Structure-based characterization of novel TRPV5 inhibitors
Figures
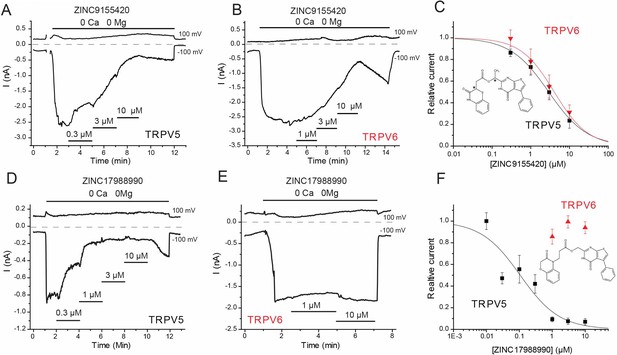
Effects of novel inhibitors on TRPV5 and TRPV6 activity.
HEK293 cells were transfected with rabbit TRPV5 or human TRPV6. Whole cell patch clamp experiments were performed as described in the methods section; monovalent currents were initiated using a divalent cation free solution. Currents are shown at 100 and −100 mV; zero currents are indicated by the dashed lines. (A–B) Representative traces for the effects of ZINC9155420 on (A) TRPV5 and (B) TRPV6 currents, the applications of various concentrations of ZINC9155420 are indicated with the horizontal lines. (C) Summary of the data, current levels after two minutes of applying the various concentrations of the compounds were divided by basal current levels before the application of the drug. The data were fitted using the Hill1 equation with the Origin 8.0 software; data are plotted as mean ± SEM (n = 6 for each concentration for TRPV6 and n = 8–11 for each concentration for TRPV5). Inset shows the chemical formula for ZINC9155420. Asterisks denote chiral centers. (D–E) Representative traces for the effects of ZINC17988990 on (D) TRPV5 and (E) TRPV6 currents, the applications of various concentrations of ZINC17988990 are indicated with the horizontal lines. (F) Summary of the data, analyzed and plotted as in panel C (n = 5–15 for each concentration). Inset shows the chemical formula for ZINC17988990.
-
Figure 1—source data 1
SBVS compound hits.
ZINC IDs and 2D chemical structures for the 65 unique chemical scaffolds identified in the in silico compound screen.
- https://doi.org/10.7554/eLife.49572.004
-
Figure 1—source data 2
In silico hits that had no effect on TRPV5 activity.
Listed are compounds identified in the SBVS that showed no effect on rbTRPV5 current in our system. The effect of each compound was tested at the listed concentration. N indicates the number of replicates tested.
- https://doi.org/10.7554/eLife.49572.005
-
Figure 1—source data 3
ZINC9155420 derivatives that had no effect on TRPV5 activity.
ZINC IDs and 2D chemical structures for the ZINC9155420 derivatives that did not alter TRPV5 activity. The effect of each compound was tested at the listed concentration. N indicates the number of replicates tested.
- https://doi.org/10.7554/eLife.49572.007
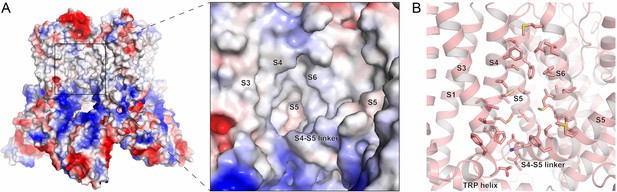
TRPV5 inhibitor binding pocket from in silico screen.
(A) Electrostatic map of tetrameric lipid-bound TRPV5 (PDB: 6DMR) used for SBVS (left). Electrostatic map of the inhibitor binding pocket in the TMD of TRPV5 (right). (B) Cartoon representation of inhibitor binding pocket of lipid-bound TRPV5. Residues involved in the screen are shown as sticks.
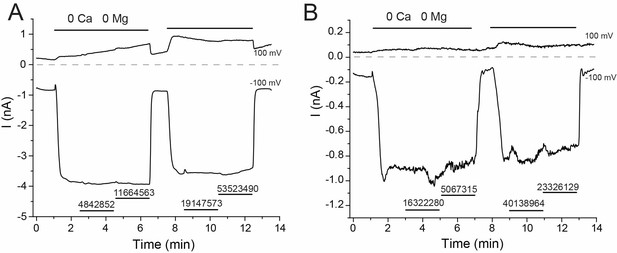
Representative traces for tested compounds that had no effect on rbTRPV5 activity.
Compounds in (A) were tested at 10 μM, compounds in (B) were tested at 3 μM. Compounds are labeled with their ZINC ID. Experiments were performed and displayed as in Figure 1.
