Ribosome biogenesis restricts innate immune responses to virus infection and DNA
Figures
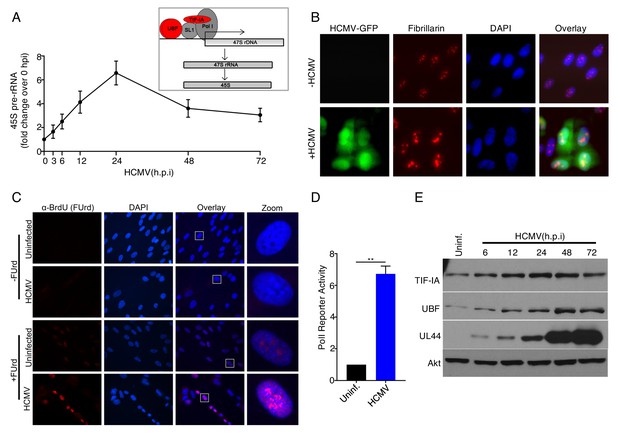
Regulation of RNA polymerase I activity by HCMV in human fibroblasts.
(a) Growth arrested NHDFs were mock- or HCMV-infected (MOI = 3 PFU/cell). Total RNA was isolated at the indicated time points, and RT-qPCR was performed using primers specific for 45S pre-rRNA (n = 4) Inset box: Production of 45S pre-rRNA. Cellular factors involved in RNA polymerase I (Pol I) transcription of DNA that encodes the full length 47S rRNA precursor are depicted (47S rDNA; horizontal arrow indicates direction of transcription). Pol I specific transcription factors (TIF-IA, UBF) referred to throughout the manuscript appear in red. The resulting 47S full length rRNA precursor product which is processed into 45S pre-rRNA are shown below. (b) NHDFs were mock-infected or HCMV infected (MOI = 3 PFU/cell) and fixed in 4% PFA 48hpi. Immunofluorescence staining was performed using an antibody specific for fibrillarin. Signal in the FITC channel represents the HCMV produced eGFP reporter (n = 2). (c) 48hpi, mock- or HCMV- infected NHDFs were labeled with 2 mM 5’-Fluorouridine for 20 min. prior to fixation in 4% PFA. Immunofluorescence staining was performed using an antibody specific for BrdU (n = 2). Rectangle in overlay panel indicates nucleus shown in zoom panel. (d) Growth arrested NHDFs were transfected with pHrP2-BH reporter plasmid. After 24 hr, cells were mock- or HCMV-infected. 24hpi, total RNA was isolated, and RT-qPCR was performed using primers specific for the pHrP2-BH reporter transcript. The error bars indicate SEM. **p≤0.01; Student's t test. (e) Total protein from NHDFs mock infected or infected with HCMV (MOI = 3 PFU/cell) was collected at the indicated times, fractionated by SDS-PAGE, and analyzed by immunoblotting using antibodies specific for TIF-IA, UBF, UL44, and Akt (loading control) (n = 2).
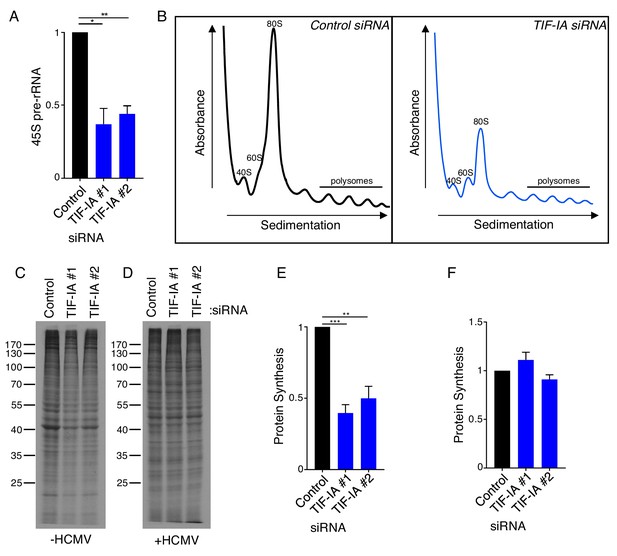
Ribosome biogenesis and protein synthesis are uncoupled in HCMV infected cells.
