Dendritic NMDA receptors in parvalbumin neurons enable strong and stable neuronal assemblies
Figures
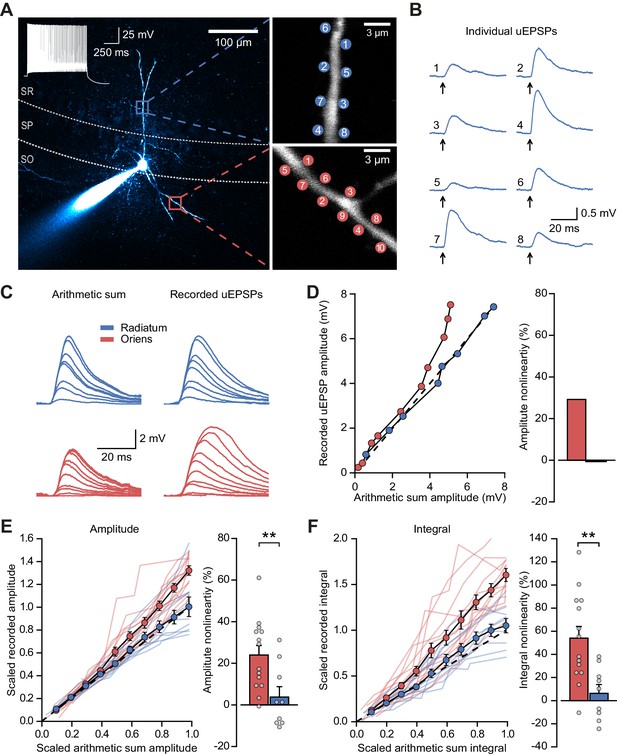
Differential input integration at stratum oriens and stratum radiatum dendrites of PV+ interneurons.
(A) Two-photon z-projection image of a PV+ interneuron recorded via a patch pipette in stratum pyramidale (SP) and filled with Alexa-594 (left, inset: firing pattern in response to current injection), with two dendritic regions of interest at higher magnification (right: top, stratum radiatum, SR; bottom, stratum oriens, SO), showing glutamate uncaging locations (numbered). (B) Individual uEPSP responses from radiatum dendritic locations shown in (A). (C) Comparison of arithmetic sum of individual uEPSPs and recorded uEPSPs evoked by near-synchronous uncaging at multiple locations in stratum radiatum (blue) and oriens (red). (D) Peak amplitudes of recorded uEPSPs plotted against arithmetically summed waveforms for the two regions shown in (A). Dashed line shows line of identity. Right: bar chart showing percentage amplitude nonlinearity. Red: oriens, blue: radiatum. (E) Summary of scaled peak amplitude comparisons for all cells (oriens locations: n = 14, radiatum locations: n = 9). Filled circles and error bars indicate mean ± SEM. Right: bar chart showing quantification of amplitude nonlinearity. (F) Time-integral nonlinearity plotted as for (E). **: p<0.01.
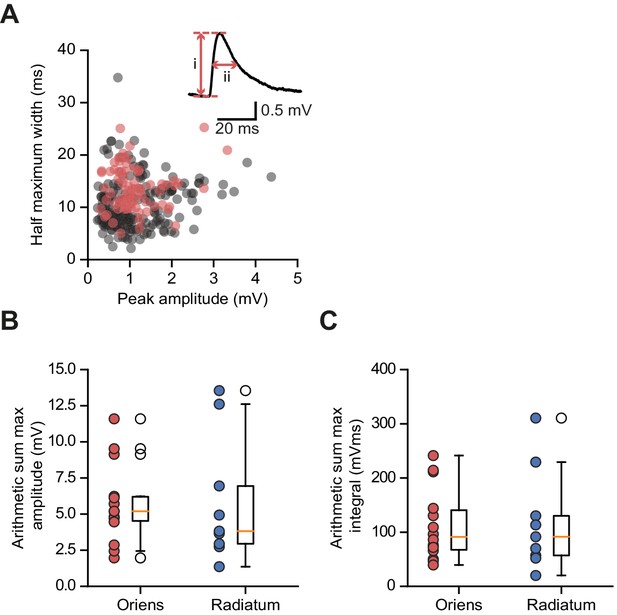
Somatic glutamate uncaging-evoked membrane responses.
(A) Comparison of uEPSPs to spontaneous EPSPs. Half-maximum width (ms) plotted against peak amplitude (mV) for n = 210 spontaneous events (gray) and n = 78 uEPSPs (red); data from 4 cells. Inset: peak amplitude (i) and half-width (ii) for one event. (B) Distribution of arithmetic sum of peak amplitudes for oriens and radiatum uncaging locations. (C) As (B) but for arithmetic sum of time-integrals.