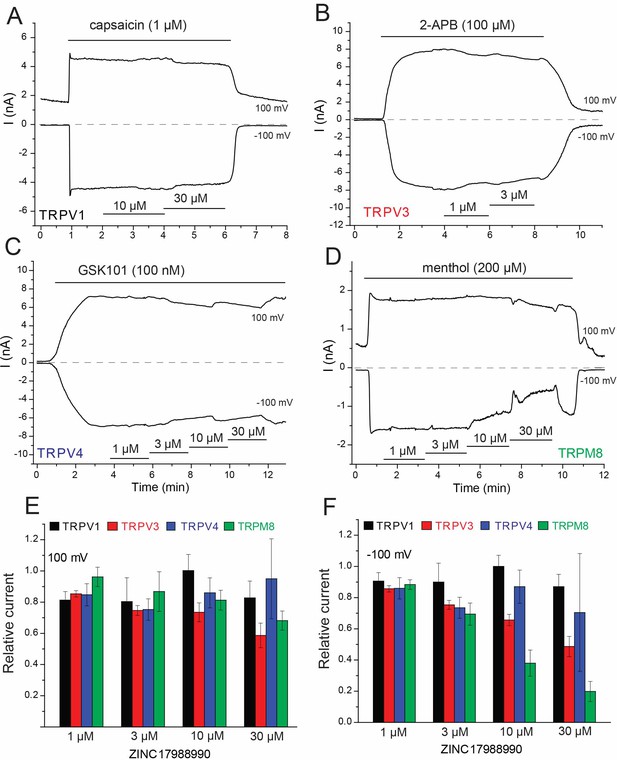
Effect of ZINC17988990 on other TRP channels.
HEK293 cells were transfected with the mouse TRPV1, mouse TRPV3, the rat TRPV4 and the rat TRPM8 clones. Whole cell patch clamp experiments were performed as described in the methods section. (A–D) Representative traces for (A) TRPV1, (B) TRPV3, (C) TRPV4 and (D) TRPM8. The applications of the various channel agonists are shown with the horizontal lines above the current traces, the application of the different concentrations of ZINC17988990 are indicated by horizontal lines at the bottom. (E–F) Summary of the data at (E) 100 mV and (F) −100 mV. Current levels after two minutes of applying the various concentrations of the compounds were divided by agonist-induced current levels before the application of the drugs. Data show mean ± SEM, n = 3–4 for TRPV1, n = 3–6 for TRPV3, n = 4–6 for TRPV4 and n = 5–7 for TRPM8.
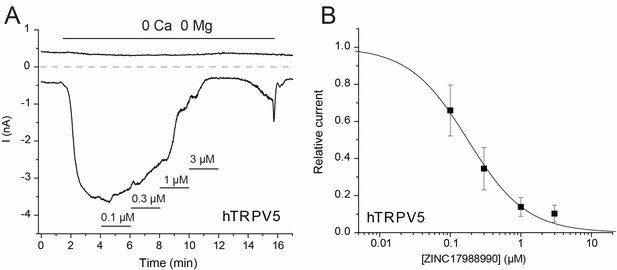
The effect of ZINC17988990 on human TRPV5.
(A) Representative current trace; the applications of various concentrations of the compound are indicated with the horizontal lines. (B) Summary of the data, current levels after two minutes of applying the various concentrations of the compounds were divided by basal current levels before the application of the drugs. The data were fitted using the Hill1 equation with the Origin eight software; data are plotted as mean ± SEM (n = 4–6 for each concentration).
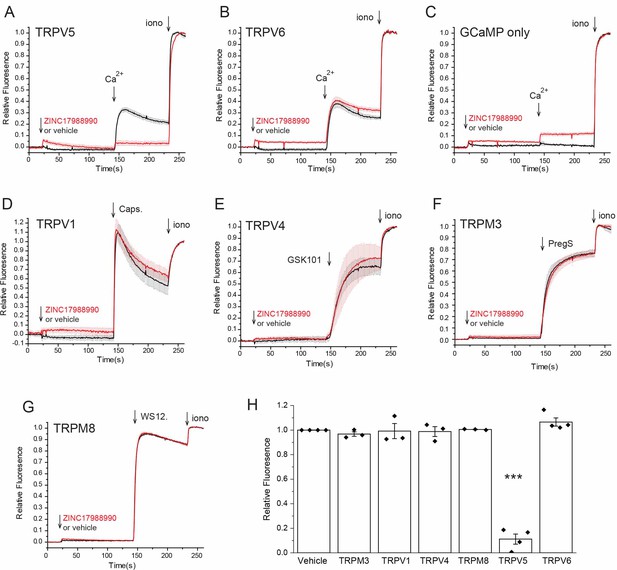
Effect of ZINC17988990 on TRP channel mediated calcium flow.
Intracellular Ca2+ measurements were performed as described in the methods section, using a Flexstation three plate reader. HEK293 cells were transiently transfected with the genetically encoded Ca2+ indicator GCaMP6 and rbTRPV5 (A), hTRPV6 (B), pCDNA3 (C), rTRPV1 (D), rTRPV4 (E), mTRPM3α2 (F) and rTRPM8 (G). For all panels the baseline fluorescence was subtracted first, then all traces were normalized to the maximal fluorescence after the application of ionomycin; the traces show mean ± SEM from three, or four different plates from independent transfections. Application of 10 μM ZINC17988990 or equivalent volume of DMSO (0.033%) at 20 s is indicated by the first arrow. For panels A-C the cells were initially kept in a divalent free solution, and the second arrow indicates the application of 2 mM Ca2+. For panels D-G the cells were kept in a solution containing 2 mM Ca2+, and the second arrow indicates the application of the agonist of the TRP channel tested: 1 μM capsaicin for TRPV1 (D), 100 nM GSK1016790A for TRPV4 (E), 50 μM pregenolone sulfate for TRPM3 (F) and 5 μM WS12 for TRPM8 (G). The third arrow in each panel show the application of the Ca2+ ionophore ionomycin (2 μM). (H) Statistical summary normalized to the relative fluorescence increase induced by the TRP channel agonist or Ca2+ in vehicle treated cells. ***<0.001 (t-test).
