Noninvasive quantification of axon radii using diffusion MRI
Figures
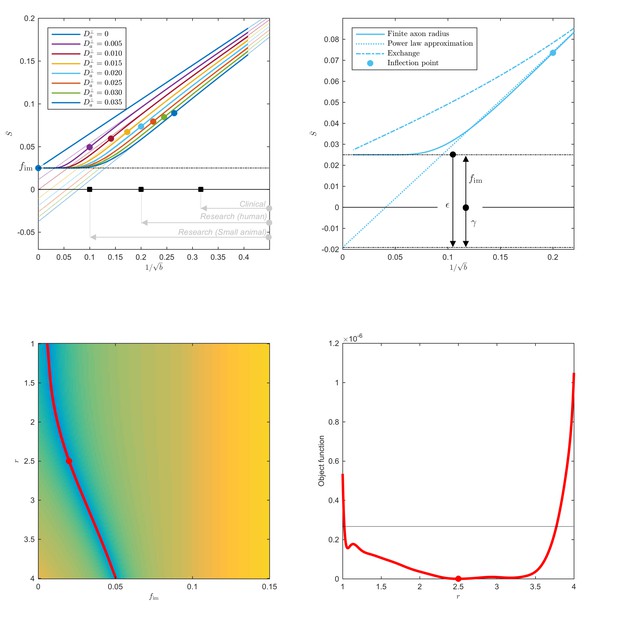
Breakdown of power law scaling: Top left: A nonzero would result in a truncated power-law signal decay.
Although the resulting signal nonlinearity might be too subtle to be discerned within the achievable -range, even for (pre-)clinical systems with strong diffusion-weighting gradients, the concavity of the curves plotted as function of for means that even the smallest will result in an extrapolated intercept when the power law, Equation 1, is used to approximately describe the signal in the delineated -ranges. The intercept is maximally negative at the inflection point (colored dots), beyond which each curve becomes convex, and the negative intercept of the linear extrapolation starts to decrease. In all plots here, diffusivities and -values are expressed in µm2/ms and ms/µm2, respectively. Top right: One representative curve () is shown to highlight the differences between the physically plausible dot compartment , and the intercept . The dot compartment is a positive signal fraction of a biophysical compartment, whereas the intercept is a parameter of the power-law approximation, Equation 1. Their difference depends on various parameters, including the axonal signal fraction, diffusivities, the axon radius, and the scan protocol. The predicted signal decay for the exchange model (dash-dotted; Equation 4) is convex in the entire -range, where the signal decay for the finite axon radius model (dotted; Equation 2) is concave until the inflection point. Bottom: The optimization landscape of Equation 3 shows a shallow valley, relative to the noise floor, for a simulation that mimics the human component of the study. (Bottom left) The valley is shown in a 2D projection of the landscape (shown as a function of radius instead of , see Equation 10). (Bottom right) The fit objective function along the valley is shown (red line) in comparison to the noise floor (dashed line) with an unrealistically high SNR of 250 for the non-DW signal. The red dot indicates the ground truth value.
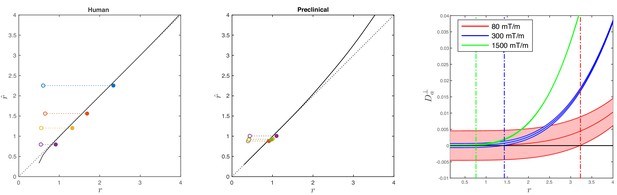
Simulations on accuracy and precision of MR-based axon radius mapping.