(a) NHDFs were transfected with non-silencing (ns) control or TIF-IA siRNAs and infected with HCMV (MOI = 3 PFU/cell). At two dpi, total RNA was isolated, and RT-qPCR was performed using primers specific for 45S pre-rRNA. The error bars indicate SEM. *p≤0.05; **p≤0.01; Student's t test (n = 3). (b) As in (a), except cytoplasmic lysate was fractionated over a 10–50% sucrose gradient and the absorbance at 254 nm was recorded. The top of the gradient is on the left. (c–f) NHDFs were transfected with ns or TIF-IA siRNAs and (c,e) mock infected or (e,f) infected with HCMV (MOI = 3 pfu/cell) two dpi, cells were radiolabeled with 35S-amino acids and radioactive amino acid incorporation was (c,d) visualized by SDS-PAGE followed by autoradiography and (e,f) quantified by counting in liquid scintillant (n = 4). Error bars indicate SEM. **p≤0.01; ***p≤0.001; Student's t test.
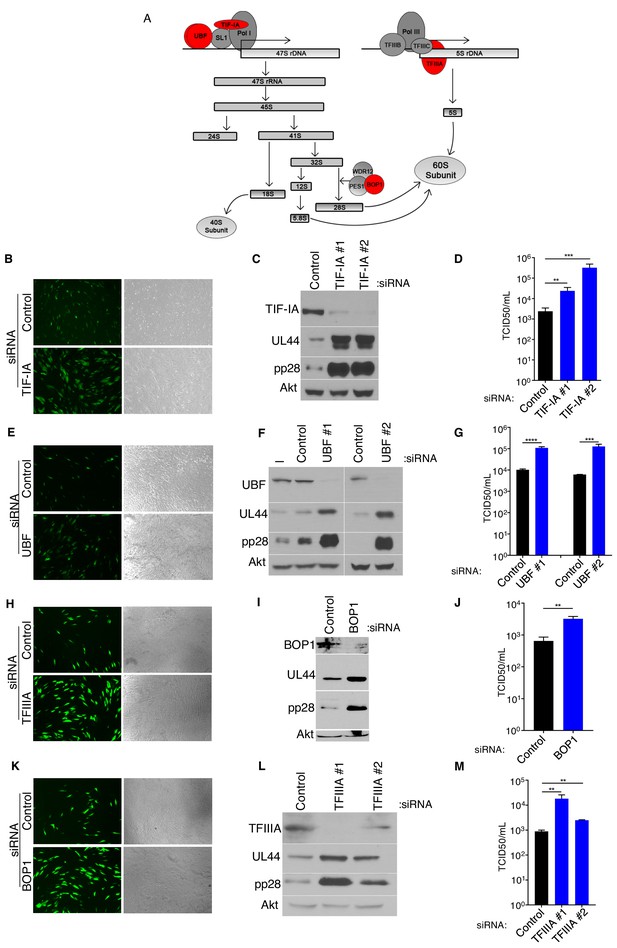
Inhibition of ribosome biogenesis enhances HCMV replication.
(a) Diagram of rRNA biogenesis. Factors targeted in this study are shown in red. (b–k) NHDFs were transfected with non-silencing (ns) control, TIF-IA, UBF, TFIIIA, or BOP1 siRNAs, infected with HCMV (MOI = 0.05 pfu/cell) and (b,e,h,k) six dpi, photographs of eGFP fluorescence were captured to visualize virus replication, (c,f,i,l) total protein was collected, fractionated by SDS-PAGE, and analyzed by immunoblotting using antibodies specific for UL44, pp28, Akt (loading control), and (c) TIF-IA, (f) UBF, (i) BOP1, (l) TFIIIA. (d,g,j,m) Infectious virus was quantified from supernatants using a TCID50 assay. The error bars indicate SEM. *p≤0.05; **p≤0.01; ***p≤0.001; Student's t test. (n = 3).