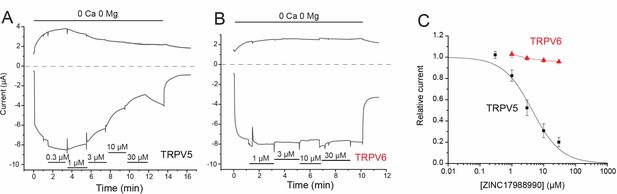
Effects of ZINC17988990 on rbTRPV5 and hTRPV6 expressed in Xenopus oocytes.
TEVC experiments were performed as described in the methods section. (A–B) Shows representative traces for rbTRPV5 and hTRPV6, respectively. (C) Shows data summary. Data were analyzed the same way as for HEK293 cell experiments.
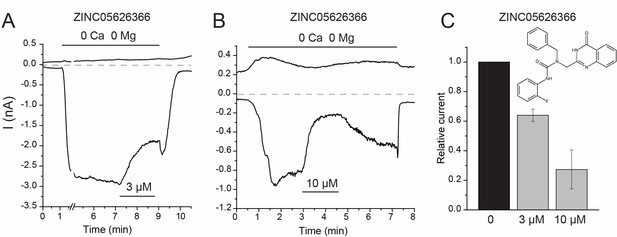
The effects of ZINC05626366 on rbTRPV5.
(A–B) Representative traces for whole cell patch clamp experiments in HEK293 cells expressing rbTRPV5. (C) Data summary, mean + /- SEM n = 3–4. Inset shows the chemical structure of ZINC05626366.
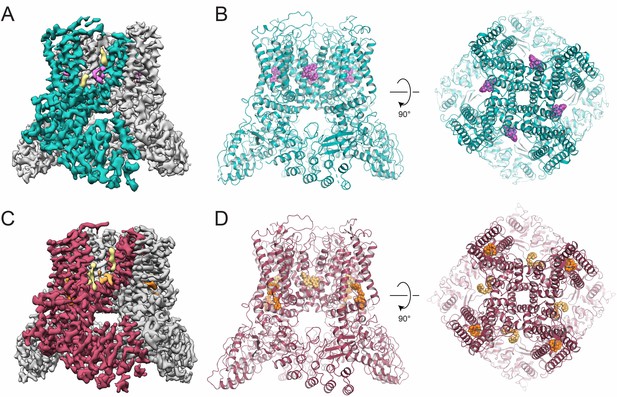
Inhibitor-bound TRPV5 cryo-EM structures.
(A) Cryo-EM density map of ZINC9155420-bound TRPV5 in nanodiscs. A single TRPV5 monomer is colored in teal while the remaining are shown in gray. Density attributed to bound ZINC9155420 is shown in pink and lipids are shown in khaki. (B) Atomic model of ZINC9155420-bound TRPV5 in nanodiscs. One potential pose of the bound ZINC9155420 molecule is shown as pink spheres. (C) Cryo-EM density map of ZINC17988990-bound TRPV5 in nanodiscs. A single TRPV5 monomer is colored in red while the remaining are shown in gray. Densities attributed to bound ZINC17988990 are shown in orange and light orange. Lipids are shown in khaki. (D) Atomic model of ZINC17988990-bound TRPV5 in nanodiscs. Potential poses for bound ZINC17988990 in each binding location are shown as orange spheres.
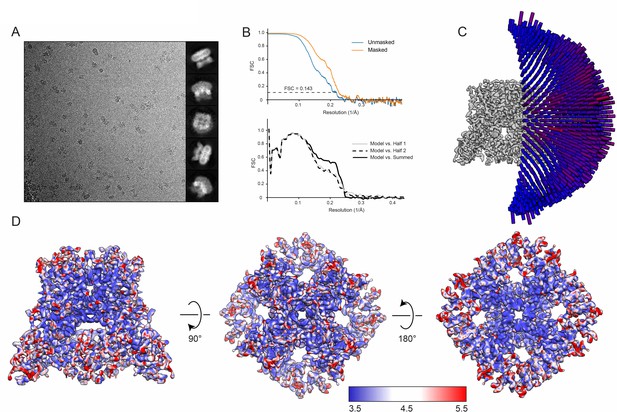
Cryo-EM overview of ZINC9155420-bound TRPV5 in nanodiscs.
(A) (left) Representative 2D micrograph of rbTRPV5 incubated with 10 µM ZINC9155420 in nanodiscs. (right) 2D classes of ZINC9155420-bound TRPV5 in nanodiscs. (B) FSC curves and model validation curves of ZINC9155420-bound TRPV5 in nanodiscs. (C) Angular distribution of particles used to reconstruct the final 3D structure of ZINC9155420-bound TRPV5 in nanodiscs. Tall red cylinders indicate a larger number of particles and short blue cylinders indicate fewer particles. (D) Multiple views of the local resolution of ZINC9155420-bound TRPV5 in nanodiscs. Regions colored blue have local resolutions of 3.5 Å, white regions are 4.5 Å and red regions are 5.5 Å.