First, the left and middle panel show the difference between the estimated, , and theoretical, , effective MR radius associated with various realistic axon caliber distributions (solid dots with different color for different distributions) for the clinical and preclinical setups, respectively. Axon caliber distributions were adopted from Aboitiz et al. (1992) and Innocenti et al. (2015) for the clinical simulations (see Figure 7), whereas various axon distributions (see Figure 4) derived from our own histology were used for the preclinical simulation. The average radii, , of the axon caliber distribution are shown for comparison (open dots). Additionally, the accuracy of the framework for a system with single cylinder with radius is shown (black line). Second (right figure), the feasibility to measure with statistical significance in case of scan settings and SNR for the Connectom (300 mT/m; blue), Aeon (1500 mT/m; green) protocol, respectively. For comparison, we also assessed the feasibility for the Prisma protocol as described in Veraart et al. (2019) (80mT/m; red). The shaded areas illustrate the 95% confidence intervals derived from Cramér-Rao lower bound analysis of model, Equation 2 with . The corresponding minimal cylinder radius that allows for the observation of significant , , 1.41 µm and 3.24 µm for Aeon, Connectom, and Prisma, respectively, is indicated by the vertical lines. In all plots, diffusivities and radii are expressed in and , respectively.
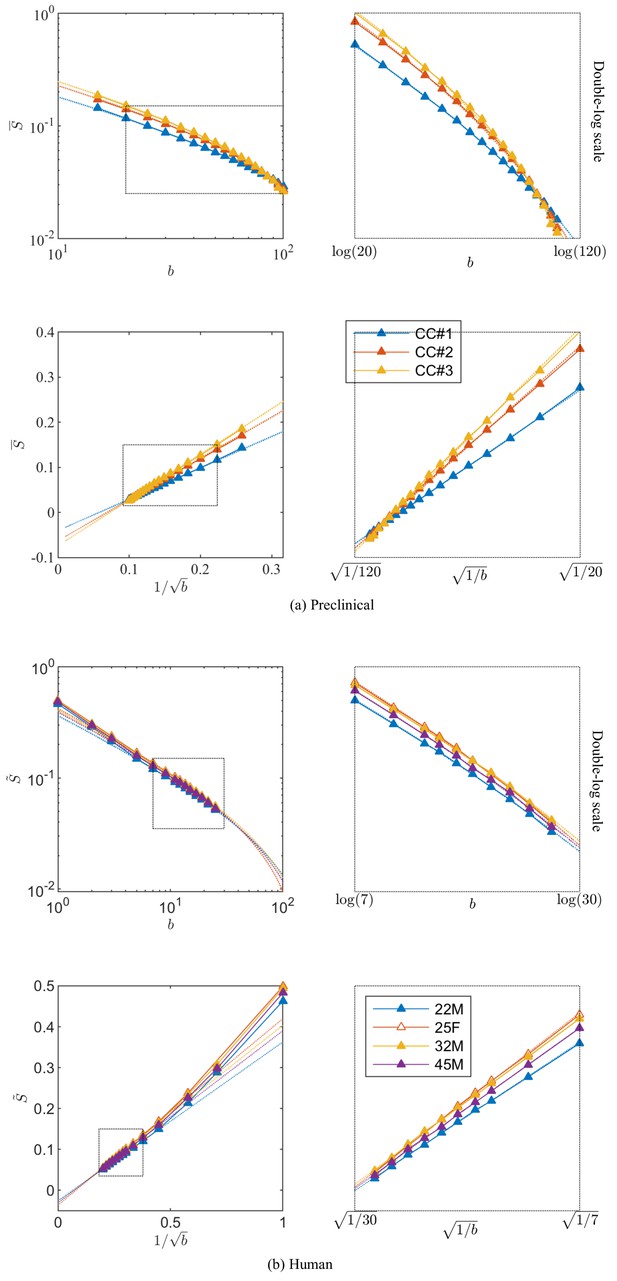
Breaking of the power law.
The ROI- and spherically averaged signal decay is shows for the different fixed samples of the rat CC (a) and human subjects (b) and as a function of and on a double logarithmic scale. The data deviate from the power law scaling with exponent 1/2 that is predicted by the stick model (i.e. nonlinear signal decay in log-log plot), thereby demonstrating sensitivity of the signal to the radial intra-axonal signal. The fits of Equation 1 are shown in dashed lines. In all plots, is expressed in .
