry #0062 | RRID:CVCL_9773 |
| Genetic reagent (mouse) | OclnΔN/+ (previously Ocln+/–) | Saitou et al., 2000 DOI: 10.1091/ mbc.11.12.4131 | C57BL/6 background | Gift of Dr. Margaret Neville (University of Colorado, Denver) | RRID:MGI:3716350 |
| Antibody | Anti-Occludin (Rb polyclonal) | Abcam | Cat# ab31721 | IHC(1:4000)* IF(1:4000)* WB(1:2000) *Tyramide signal amplification used | RRID:AB_881773 |
| Antibody | Anti-Occludin (Ms monoclonal) | BD Biosciences | Cat# 611091 | IHC(1:4000)* *Tyramide signal amplification used | RRID:AB_398404 |
| Antibody | Anti-Occludin (Ms monoclonal) | LifeSpan Biosciences | Cat#: LS-B2320-50 | IF(1:2000)* *Tyramide signal amplification used | RRID:AB_1651895 |
| Antibody | Anti-Occludin (Rb monoclonal) | Abcam | Cat#: ab167161 | IHC (1:4000) | RRID:AB_2756463 |
| Antibody | Anti-Pericentrin (Ms monoclonal) | BD Biosciences | Cat#: 611814 | IHC (1:2000) | RRID:AB_399294 |
| Antibody | Anti-Gamma-Tubulin (Ms monoclonal) | Abcam | Cat#: ab27074 | IHC, IF (1:2000) | RRID:AB_2211240 |
| Antibody | Anti-Gamma-Tubulin (Rb polyclonal) | Abcam | Cat#: ab11317 | IHC, IF (1:2000) | RRID:AB_297921 |
| Antibody | Anti-Phospho-Beta-Catenin (Rb polyclonal) | Cell Signaling | Cat#: 9561 | WB (1:1000) | RRID:AB_331729 |
| Antibody | Anti-PhosphoH3 (Rb polyclonal) | EMD Millipore | Cat#: 06–570 | WB (1:2000) | RRID:AB_310177 |
| Antibody | Anti-PhosphoH3 (Ms monoclonal) | EMD Millipore | Cat#: 05–806 | IHC, IF (1:3000) | RRID:AB_310016 |
| Antibody | Anti-Transferrin (Ms monoclonal) | Thermo Fisher | Cat#: 13–6800 | WB (1:2000) | RRID:AB_86623 |
| Antibody | Anti-HDAC2 (Rb polyclonal) | Cell Signaling | Cat#: 2540 | WB (1:2000) | RRID:AB_2116822 |
| Antibody | Anti-GAPDH (Ms monoclonal) | Santa Cruz | Cat#: sc-365062 | WB (1:1000) | RRID:AB_10847862 |
| Antibody | Anti-PAX6 (Rb polyclonal) | Biolegend | Cat#: 901301 | IHC, IF (1:1000) | RRID:AB_2565003 |
| Antibody | Anti-TUJ1 (Ms monoclonal) | Biolegend | Cat#: 801201 | IHC, IF (1:3000) | RRID:AB_2313773 |
| Antibody | Anti-TBR2 (Rb polyclonal) | Abcam | Cat#: ab23345 | IHC (1:2000) | RRID:AB_778267 |
| Antibody | Anti-Ki67 (Ms monoclonal) | Thermo Fisher | Cat#: MA5-14520 | IHC (1:1000) | RRID:AB_10979488 |
| Antibody | Anti-CTIP2 (Rat monoclonal) | Abcam | Cat#: ab18465 | IHC (1:1000) | RRID:AB_2064130 |
| Antibody | Anti-CUX1 (Rb polyclonal) | Santa Cruz | Cat#: sc-13024 | IHC (1:1000) | RRID:AB_2261231 |
| Antibody | Anti-Activated Caspase-3 (Rb monoclonal) | Cell Signaling | Cat#: 9664 | IHC, IF (1:2000) | RRID:AB_2070042 |
| Antibody | Anti-ZO-1 (Ms monoclonal) | Thermo Fisher | Cat#: 33–9100 | IF (1:2000) | RRID:AB_2533147 |
| Antibody | Anti-anti-HOPX (Rb polyclonal) | Sigma | Cat#: HPA030180 | IF (1:2000) | RRID:AB_10603770 |
| Antibody | Anti-NeuroD1 (Ms monoclonal) | Abcam | Cat#: ab60704 | IF (1:2000) | RRID:AB_943491 |
| Antibody | Anti-NuMA (Rb polyclonal) | Abcam | Cat#: ab84680 | IF (1:2000) | RRID:AB_2154610 |
| Antibody | Anti-Alpha-Tubulin (Rb monoclonal) | Abcam | Cat#: ab52866 | IF (1:3000) | RRID:AB_869989 |
| Antibody | Anti-Ran (Ms monoclonal) | BD Biosciences | Cat#: 610341 | WB: (1:2000) IF (1:4000)* *Tyramide signal amplification used | RRID:AB_397731 |