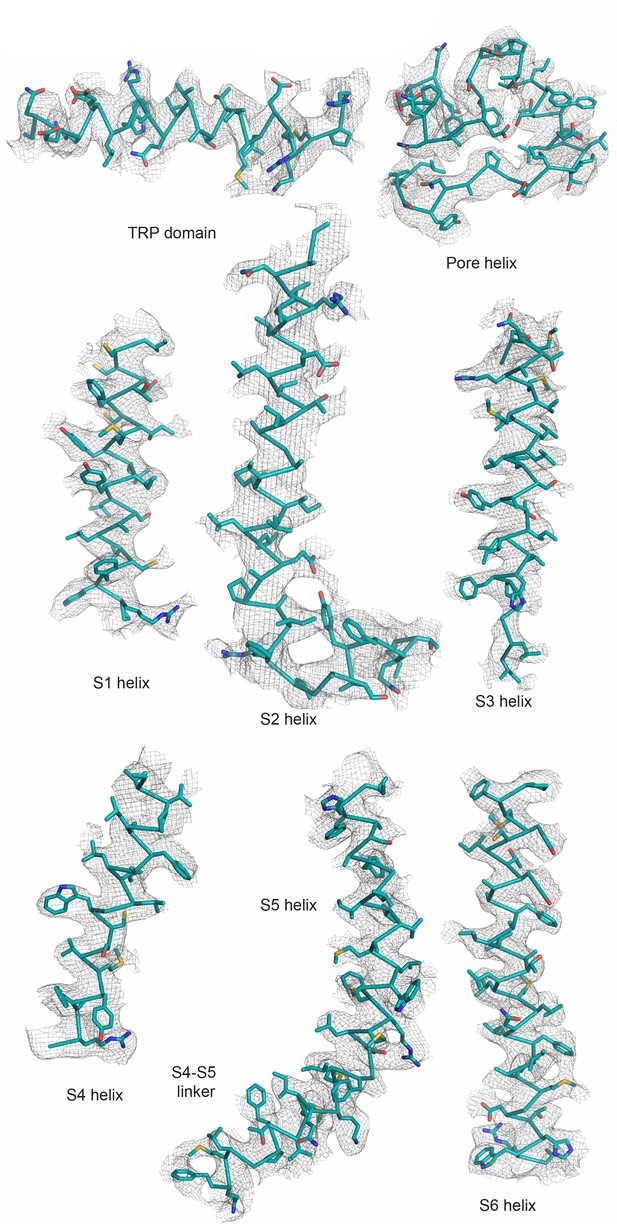
Data quality of ZINC9155420-bound TRPV5 in nanodiscs.
Various helices of ZINC9155420-bound TRPV5 shown as atomic models (teal ribbons) with the corresponding cryo-EM density (gray mesh).
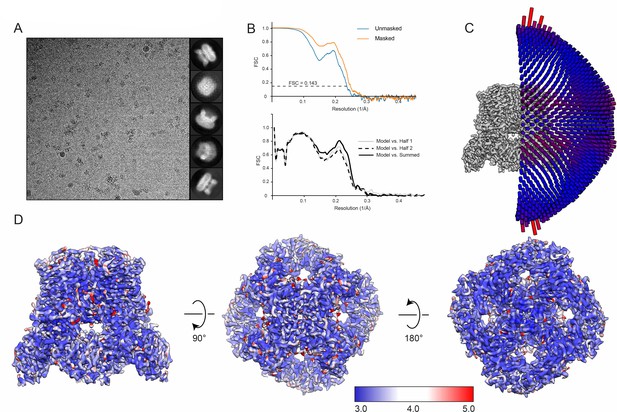
Cryo-EM overview of ZINC17988990-bound TRPV5 in nanodiscs.
(A) (left) Representative 2D micrograph of rbTRPV5 incubated with 10 µM ZINC17988990 in nanodiscs. (right) 2D classes of ZINC17988990-bound TRPV5 in nanodiscs. (B) FSC curves and model validation curves of ZINC17988990-bound TRPV5 in nanodiscs. (C) Angular distribution of particles used to reconstruct the 3D structure of ZINC17988990-bound TRPV5 in nanodiscs. Tall red cylinders indicate a larger number of particles and short blue cylinders indicate fewer particles. (D) Multiple views of the local resolution of ZINC17988990-bound TRPV5 in nanodiscs. Regions colored blue have local resolutions of 3.0 Å, white regions are 4.0 Å and red regions are 5.0 Å.
Data quality of ZINC17988990-bound TRPV5 in nanodiscs.
Various helices of ZINC17988990-bound TRPV5 shown as atomic models (red ribbons) with the corresponding cryo-EM density (gray mesh).