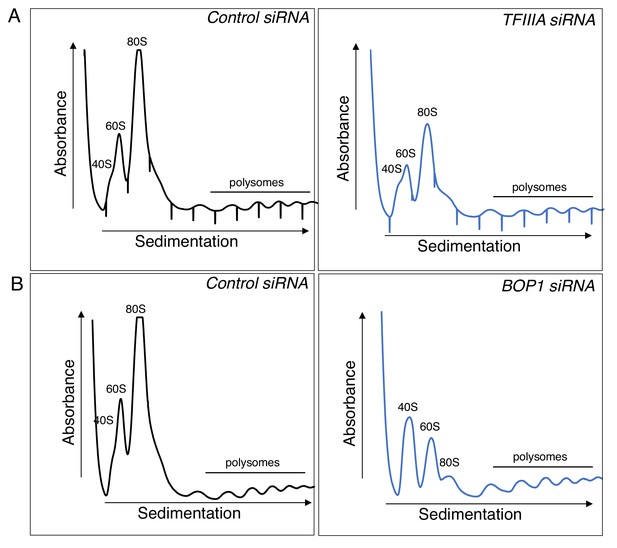
Depletion of TFIIIA or BOP1 reduce ribosome abundances In HCMV infected cells.
(a) NHDFs were transfected with non-silencing (ns) control or TFIIIA siRNA and infected with HCMV (MOI = 3 PFU/cell). At two dpi, cytoplasmic lysate was fractionated over a 10–50% sucrose gradient to resolve ribosomal subunits, monosomes, and polysomes, and the absorbance at 254 nm was recorded. The top of the gradient is on the left. (b) As in (a) except ns control and BOP1 siRNA were used.
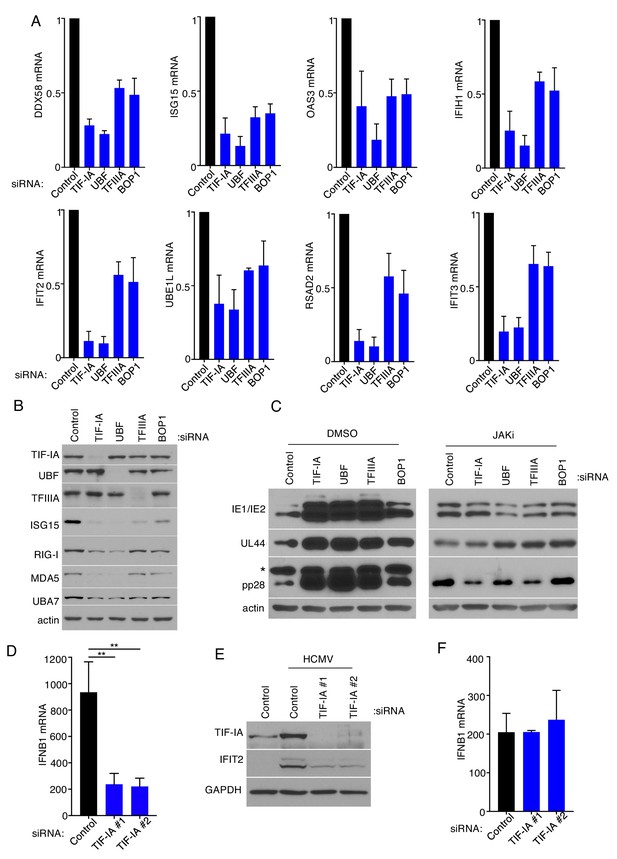
Ribosome biogenesis impairs HCMV protein accumulation by augmenting the innate immune reponse.