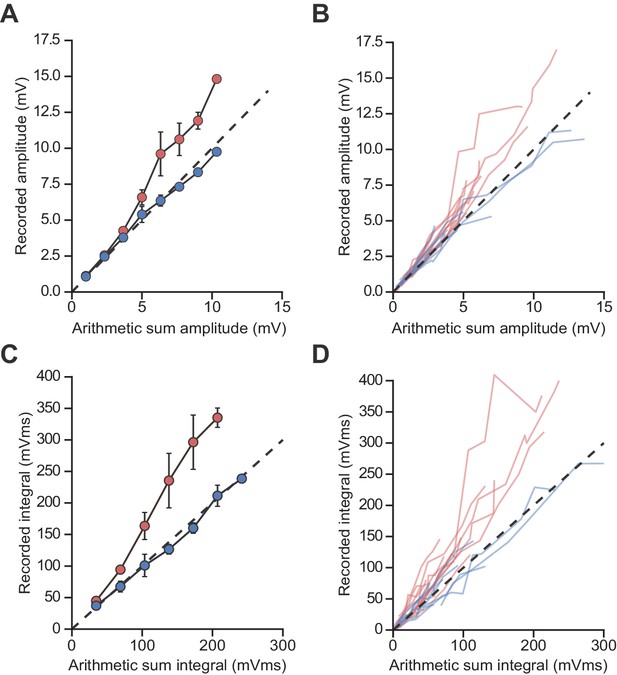
Unscaled uEPSP integration location-dependent nonlinearity.
Comparisons of the arithmetic sum of individual uEPSPs and corresponding recorded uEPSPs evoked at locations in stratum radiatum (blue, n = 9) and stratum oriens (red, n = 14). (A, B) Unscaled uEPSP amplitudes, average (A) and raw data (B). (C, D) Unscaled uEPSP integrals, average (C) and raw data (D). For panels (A) and (C) linearly interpolated averages are shown ± SEM.
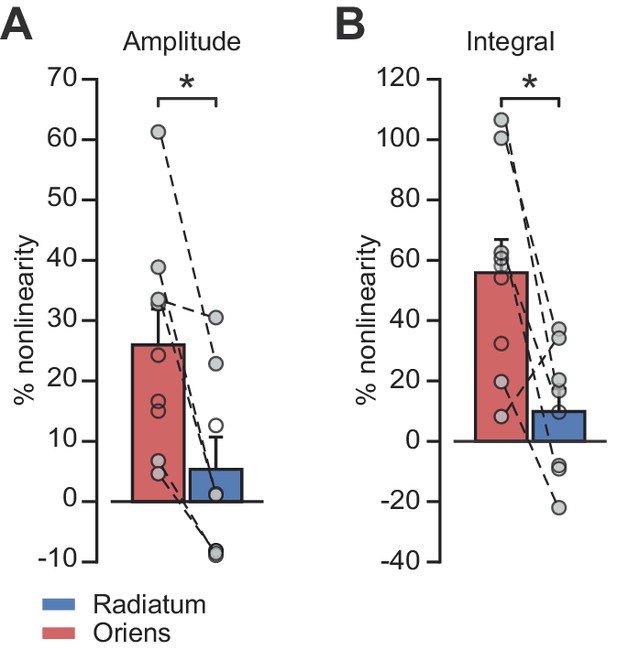
uEPSP integration location-dependent nonlinearity by cell.
(A) Peak amplitude nonlinearity comparison by dendrite location. Statistics for all data, oriens vs radiatum: 26.0 ± 6.0% (n = 9) vs 5.4 ± 5.4% (n = 8), p=0.02. Statistics for paired data oriens vs radiatum: 29.6 ± 8.7% vs 6.4 ± 6.7%, p=0.01, n = 6. (B) Time-integral nonlinearity comparison by dendrite location. Statistics for all data, oriens vs radiatum: 55.9 ± 11.0% (n = 9) vs 9.9 ± 7.6% (n = 8), p=0.004. Statistics for paired data, oriens vs radiatum: 59.3 ± 16.4% vs 13.1 ± 9.6%, p=0.035, n = 6. Closed circles: paired recordings, open circles: unpaired. Bars show mean ± SEM of all recordings.