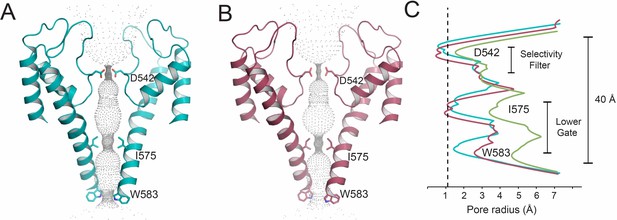
Inhibition of TRPV5 by novel compounds.
(A) Cartoon dimer pore of ZINC9155420-bound TRPV5 with constriction residues labeled and shown as sticks. (B) Cartoon dimer pore of ZINC17988990-bound TRPV5 with constriction residues labeled and shown as sticks. Gray dots indicate the diameter of the pore. (B) Graphical representation of the pore radii of ZINC9155420-bound (teal), ZINC17988990-bound (red) and PI(4,5)P2-bound (green) TRPV5. The dotted line indicates the radius of a dehydrated calcium ion. Constriction residues are labeled.
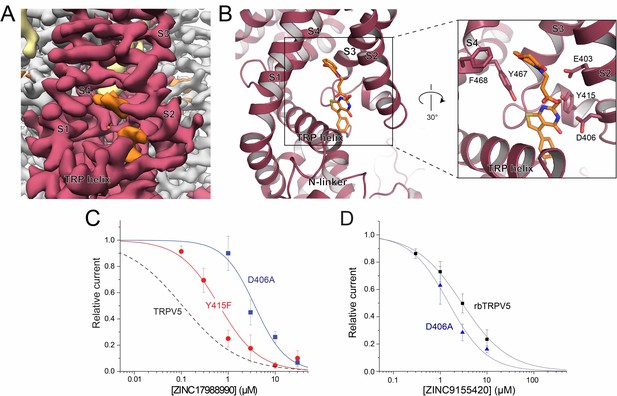
ZINC17988990 binding in the S1-S4 bundle.
(A) Cryo-EM density map of the ZINC17988990 binding pocket in the S1-S4 bundle. Density attributed to ZINC17988990 is shown in orange. Lipids are colored in khaki. The TRPV5 protein is depicted with one monomer in red and three in gray. (B) (left) Atomic model of ZINC17988990 binding pocket in the lower portion of the S1-S4 bundle. (right) Zoomed in view of the binding pocket with residues that could constitute binding labeled and shown as sticks. One potential pose of the ZINC17988990 molecule is shown as orange sticks. (C) Summary of the effects of ZINC17988990 on wild type TRPV5 (dashed line, replotted from Figure 1F) and the Y415F (red) and D406A (blue) mutants analyzed and plotted as in Figure 1. n = 3–9 and 5–11 for each concentration for D406 and Y415F, respectively. (D) Summary of whole cell patch clamp experiments in HEK293 cells expressing wild type rbTRPV5 (black, replotted from Figure 1C) and D406A rbTRPV5 (blue, n = 6 for each concentration) in the presence of ZINC9155420 analyzed and plotted as in Figure 1.
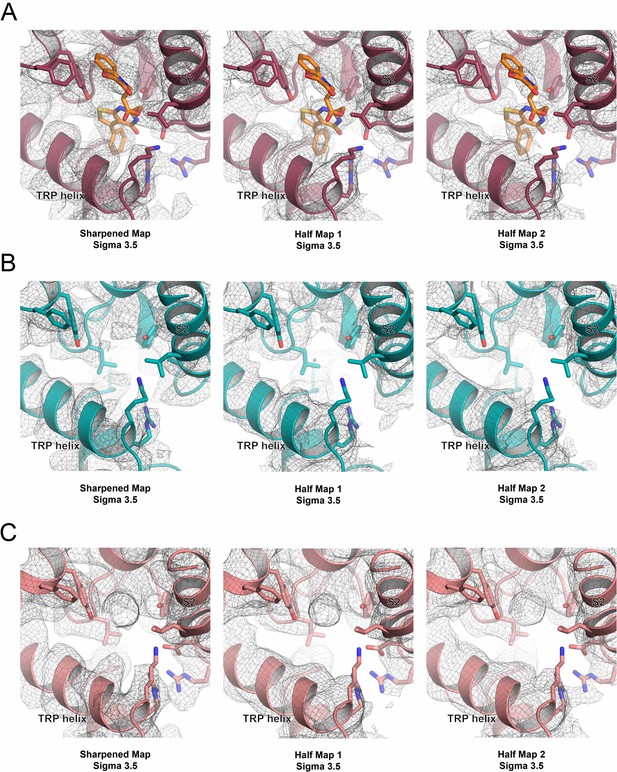
Cryo-EM half maps of intracellular S1-S4 bundle binding pocket.
(A) The intracellular S1-S4 bundle binding pocket in the sharpened map (left), half map 1 (center) and half map 2 (right) of ZINC17988990-bound TRPV5. One potential pose of bound ZINC17988990 is shown as orange sticks. (B) The intracellular S1-S4 bundle binding pocket in the sharpened map (left), half map 1 (center) and half map 2 (right) of ZINC9155420-bound TRPV5. (C) The intracellular S1-S4 bundle binding pocket in the sharpened map (left), half map 1 (center) and half map 2 (right) of lipid-bound TRPV5 (PDB: 6DMR, EMB-7965). The density maps are shown as gray mesh.