(a) NHDFs were treated with non-silencing (ns) control, TIF-IA, UBF, TFIIIA, or BOP1 siRNAs and infected with HCMV (MOI = 3 PFU/cell) two dpi total RNA was isolated and RT-qPCR analysis was performed for DDX58, IFIT2, ISG15, IFIT3, RSAD2, OAS3, UBA7, and IFIH1 mRNA (n = 3). (b) As in (a) except total protein was collected, fractionated by SDS-PAGE, and analyzed by immunoblotting using antibodies specific for TIF-IA, UBF, TFIIIA, ISG15, RIG-I, MDA5, UBA7, and actin (loading control) (n = 2). (c) NHDFs were transfected with the indicated siRNAs. 3 d post-transfection cells were treated with DMSO or JAKi (10 μM) prior to infection with HCMV (MOI = 0.05 pfu/cell) 5 d post-infection total protein was collected, and immunoblotting was performed for IE1/IE2, UL44, pp28, and actin (loading control). *Denotes a non-specific band frequently observed on overexposed blots using anti-pp28 (n = 2). (d,e) NHDFs were transfected with ns control or TIF-IA siRNAs and infected with HCMV(MOI = 3 pfu/cell). (d) Six hpi total RNA was isolated and RT-qPCR was performed using primers specific for IFNB1 mRNA. The error bars indicate SEM. **p≤0.01; Student's t test (n = 3). (e) 48 hpi total protein was collected, fractionated by SDS- PAGE, and analyzed by immunoblotting using antibodies specific for TIF-IA, IFIT2, and GAPDH (loading control) (f). As in (d) except cells were infected with VSV (MOI = 3 PFU/Cell).
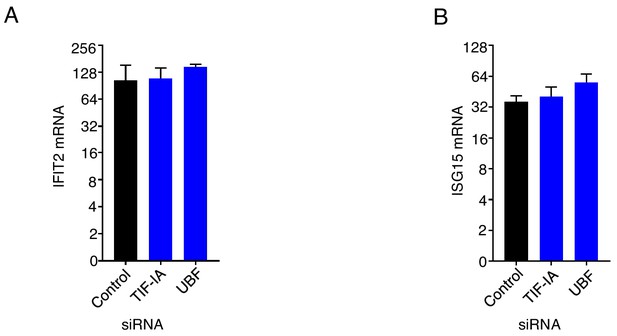
Depletion of RNA polymerase I factors does not impact interferon signaling.
(a,b) NHDFs were transfected with non-silencing (ns) control, TIF-IA, or UBF siRNA. After three days, cells were treated with recombinant IFNα for 6 hr, total RNA was isolated, and RT-qPCR was performed using primers specific for (a) IFIT2 mRNA and (b) ISG15 mRNA (n = 3).
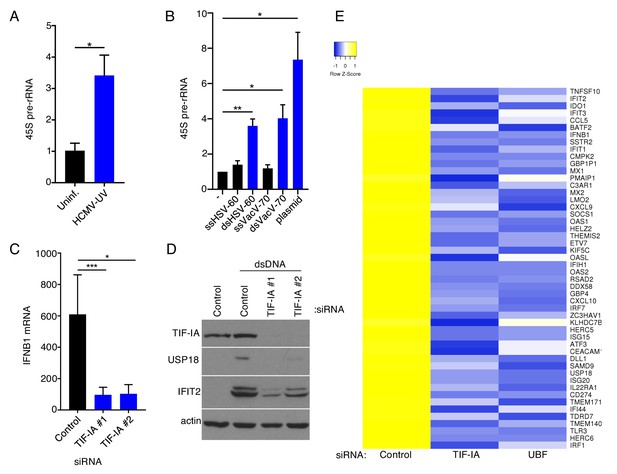
RNA polymerase I factors control accumulation of dsDNA-responsive genes.