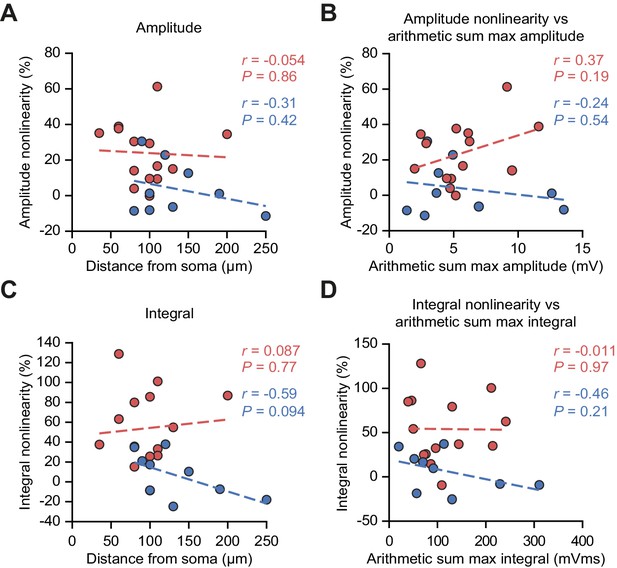
uEPSP nonlinearity does not depend on uncaging location distance from soma or on the size of the arithmetic sum of uEPSPs.
(A) uEPSP amplitude nonlinearity plotted against glutamate uncaging site distance from soma. (B) uEPSP amplitude nonlinearity plotted against maximal arithmetic sum of uEPSP amplitudes. (C) As (A) but for time-integrals. (D) As (B) but for time-integrals. Statistics: r = Pearson correlation coefficient, p=associated two-tailed significance.
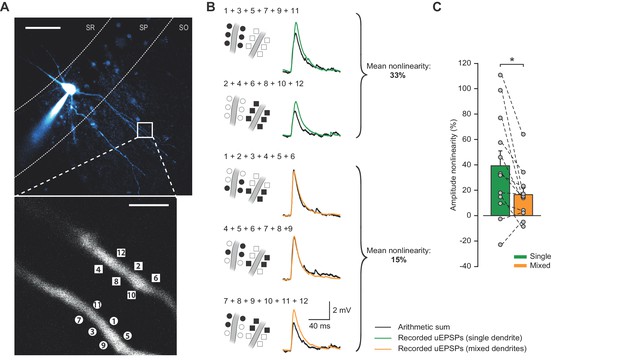
Compound uEPSPs from uncaging locations clustered on a single dendrite display larger nonlinearities than when distributed across two dendrites.
(A) Top: Two-photon z-stack of PV+ interneuron in CA1 region of hippocampus. White box marks region of interest shown below. Scale bar: 50 µm. Bottom: dendritic regions of interest at higher magnification showing glutamate uncaging locations (numbered) on two dendrites in the same focal plane. Scale bar: 3 µm. (B) Comparison of arithmetic sum of six individual uEPSPs (black) and the recorded compound uEPSP elicited by uncaging glutamate on single dendrites (green) or across the two dendrites (orange). (C) Summary of single- vs mixed-dendrite peak amplitude nonlinearity for all recordings. Statistics: paired t-test, single vs mixed nonlinearity: 39.3 ± 11.7% vs 16.5 ± 5.6%, p=0.016, n = 12 dendrite pairs, n = 6 cells. Error bars indicate mean ± SEM.
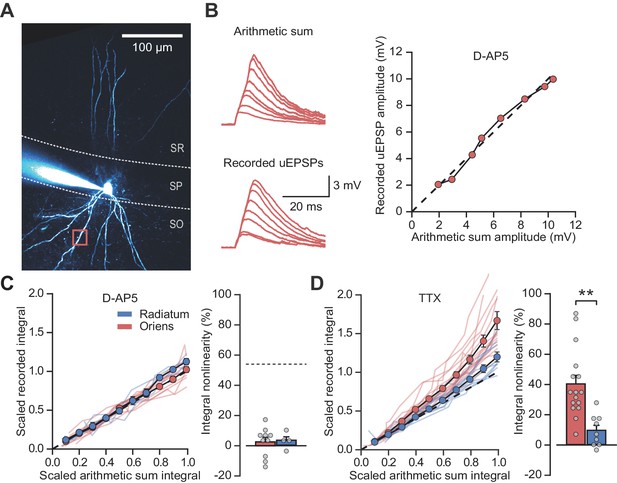
NMDARs mediate stratum oriens dendrite synaptic integration supralinearity.
(A) Two-photon z-stack of PV+ interneuron in CA1 region of hippocampus. Red box marks glutamate uncaging location. (B) Comparison of arithmetic and recorded uEPSP summation waveforms in the presence of D-AP5. Right: peak recorded amplitude vs peak arithmetic amplitude. (C) Summary data of time-integrals plotted against arithmetic sum time-integrals for 14 dendritic locations recorded in D-AP5 (oriens locations: n = 10, radiatum locations: n = 4). Right: quantified synaptic integration nonlinearity. The dashed line marks the average magnitude of oriens nonlinearity from Figure 1F. (D) Summary data for 25 dendritic locations recorded in TTX (oriens locations: n = 16, radiatum locations: n = 9). Right: quantification of synaptic integration nonlinearity. Filled circles and error bars indicate mean ± SEM.