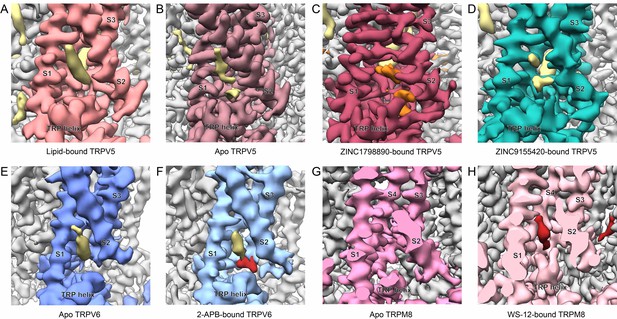
Ligand binding in the intracellular S1-S4 bundle.
Cryo-EM density maps of the intracellular S1-S4 bundle of (A) lipid-bound TRPV5 (EMB-7965) with the lipid shown in khaki, (B) apo TRPV5 (EMB-0594) with lipids shown in khaki, (C) ZINC17988990-bound TRPV5 with ZINC17988990 shown in orange, (D) ZINC9155420-bound TRPV5 with non-protein density shown in khaki (E) apo TRPV6 (EMB-7824) with the lipid shown in khaki, (F) 2-APB-bound TRPV6 (EMB-7825) with the lipid shown in khaki and 2-APB shown in red, (G) apo TRPM8 (EMB-7127) and (H) WS-12-bound TRPM8 (EMB-0487) with WS-12 shown in red.
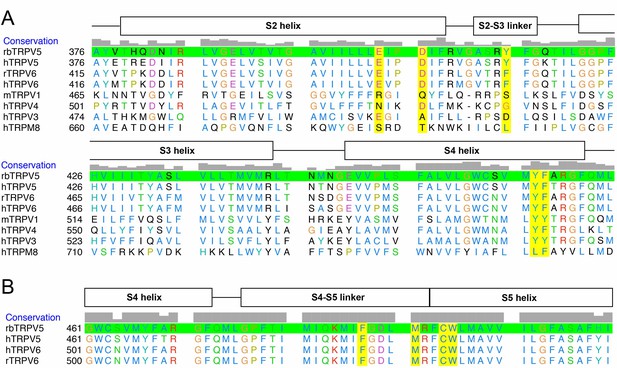
Select sequence alignments.
(A) Sequence alignments for the S1-S4 bundle for various TRP family channels. rbTRPV5 is highlighted in green. (B) Sequence alignments for the S4-S5 linker of multiple orthologs of TRPV5 and TRPV6. Residues that constitute the compound binding sites are highlighted in yellow. Sequence conservation is shown above the alignments and represented as gray boxes.
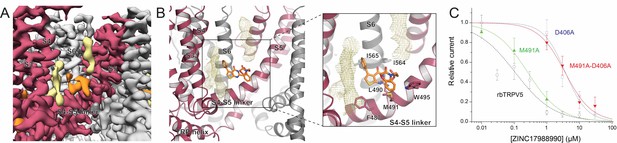
ZINC17988990 interaction with the S4-S5 linker.
(A) Cryo-EM density map of the ZINC17988990 binding pocket in the S4-S5 linker. Density attributed to ZINC17988990 is shown in orange. Lipids are colored in khaki. The TRPV5 protein is depicted with one monomer in red and three in gray. (B) (left) Atomic model of the ZINC17988990 binding pocket in the S4-S5 linker. (right) Zoomed in view of the ZINC17988990 binding pocket with residues that could constitute binding labeled and shown as sticks. One potential pose of the ZINC17988990 molecule is shown as orange sticks and annular lipids are shown as khaki mesh. (C) Summary of the effects of ZINC17988990 on wildtype TRPV5 (black, replotted from Figure 1F), D406A TRPV5 (blue, replotted from Figure 4C), M491A TRPV5 (green, n = 5–6) and M491A-D406A TRPV5 (red, n = 6) analyzed and plotted as in Figure 1.
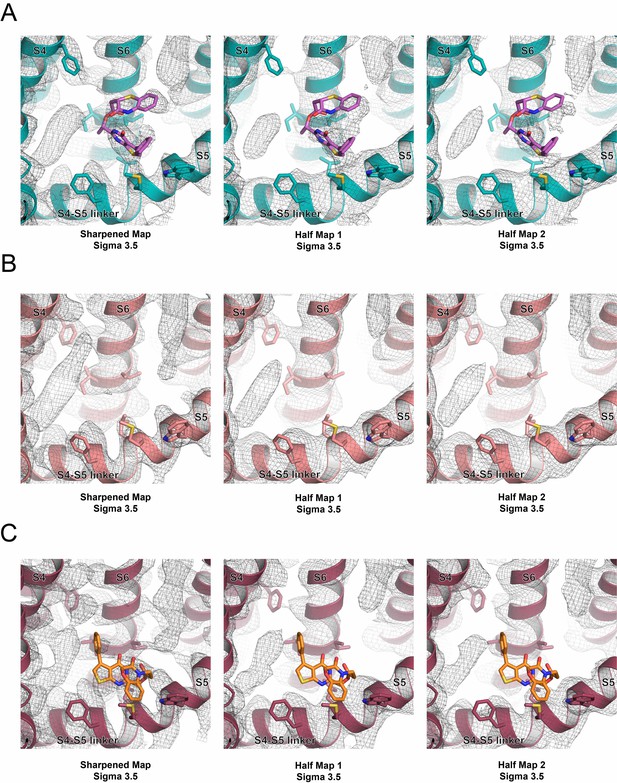
Cryo-EM half maps of the S4-S5 binding pocket.