(a) Growth arrested NHDFs were uninfected or infected with UV-inactivated HCMV (MOI = 3 PFU/Cell) 24 hr post-transfection total RNA was isolated and RT- qPCR analysis was performed for 45S pre-rRNA. The error bars indicate SEM. *p≤0.05; Student's t test (n = 3). (b) Growth arrested NHDFs were transfected with the indicated DNA. 24 hr post-transfection total RNA was isolated and RT-qPCR analysis was performed for 45S pre-rRNA. Error bars indicate SEM. *p≤0.05; **p≤0.01; Student's t test (n = 4). (c) NHDFs were transfected with non-silencing (ns) control or TIF-IA specific siRNAs. After 3 d, cultures were transfected with either no DNA or dsVacV-70 (dsDNA.) 6 hr post-transfection total RNA was isolated and RT-qPCR was performed using primers specific for IFNB1 mRNA. (n = 3) Error bars indicate SEM. *p≤0.05; ***p≤0.001; Student's t test. (d) As in (c), except 24 hr post-transfection total protein was collected, fractionated by SDS-PAGE, and analyzed by immunoblotting using antibodies specific for TIF-IA, USP18, IFIT2, and actin (loading control). (e) NHDFs were transfected with ns, TIF-IA specific, or UBF specific siRNA. After 3 d, cells were transfected with no DNA or dsDNA for 6 hr after which total RNA was isolated and poly(A) selected. RNA-seq was performed and a heatmap showing differential regulation in TIF-IA and UBF siRNA treated cells of the top 50 genes induced by dsDNA in ns siRNA treated cells was generated.
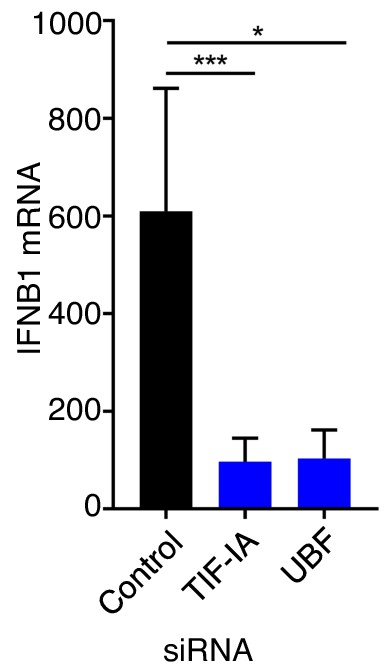
UBF is required for efficient dsDNA-induced accumulation of IFNB1 mRNA.
NHDFs were transfected with non-silencing (ns), TIF-IA, or UBF siRNA. After three days, cells were transfected with dsDNA for 6 hr, total RNA was isolated, and RT-qPCR was performed using primers specific for IFNB1 mRNA (n = 3).
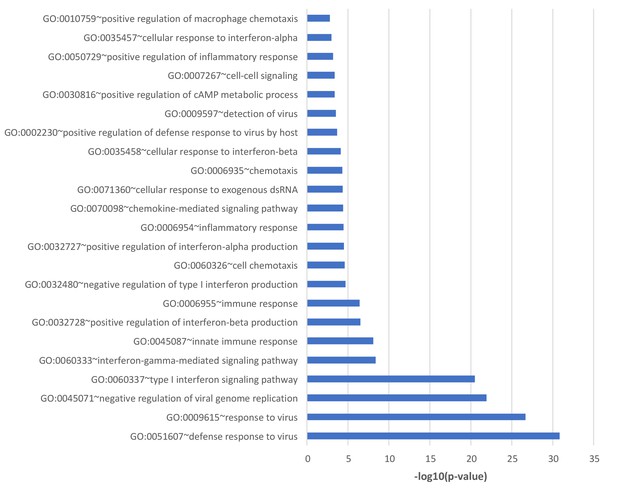
Genome-Wide responses to dsDNA.