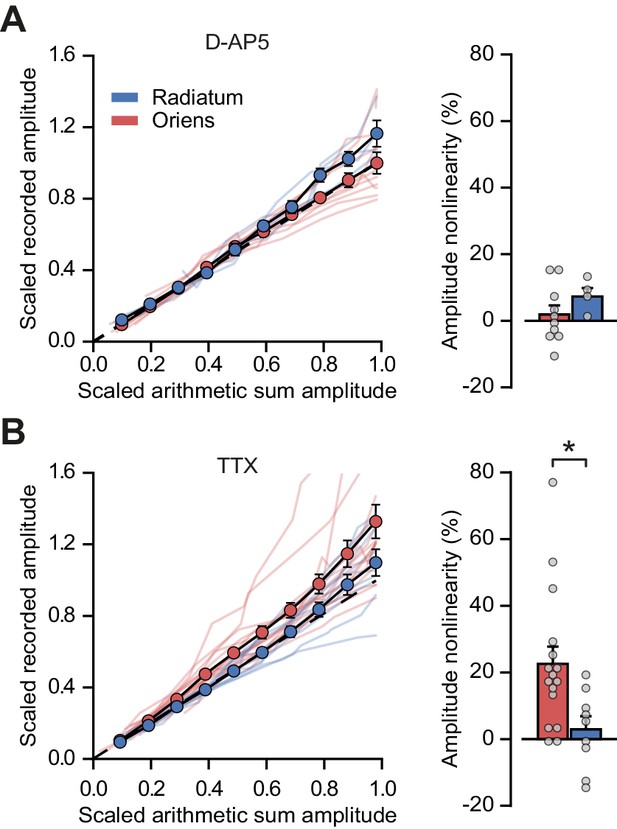
Peak amplitude nonlinearity in oriens dendrites is abolished by blocking NMDARs with D-AP5 but not by blocking sodium channels with TTX.
(A) Summary scaled peak amplitude uEPSP data for 14 dendritic locations recorded in D-AP5, n = 10 oriens; n = 4 radiatum. Right: quantified synaptic integration nonlinearity. Oriens vs radiatum: 1.9 ± 2.7% vs 7.3 ± 2.5%, p=0.27. Oriens D-AP5 vs oriens control, p=0.001. Radiatum D-AP5 vs radiatum control, p=0.66. (B) Summary scaled peak amplitude uEPSP data for 25 dendritic locations recorded with TTX in the ACSF, n = 16 oriens; n = 9 radiatum. Right: quantified synaptic integration nonlinearity. Oriens vs radiatum: 22.6 ± 5.2% vs 2.9 ± 3.9%, p=0.016. Oriens TTX vs oriens control, p=0.84. Radiatum TTX vs radiatum control, p=0.9.
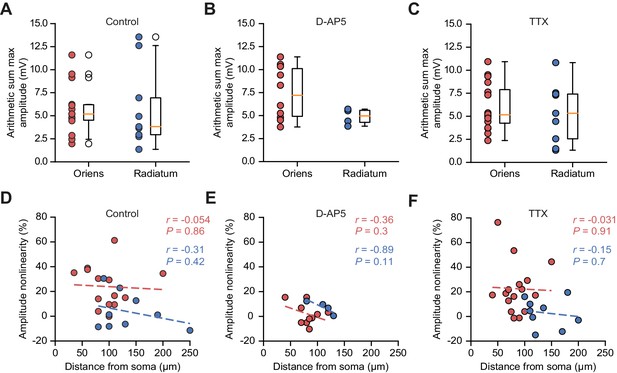
Arithmetic sum maximum uEPSP amplitudes, and integration nonlinearity vs uncaging location distances, across pharmacological conditions.
(A – C) Peak arithmetic uEPSP amplitudes by uncaging dendrite location in control conditions (A, replotted from Figure 1—figure supplement 1B for comparison), D-AP5 (B), and TTX (C). (D – F) uEPSP peak amplitude nonlinearity vs uncaging location distance from soma in control conditions (D, replotted from Figure 1—figure supplement 4 for comparison), D-AP5 (E), and TTX (F).