(A) The S4-S5 binding pocket in the sharpened map (left), half map 1 (center) and half map 2 (right) of ZINC9155420-bound TRPV5. One potential pose of bound ZINC9155420 is shown as pink sticks. (B) The S4-S5 binding pocket in the sharpened map (left), half map 1 (center) and half map 2 (right) of lipid-bound TRPV5 (PDB: 6DMR, EMB-7965). (C) The S4-S5 binding pocket in the sharpened map (left), half map 1 (center) and half map 2 (right) of ZINC17988990-bound TRPV5. One potential pose of bound ZINC17988990 is shown as orange sticks. The density maps are shown as gray mesh.
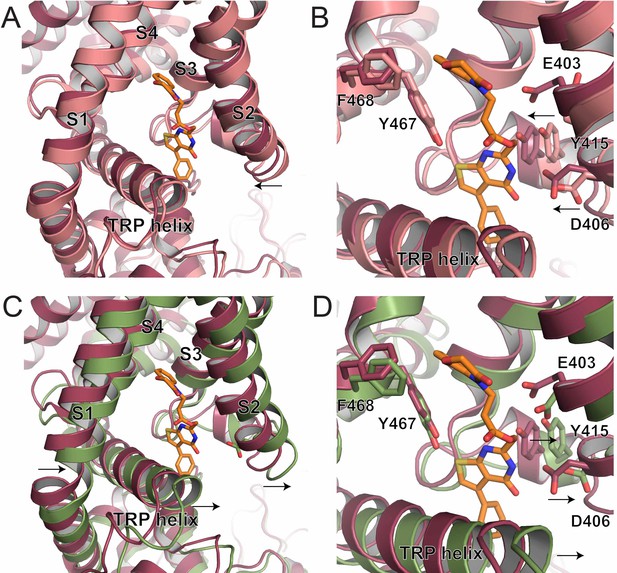
S1-S4 bundle mediated inhibition of TRPV5.
(A) Overlay of the lower S1-S4 bundle in ZINC17988990-bound (red) and lipid-bound (salmon) TRPV5. The arrow indicates the movement attributed to ZINC17988990 binding. (B) Overlay of the inhibitor binding pocket of ZINC17988990-bound (red) and lipid-bound (salmon) TRPV5. The arrows indicate the movement attributed to ZINC17988990 binding. Residues that could constitute ZINC17988990 binding are labeled and shown as sticks. (C) Overlay of the lower S1-S4 bundle in ZINC17988990-bound (red) and PI(4,5)P2-bound (green) TRPV5. The arrows indicate movement necessary for PI(4,5)P2 activation. (D) Overlay of the inhibitor binding pocket of ZINC17988990-bound (red) and PI(4,5)P2-bound (green) TRPV5. The arrows indicate movement necessary for PI(4,5)P2 activation. Labels of residues that could constitute ZINC17988990 binding are labeled and shown as sticks.
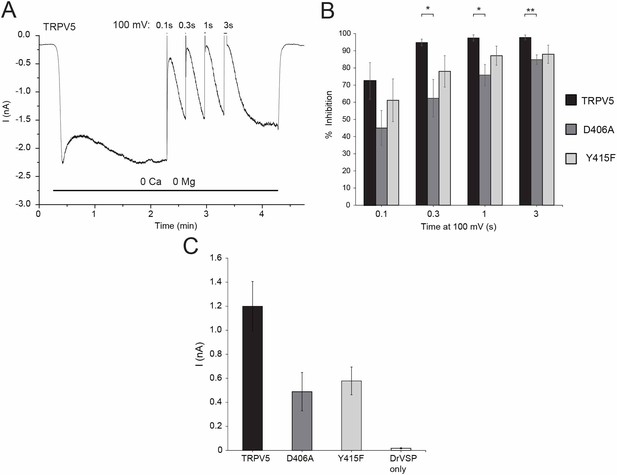
The effect of reducing PI(4,5)P2 levels with a voltage sensitive phosphatase on wild type and mutant TRPV5 channels.
HEK293 cells were transfected with drVSP and TRPV5 or its mutants. Whole cell patch clamp recordings were performed as described in the methods section; the membrane potential was clamped at −80mV. drVSP is inactive at −80 mV, once it is activated by depolarizing membrane potentials, it removes the 5’ phosphate from PI(4,5)P2. (A) Representative current trace in cells transfected with drVSP and wild type rbTRPV5, depolarizing pulses to 100 mV were applied to activate drVSP. (B) Summary of inhibition by the different length depolarization pulses for wild type and the D406A and the Y415F mutants of TRPV5. Statistical significance was calculated with two-way analysis of variance, inhibition of the wild type TRPV5 was significantly different from that of the D406A mutant (p=0.00037), but not from the Y415F mutant (p=0.137) with Tukey’s post hoc test. In the figure, statistical difference was also calculated by uncorrected t-test at individual depolarization lengths, *p<0.05, **p<0.01, (C) Summary of raw current amplitudes (n = 6–8).