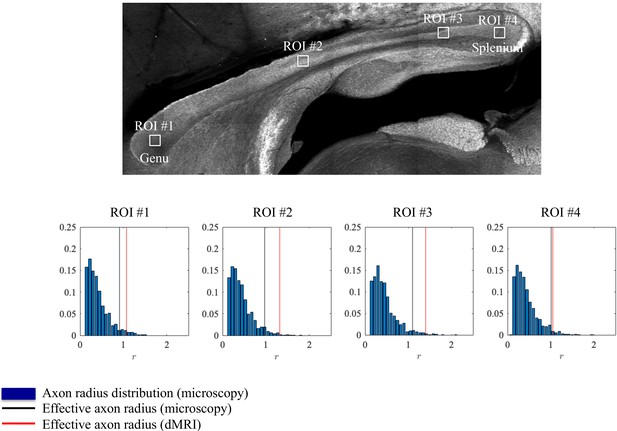
Histological validation, part I.
The axon radius distributions for different ROIs of rat CC#1 are shown (blue bars).The associated tail-weighted effective radii are shown in the black lines, whereas the corresponding MR estimates are shown by the red lines. In all plots, is expressed in .
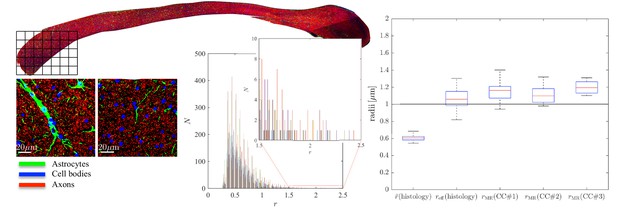
Histological validation, part II.
(left) For a second fixed brain sample, CC#2, the confocal microscopy images, stained for neurofilaments (red), astrocytes (green), and cell bodies (blue), are shown for two representative -patches that are positioned within the Genu (microscopy image of CC shown for ROI positioning). The abundance of astrocytes and cell bodies, both representing 5% of the volume, is clear in both patches. The astrocytic processes can have a large diameter, up to 7 µm in the first patch. A detailed analysis of the radius distribution of the astrocytic processes is not possible due to their random orientation w.r.t. the image plane. (middle) Axon radius distributions for all 16 patches of the Genu (each patch has different color in the bar plot). (right) Boxplots represent the distribution of the average and effective radius of the axon radii distribution that were extracted from each of the 16 patches within the genu. The effective radius is larger than the average , respective medians are 1.06 and 0.61 µm. The boxplots for the MR-derived axon radius measurements for 16 MR voxels within the genu for the three fixed CC samples are also shown. In all plots, radii are expressed in .
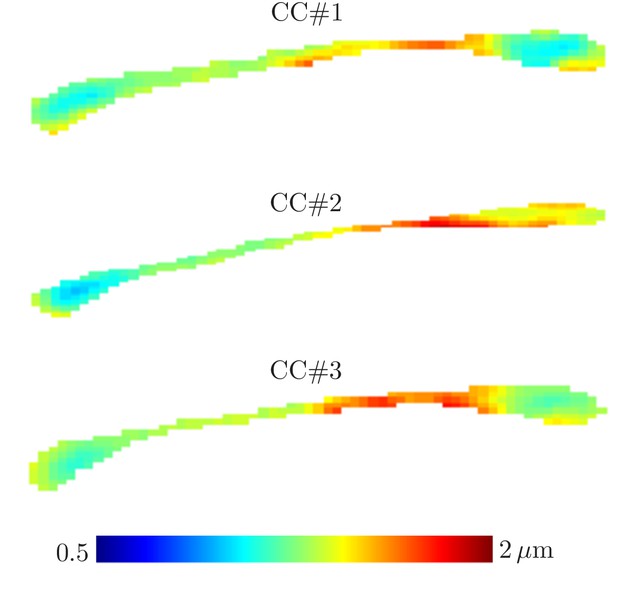
Effective radii in the CC.
Maps of the effective radii derived from diffusion MR data, for the 3 samples of the rat CC.