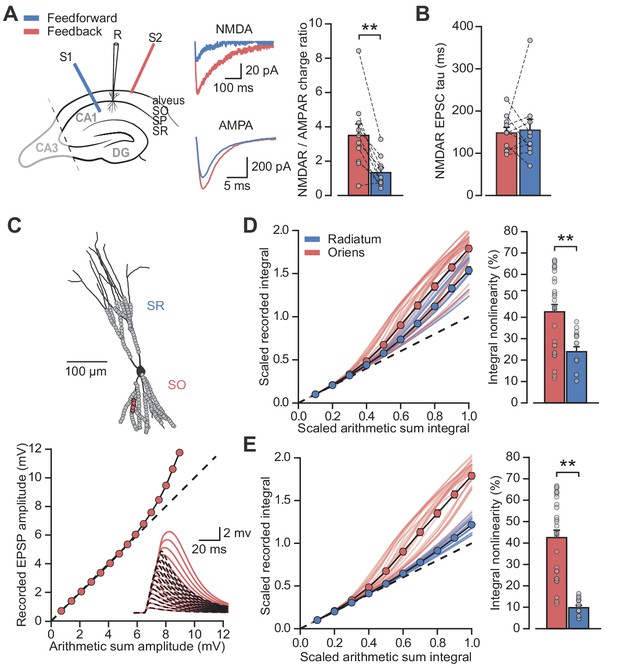
Differential NMDAR expression and dendrite morphology explain stratum-dependent synaptic integration difference.
(A) Schematic describing stimulation of feedforward (S1, blue) and antidromic stimulation of feedback (S2, red) axons. Middle: example paired AMPAR and NMDAR EPSC components in low [Mg2+]. Right: NMDAR/AMPAR charge ratios (n = 10). (B) Decay time constants of NMDAR-mediated EPSCs recorded in the same PV+ neurons in response to stimulation of feedforward (blue) and feedback (red) axons (n = 10). (C) Top: reconstruction of a PV+ interneuron (axon not shown). Simulated synaptic locations are shown in gray. Bottom: example simulated uncaging experiment at the synapses marked with red circles; graph shows recorded EPSP amplitudes vs arithmetic sum of EPSP amplitudes. Inset: red solid lines, recorded summation; dashed black lines, arithmetic summation; waveforms calculated from individual synaptic responses. (D) Scaled recorded time-integrals vs scaled arithmetic sum of time-integrals at all locations with equal NMDAR conductance (oriens locations: n = 28, radiatum locations: n = 16). Right: quantified synaptic integration nonlinearity. (E) As (D), but with reduced NMDAR/AMPAR conductance ratio at radiatum dendrites. Oriens data replotted from (D).
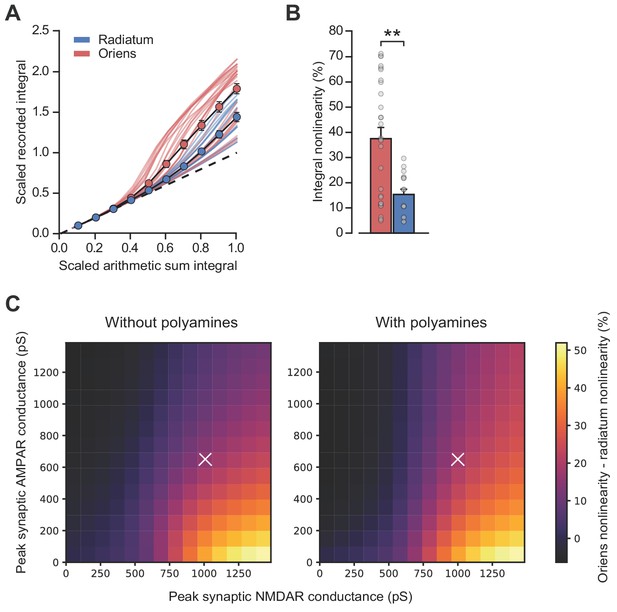
Simulations including polyamine modulation of AMPARs show synaptic integration differences between strata oriens and radiatum dendrite locations.
(A) Scaled recorded time-integrals vs scaled arithmetic sum of time-integrals at all locations with equal NMDAR conductance (oriens locations: n = 28, radiatum locations: n = 16). As Figure 3D, but with 100 µM intracellular spermine. (B) Quantified synaptic integration nonlinearity, oriens 37.5 ± 4.4% vs radiatum 15.3 ± 2.0. (C) Mean percentage integral nonlinearity difference between oriens and radiatum dendrites as peak synaptic AMPA and NMDA receptor conductance is varied. White crosses denote parameter values for simulations shown in panel A and Figure 3D. Mean difference between simulations over parameter range was 2.77 ± 0.17%. Polyamine modulation was simulated using a kinetic scheme based on Bowie et al. (1998). Values for peak synaptic conductance were calculated at −60 mV and +60 mV for AMPA and NMDA receptors respectively. n = 16 radiatum, n = 28 oriens.