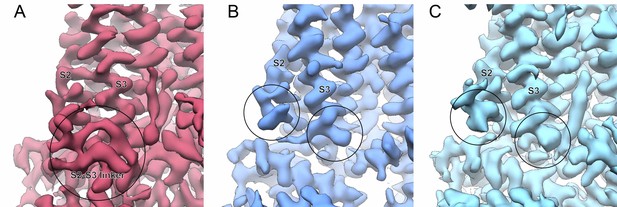
Structural divergence between TRPV5 and TRPV6 at the S2-S3 linker.
(A) Cryo-EM density of ZINC17988990-bound TRPV5 in nanodiscs. The S2-S3 linker is circled and labeled. (B–C) Cryo-EM densities of (B) rat TRPV6 in nanodiscs (EMB-7123) and (C) human TRPV6 in nanodiscs (EMB-7120). The end of the resolved cryo-EM density for both the S2 and S3 helices are circled.
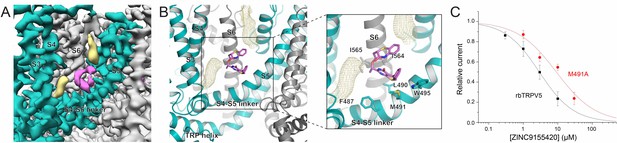
ZINC9155420 binding and inhibition.
(A) Cryo-EM density map of the ZINC9155420 binding pocket. Density attributed to ZINC9155420 is shown in pink. Lipids are colored in khaki. The TRPV5 protein is depicted with one monomer in teal and three in gray. (B) (left) Atomic model of the ZINC9155420 binding pocket. (right) Zoomed in view of the ZINC9155420 binding pocket with residues that could constitute binding labeled and shown as sticks. One potential pose of the ZINC9155420 molecule is shown as pink sticks and densities attributed to lipids are shown as khaki mesh. (C) Summary of the effects of ZINC9155420 on wildtype TRPV5 (black, replotted from Figure 1C) and M491A TRPV5 (red, n = 4–7) analyzed and plotted as in Figure 1.
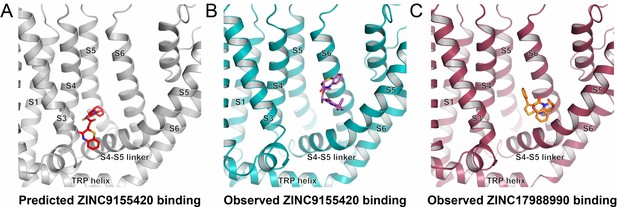
Predicted vs. observed binding.
(A) Predicted ZINC9155420 binding mode at the econazole binding site. (B–C) Observed binding site on the S4-S5 linker based on the cryo-EM data for (B) ZINC9155420 shown as pink sticks and (C) ZINC17988990 shown as orange sticks.
Tables
Cryo-EM data collection and model statistics
https://doi.org/10.7554/eLife.49572.019| ZINC9155420-Bound TRPV5 in nanodiscs (PDB: 6PBF, EMB-20292) | ZINC17988990-bound TRPV5 in nanodiscs (PDB: 6PBE, EMB-20291) | |
|---|---|---|
| Data collection and processing | ||
| Magnification | ~45,500 | ~45,500 |
| Voltage (kV) | 300 | 300 |
| Defocus range (μm) | 1.0–2.5 | 0.8–2.5 |
| Pixel size (Å) | 1.064 | 1.064 |
| Symmetry imposed | C4 | C4 |
| Initial particle images (no.) | 510,500 | 670,057 |
| Final particle images (no.) | 21,802 | 98,516 |
| Map resolution (Å) FSC threshold | 4.2 0.143 | 3.78 0.143 |
| Map resolution range (Å) | 3.5–5.5 | 3.0–5.0 |
| Refinement | ||
| Model resolution cut-off (Å) FSC threshold | 4.3 0.143 | 3.78 0.143 |
| Map sharpening B factor (Å2) | −330 | −253 |
| Model composition | 0 | 0 |
| Nonhydrogen atoms | 2392 | 2416 |
| Protein residues | 4 | 4 |
| Ligands | ||
| R.m.s. deviations | 0.005 | 0.006 |
| Bond lengths (Å) | 0.897 | 0.935 |
| Bond angles (°) | ||
| Validation | 1.70 | 1.72 |
| MolProbity score | 5.02 | 3.88 |
| Clashscore | 0.22 | 1.18 |
| Poor rotamers (%) | ||
| Ramachandran plot | 93.20 | 91.83 |
| Favored (%) | 6.80 | 8.17 |
| Allowed (%) | 0.00 | 0.00 |
| Disallowed (%) |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.49572.031