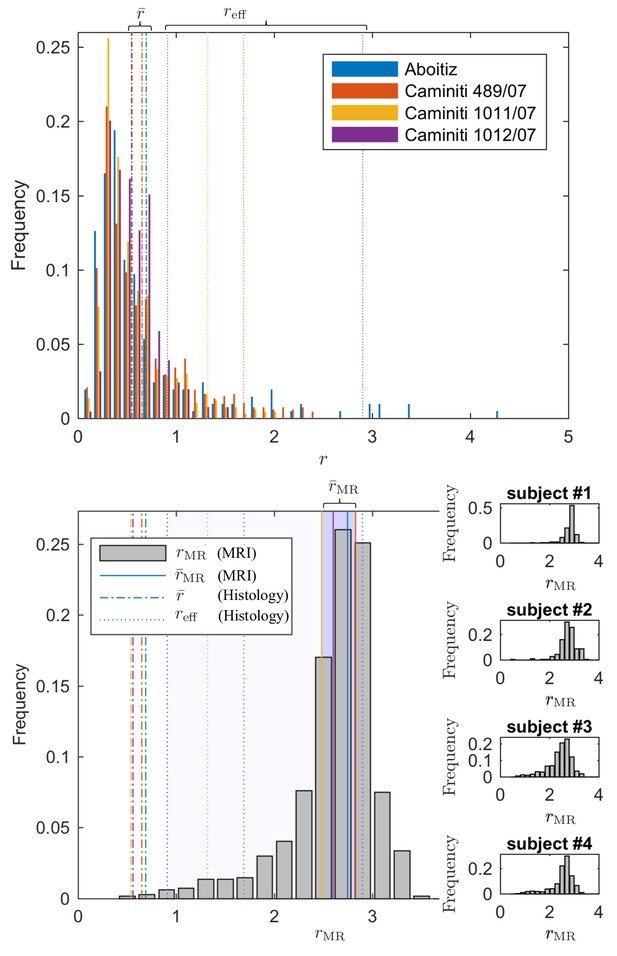
Comparing the effective radius from histology and in vivo dMRI.
(top - histology) Axon radius distributions of multiple histological studies and human CC samples show a good correspondence for the bulk of the distribution, represented by the average radius (dashed-dotted lines). Due to mesoscopic fluctuations of the large axons in histological samples, the corresponding effective radius dominated by large axons, shows strong variability (dotted lines). (bottom - MRI) The four Connectom subjects show good correspondence in terms of . The distribution describing for all voxels in the midbody of the CC for all four subjects falls almost entirely within the range spanned by values predicted by histology, with no need to account for potential shrinkage (Horowitz et al., 2015) during tissue preparation. In all plots, radii are expressed in .
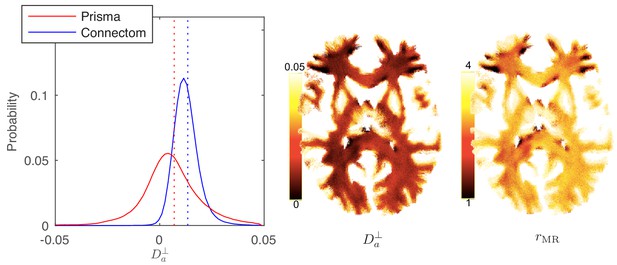
Distribution and maps of and .
(left) The distribution of estimated via Equation 3 for all WM voxels (all scanner-specific subjects pooled) shown for both scan set-ups. In agreement with Figure 2, Prisma (80mT/m) data shows a much lower precision for the estimator of . Despite the small yet positive mean value and the associated negative offset in Figure 3, a large number of WM voxels yield biophysically implausible values. Precision drastically improves on the Connectom scanner (300 mT/m). (right) Maps of , and of the effective MR radius heavily weighted by the tail of axonal distribution (Figure 7), for a single subject. Here, is derived from via Equation 10. In all plots, diffusivities and radii are expressed in and , respectively.
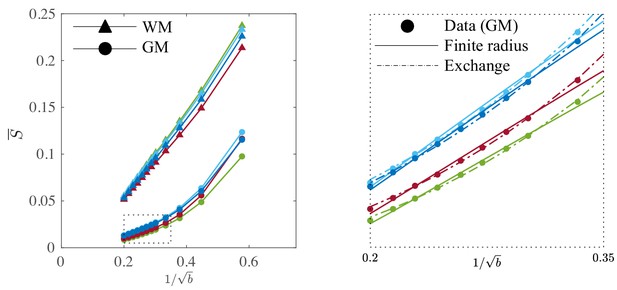
Signal decay in the GM.