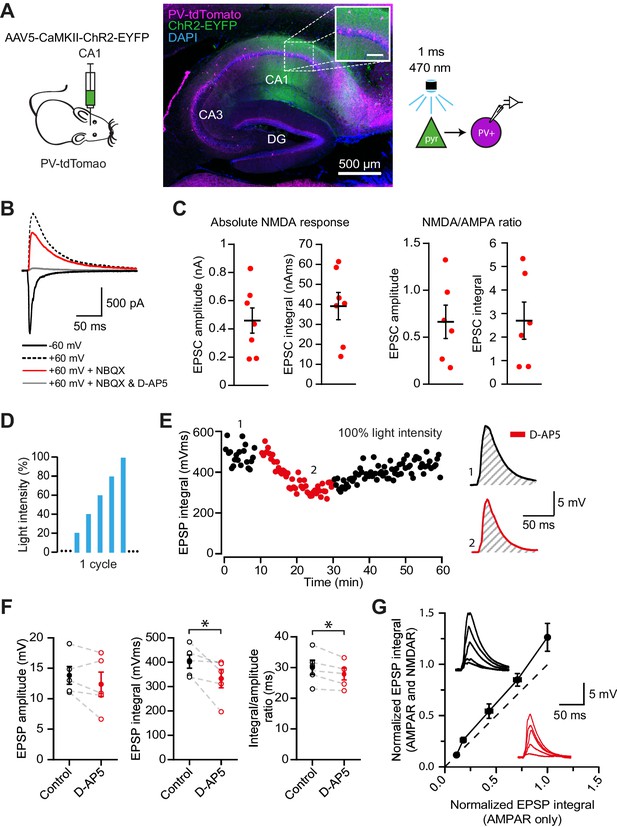
NMDAR recruitment at CA1 pyramidal cell feedback connections onto PV+ interneurons.
(A) Schematic of viral injections into dorsal CA1 of PV-tdTomato mice (left), and confocal image of a sagittal hippocampal slice showing selective ChR2-EYFP expression in CA1 pyramidal cells (middle, inset scale: 100 µm). Right: schematic of optogenetic patch clamp experiments. (B) Example traces of light-evoked feedback EPSCs in a PV+ interneuron held at −60 mV (black), +60 mV (black dashed), +60 mV with application of NBQX (red) or +60 mV with NBQX and D-AP5 in voltage clamp. (C) Quantification of absolute NMDAR-mediated feedback EPSCs (amplitude and integral, left, n = 7) and NMDAR/AMPAR ratios (amplitude and integral, right, n = 6), measured from voltage clamp experiments as in (B). Black bars indicate mean ± SEM. (D) Schematic of optogenetic stimulation protocol for current clamp experiments: light power was cycled from 20% to 100% of power for maximal response (see Materials and methods). (E) EPSP integral of maximal response over time, with 20 min application of D-AP5 (red). (F) EPSP amplitude, EPSP integral and integral/amplitude ratio in the presence (red) or absence (black) of D-AP5 (n = 5, one-tailed t-tests; control = average of baseline and wash). Filled circles and error bars indicate mean ± SEM. (G) Normalized EPSP integrals (black example traces) vs normalized EPSP integrals in the presence of D-AP5 (red example traces), for all stimulation intensities (n = 5).
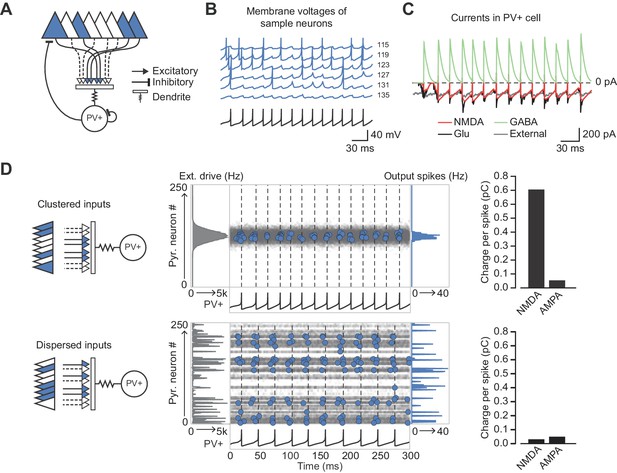
Network architecture and NMDAR recruitment at feedback connections.
(A) Schematic of network structure. (B) Voltage traces of interneuron (black) and principal cells (blue, cell # at right) during network simulation. The network was driven by an asynchronous barrage of spikes, maximal in cell #125 (‘clustered’ input). (C) Corresponding currents in interneuron. Red: NMDAR currents from principal cells; green: GABAR currents from autaptic PV+ cell connections; black: sum of NMDAR and AMPAR currents from principal cells; gray: AMPAR currents from external drive. (D) Left: schematic showing cell assemblies receiving clustered (top) or dispersed (bottom) external inputs, and middle: corresponding summary plots of network simulation showing external drive input distribution (gray), pyramidal cell firing (blue, circles), and interneuron firing (black, and vertical dashed lines). Right: average NMDAR and AMPAR charge in interneuron per principal neuron spike. (Autaptic and feedback connections from PV+ cells are omitted from the schematic for clarity.).