Gene ontology analysis (GOTERM_BP_DIRECT) of genes upregulated more than log(2) in non-silencing (ns) control sIRNA treated dsVacV-70 transfected cells compared to ns siRNA treated cells transfected with no DNA was performed using DAVID Functional Annotation Clustering Tool.
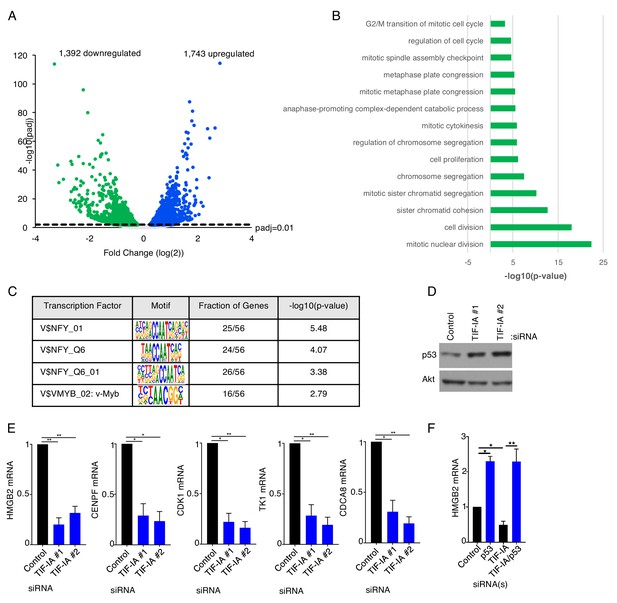
TIF-IA controls the abundances of nuclear factor Y and DREAM complex transcriptional targets.
(a,b) NHDFs were transfected with non-silencing (ns) control siRNA or a TIF-IA siRNA. Three days post transfection total RNA was isolated and RNA-seq was performed and (a) a volcano plot showing differentially expressed genes identified from RNA-seq of cells treated with TIF-IA siRNA (adjusted p-value<0.01) was generated and (b) Gene ontology analysis (GOTERM_BP_DIRECT) of genes downregulated more than four-fold in TIF-IA depleted cells compared to ns siRNA treated cells was performed using DAVID Functional Annotation Clustering Tool (n = 2). (c) TRANSFAC analysis of genes downregulated more than log(2) in TIF-IA depleted cells compared to ns siRNA treated cells was performed using GATHER to identify common modes of transcriptional regulation. (d) 3 d post-transfection of NHDFs with ns or TIF-IA targeting siRNAs, total protein was collected, separated by SDS-PAGE, and analyzed by immunoblotting using antibodies specific for p53 and Akt (loading control) (n = 3). (e) 3 d post-transfection of NHDFs with ns or TIF-IA targeting siRNAs, total RNA was isolated and RT-qPCR was performed for HMGB2 mRNA, CENPF mRNA, CDK1 mRNA, TK1 mRNA and CDCA8 mRNA. Error bars indicate SEM. *p≤0.05; **p≤0.01; Student's t test (n = 3). (f) As in d using the indicated siRNAs and measuring HMGB2 mRNA levels.
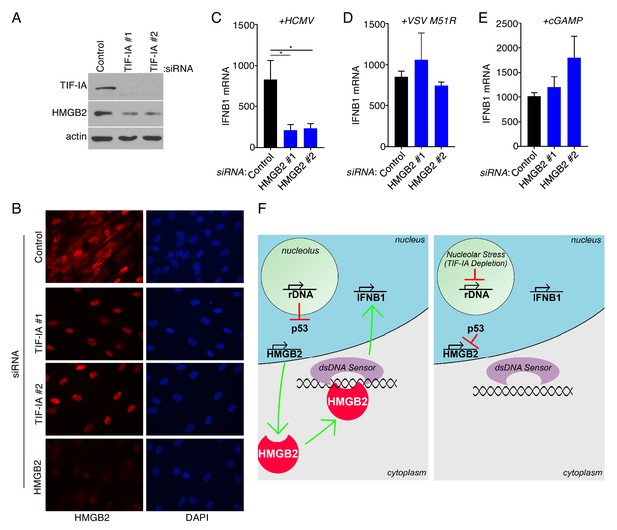
Cytoplasmic HMGB2 abundance is controlled by TIF-IA and dictates the cellular responses to dsDNA.