The spherically-averaged signal decay in the WM and GM is shown for all human subjects as a function of . The consistent non-linear scaling of the signal as a function of demonstrates deviations from the ‘stick’ model in the cortical GM. In contrast to the WM, the convex signal decay in the GM is better described by an anisotropic exchange model of two compartments (Equation 4), than the finite radius model (Equation 3). In all plots, is expressed in .
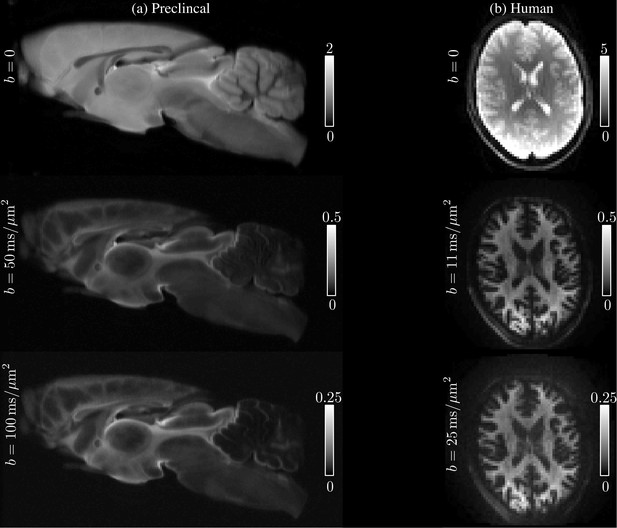
Raw data.
The spherically-averaged diffusion-weighted images, prior to any other image corrections, are shown for various low and high -values for one rat brain sample (a) and one human subject (b).
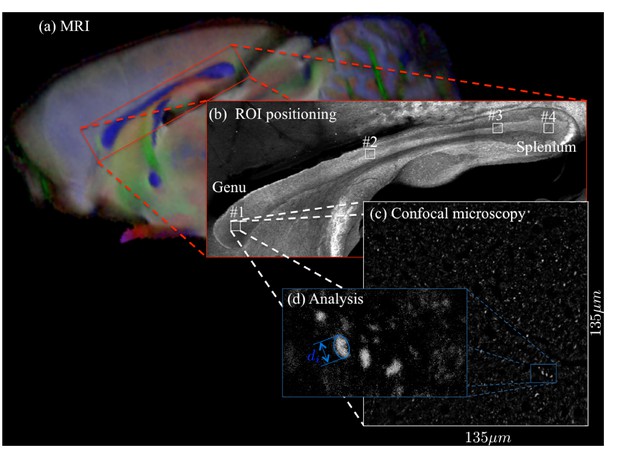
For two brain samples, MR scanning (a, color encoded FA map) was followed by low (b) and high (c) resolution confocal microscopy with staining for neurofilaments to identify the axons.
The low-resolution image was used to position various ROIs, whereas the axon caliber distributions were extracted from the high-resolution image of the corresponding ROIs. The long axes of fitted ellipsoids served as proxies for the respective axon diameters (d).
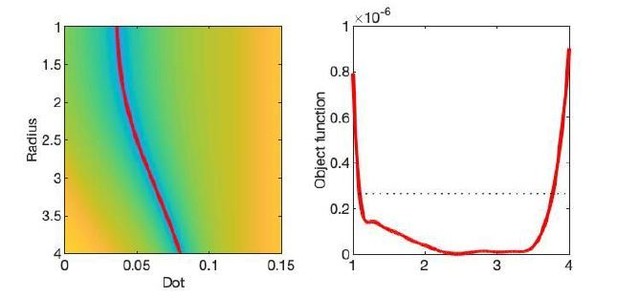
The optimization landscape of model (III) shows a shallow valley, relative to the noise floor, for a simulation that mimics the human component of the study.