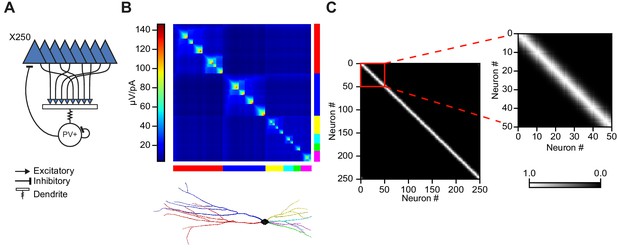
Modeling principal cell input cooperation onto feedback interneurons.
(A) Network schematic showing postsynaptic membrane locations of Izhikevich-model PV+ interneuron. (B) Steady state transfer resistance for a PV+ cell dendritic tree reconstructed using the TREES toolbox (Cuntz et al., 2010). The matrix shows the electrotonic ‘proximity’ of neighbouring patches of membrane on the dendritic tree (color scale: µV/pA). The main branches of the dendritic tree are indicated at the bottom. (C) Input cooperativity matrix used in simulations. Inset shows zoomed view of cooperativity matrix for the first 50 principal neurons.
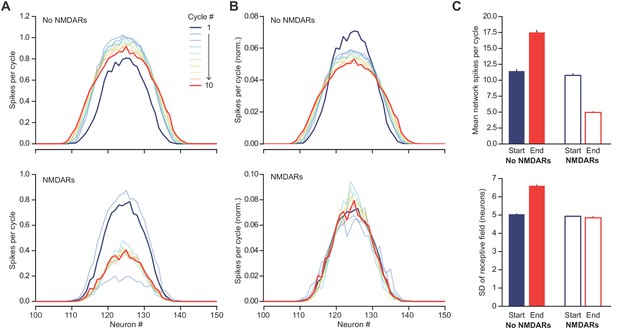
NMDARs help to maintain a sparse and sharp representation of a ‘hump’ of excitation to the feedback circuit shown in Figure 5D.
(A) Distribution of firing rate by neuron during each PV+ interneuron firing cycle, averaged across 500 simulations. Top – without NMDARs, bottom – with NMDARs. The hump of excitation was centered on neuron #125. Note the similarity of the first cycle between the two conditions. The behaviors of the networks diverge as the NMDARs are engaged. (B) As (A) but normalized by total firing rates across neurons, for better comparison of distribution dispersion. (C) Comparison of first and tenth simulation cycle: top – mean spikes per cycle, bottom - standard deviation of average network ‘receptive field’.
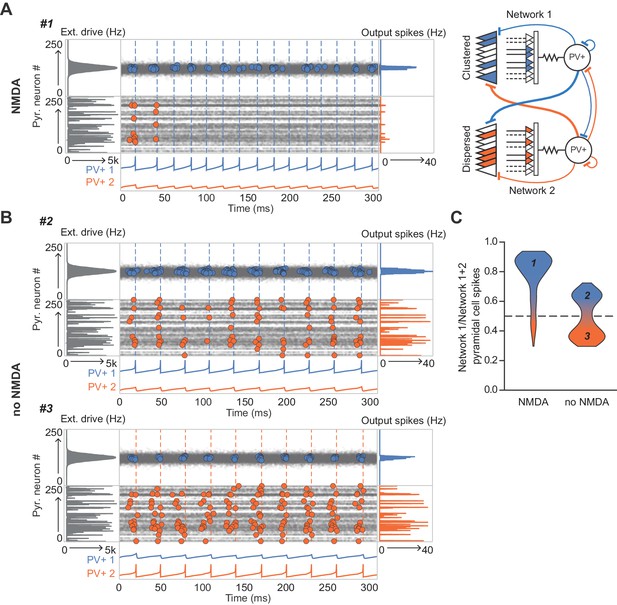
The role of NMDARs at feedback connections in cell assembly competition.
(A) Right: schematic showing competing cell assemblies with clustered (blue) or dispersed (orange) inputs; left: example simulation of lateral inhibition between these subnetworks with NMDARs at feedback connections to interneuron (input distribution: gray; pyramidal cell firing: blue/orange, circles; interneuron firing: blue/orange spikes and vertical dashed lines). The network receiving the clustered input out-competed the network receiving the dispersed input. (B) Same as (A) but without NMDARs at feedback connections, showing, in one case the network receiving clustered input firing more than the network receiving dispersed input (top), and in the other case the network receiving dispersed input winning (bottom). (C) Summary of 250 simulations showing ratio of principal cell spikes for each subnetwork with and without NMDARs at feedback inputs onto interneuron. Numbers correspond to simulations illustrated in (A) and (B).