| Antibody | Anti-Acetylated Alpha Tubulin (Ms monoclonal) | Sigma | Cat#: T6793 | IF (1:4000) | RRID:AB_477585 |
| Antibody | Anti-PKC iota (aPKC) (Ms monoclonal) | BD Biosciences | Cat#: 610175 | IHC (1:1000) | RRID:AB_397574 |
| Antibody | Anti-PARD3 (Rb polyclonal) | EMD Millipore | Cat#: 07–330 | IHC (1:1000) | RRID:AB_2101325 |
| Antibody | Anti-Beta-Catenin (Rb polyclonal) | Cell Signaling | Cat#: 9562 | IHC (1:1000) | RRID:AB_331149 |
| Recombinant DNA reagent | pSpCas9(BB)−2A-Puro | Addgene | Plasmid #62988 | RRID:Addgene_62988 | |
| Commercial assay or kit | MycoAlert mycoplasma detection kit | Lonza | Cat#: LT07-318 | ||
| Commercial assay or kit | Tyramide Signal Amplification | Thermo Fisher | Cat#: B40956 | ||
| Commercial assay or kit | Subcellular Fractionation for Cells | Thermo Fisher | Cat#: 78840 | ||
| Commercial assay or kit | Subcellular Fractionation for Tissue | Thermo Fisher | Cat#: 87790 | ||
| Commercial assay or kit | iScript cDNA synthesis kit | BioRad USA | Cat#: 1708890 | ||
| Commercial assay or kit | Pierce BCA assay | Thermo Fisher | Cat#: 23225 | ||
| Other | Accutase | StemCell Technologies | Cat#: 07920 | 1X | |
| Other | Aggrewell plates | StemCell Technologies | Cat#: 34811 | ||
| Other | DMEM/F-12 Medium | Thermo Scientific | Cat#: 11320033 | ||
| Other | B-27 serum without Vitamin A | Thermo Scientific | Cat#: 12587010 | 1X | |
| Other | N2 supplement | Thermo Scientific | Cat#: 17502048 | 1X | |
| Other | GlutaMax | Thermo Scientific | Cat#: 35050061 | 1X | |
| Other | MEM Non-essential Amino Acid | Thermo Scientific | Cat#: 11140050 | 1X | |
| Other | penicillin-streptomycin | Thermo Scientific | Cat#: 15140122 | 100 U/ml | |
| Other | Neurobasal Medium | Thermo Scientific | Cat#: 21103049 | ||
| Other | B-27 supplement without Vitamin A | Thermo Scientific | Cat#: 12587010 | 1X | |
| Other | GlutaMax | Thermo Scientific | Cat#: 35050061 | 1X | |
| Other | 2-mercaptoethanol | Thermo Scientific | Cat#: 21985023 | 1:1000 | |
| Other | SB-431542 | StemCell Technologies | Cat#: 72234 | ||
| Other | LDN193189 | StemCell Technologies | Cat#: #72147 | ||
| Other | ROCK inhibitor Y-27632 | StemCell Technologies | Cat#: 72304 | 10 μM | |
| Other | RIPA buffer | Thermo Fisher | Cat # 89900 | ||
| Peptide, recombinant protein | bFGF | Thermo Fisher | PHG0261 | 20 ng/mL | |
| Peptide, recombinant protein | EGF | Thermo Fisher | PHG0311 | 20 ng/mL | |
| Peptide, recombinant protein | BDNF | Peprotech | 450–02 | 20 ng/ml | |
| Peptide, recombinant protein | NT3 | Peprotech | 450–03 | 20 ng/ml | |
| Chemical compound | 1-bromo-3-chloropentane | Sigma | B62404 |
-
IHC=immunohistochemistry; IF=immunofluorescence; WB=Western Blot; Ms=mouse; Rb=rabbit.
Additional files
-
Supplementary file 1
Primer sequence table.
A compiled list of all primer sequences used in this study, cross-checked by experiment and figure number.
- https://cdn.elifesciences.org/articles/49376/elife-49376-supp1-v1.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/49376/elife-49376-transrepform-v1.docx