(left) The valley is shown in a 2D projection of the landscape (plot shown as a function of radius instead of Daperp). (right) The fit objective function along the valley is shown (red line) in comparison to the noise floor with an unrealistically high SNR of 250 for the non-DW signal.
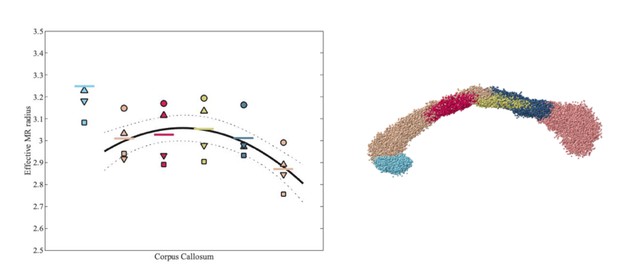
Effective MR radius for various segments of the human CC, including rostrum (light blue) and genu to splenium (from left to right), for each of the 4 subjects.
Each subject is represented by a subject-specific marker. The segmentation of the CC is shown on the right hand side.
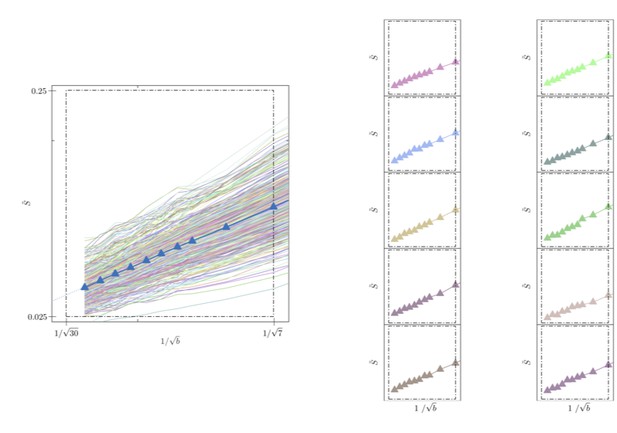
Signal decay in a single voxel.
(left) The spherically-averaged signal decay is shown as a function of for all individual voxels of the WM of one human subject (faded colored lines) and for the average across all WM voxels (blue line with markers, Figure 5B). (right) The signal decay for individual voxels is shown for 10 arbitrarily-chosen WM voxel. The dashed box is positioned the same for all graphs.
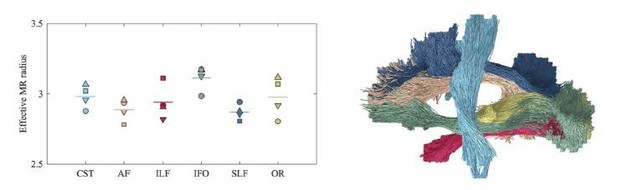
Average effective MR radius within various tracts (color encoded as shown on the right; both hemispheres were considered simultaneously) for the 4 human subjects (encoded by marker).
The line segments show the mean across the subjects. The inter-tract variability exceeds the inter-subject variability. We hypothesize that the hereby introduced technique can be used in future studies to characterize the typical, developing, or pathological brain in a wide range of species, including humans, rodents, or non-human primates.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | anti-Neurofilament 160/200 (Mouse monoclonal) | Sigma Aldrich | Cat# N2912 (clone RMdo20) | 2.5 µg/mL |
| Antibody | anti-GFAP (rabbit polyclonal) | Thermo Fisher Scientific | Cat# PA1-10019 | 1:1000 |
| software. algorithm | ImageJ | imagej.nih.gov/ij/ | RRID: SCR003070 | 1.52q |
| software. algorithm | FSL | fsl.fmrib.ox.ac.uk/fsl/ | RRID: SCR002823 | v6 |
| software. algorithm | MRtrix | www.mrtrix.org | RRID: SCR006971 | v3.0 |
| software. algorithm | FreeSurfer | surfer.nmr.mgh.harvard.edu | RRID: SCR001847 | v6.0.0 |
| Other | DAPI | Sigma Aldrich | Cat# D9542 | 500nM |