(a) 3 d post-transfection of NHDFs with non-silencing (ns) control or TIF-IA targeting siRNAs, protein was collected, separated on an SDS-PAGE gel, and analyzed by immunoblotting using antibodies specific for TIF-IA, HMGB2, and actin (loading control) (n = 2). (b) NHDFs were transfected with ns, TIF-IA, or HMGB2 siRNAs. 3 d post-transfection cells were fixed in 4% PFA and immunofluorescence staining was performed using an antibody specific for HMGB2 (n = 2). Nuclei were visualized following DAPI staining. (c–e) NHDFs were transfected with ns or HMGB2 siRNA and (c) infected with HCMV, (d) infected with VSV, or (e) treated with cGAMP and (c–e) 6 hr later, total RNA was isolated, and RT- qPCR was performed using primers specific for IFNB1 mRNA. Error bars indicate SEM. *p≤0.05; Student's t test (n = 3). (f) model depicting how ribosome biogenesis regulates cellular responses to dsDNA via HMGB2. (Left panel) Normal rDNA transcription precludes nucleolar stress-induced p53 stabilization, permitting HMGB2 transcription by RNA pol II in the nucleoplasm and HMGB2 protein (shown in red) accumulation in the cytoplasm. Upon detecting non-microbial or HCMV cytoplasmic dsDNA, HMGB2 either alone or together with a dsDNA sensor like cGAS stimulates IFNB1 mRNA accumulation. (Right panel) Inhibiting RNAPI by TIF-IA depletion results in nucleolar stress and stabilizes p53, which in turn represses HMGB2 transcription, reduces HMGB2 protein abundance and restricts IFNB1 transcription in response to dsDNA.
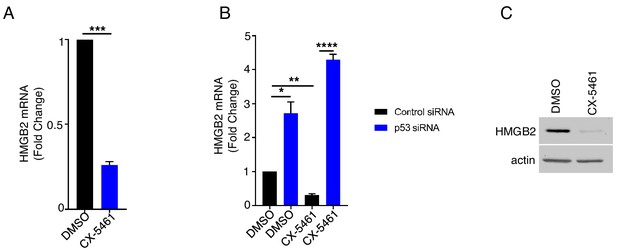
Inhibiting RNA polymerase I reduces HMGB2 abundance in a p53-dependent manner.
(a) NHDFs were treated with DMSO or 1 μM CX-5461. After 3d, total RNA was isolated and RT-qPCR performed using primers specific for HMGB2 mRNA. Error bars indicate SEM. ***p<0.001; Students t-test (n = 3). (b) NHDFs transfected with the indicated siRNA were treated as in (a). *p<0.05; **p<0.01; ****p<0.0001. (c) As in (a) except total protein was collected and analyzed by immunoblotting using the indicated antibodies (n = 2).
Additional files
-
Supplementary file 1
List of genes induced by dsDNA in NHDFs treated with non-silencing siRNA, TIF-IA siRNA, or UBF siRNA.
- https://cdn.elifesciences.org/articles/49551/elife-49551-supp1-v2.xlsx
-
Supplementary file 2
List of genes regulated by TIF-IA depletion in NHDFs.
- https://cdn.elifesciences.org/articles/49551/elife-49551-supp2-v2.xlsx
-
Supplementary file 3
List of primers sequences used for RT-qPCR in this study.
- https://cdn.elifesciences.org/articles/49551/elife-49551-supp3-v2.xlsx
-
Supplementary file 4
List of siRNA sequences used in this study.
- https://cdn.elifesciences.org/articles/49551/elife-49551-supp4-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/49551/elife-49551-transrepform-v2.docx