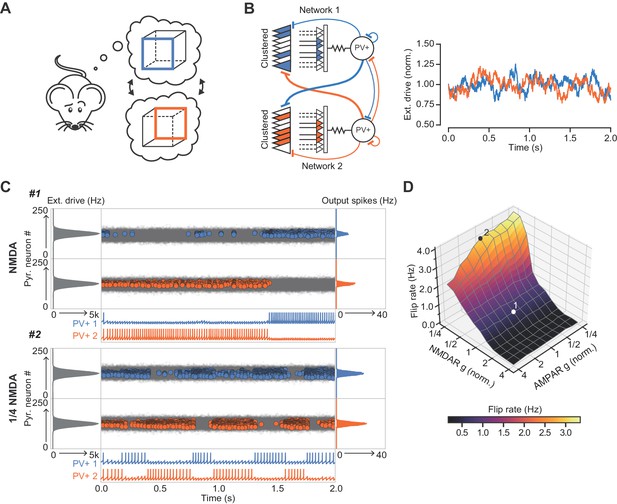
The role of NMDARs at feedback connections in cell assembly stability.
(A) Cartoon illustrating a bistable neural representation. (B) Schematic of competing subnetworks both receiving clustered inputs (left) with random fluctuations in external input strength (plotted right). (C) Example simulation of network activity with NMDARs at synapses on interneurons (top), and with NMDARs scaled down to 25% of baseline (bottom). Input distribution: gray; pyramidal cell firing: blue/orange, circles; interneuron firing: blue/orange spikes. Although the dominant networks flipped spontaneously in both cases, the frequency of flipping was much higher with the down-scaled NMDARs. (D) Plot of network flip rate vs NMDAR and AMPAR conductance. White point (1): baseline NMDAR simulation parameters; black point (2): NMDARs down-scaled to 25%.
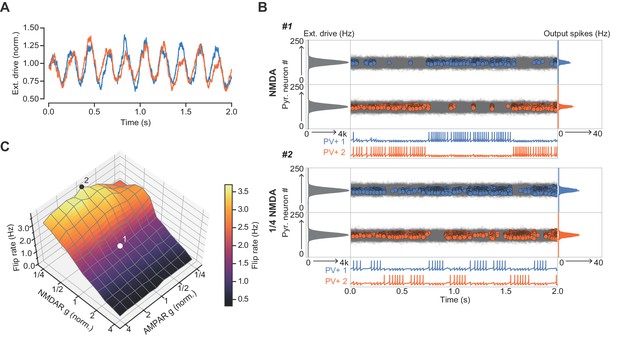
The role of NMDARs at feedback connections in cell assembly stability with theta-modulated external drive.
(A) Theta-modulated external drive to the system. (B) Example simulation of network with NMDARs at synapses on interneurons (top), and with NMDARs scaled down to 25% of baseline (bottom). External drive (modulated as shown in panel A): gray; pyramidal cell firing: blue/orange, circles; interneuron firing: blue/orange spikes. Although the dominant networks flipped spontaneously in both cases, the frequency of flipping was much higher with the down-scaled NMDARs. (C) Plot of network flip rate vs NMDAR and AMPAR conductance. White point (1): baseline NMDAR simulation parameters; black point (2): NMDARs down-scaled to 25%.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Mus musculus) | B6;129P2-Pvalbtm1(cre)Arbr/J | The Jackson Laboratory | 008069 | |
| Strain, strain background (Mus musculus) | B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze | The Jackson Laboratory | 007909 | |
| Other | AAV5-CaMKIIa-hChR2(H134R)-EYFP | UNC Vector Core | ||
| Chemical compound, drug | Picrotoxin | Sigma-Aldrich | P1675 | |
| Chemical compound, drug | CGP 55845 | Abcam | Ab120337 | |
| Chemical compound, drug | D-AP5 | Tocris | 0105 | |
| Chemical compound, drug | TTX | Tocris | 1078 | |
| Chemical compound, drug | ZD 7288 | Tocris | 1000 | |
| Chemical compound, drug | MNI-glutamate TFA | Femtonics | 1951 |
NEURON model parameters.
https://doi.org/10.7554/eLife.49872.020| Parameter | Proximal | Distal | Units |
|---|---|---|---|
| Cm | 0.9 | 0.9 | µF cm-2 |
| Raxial | 170 | 170 | Ω cm |
| Rm | 5.55 | 55.5 | kΩ |
| eleak | -65 | - | mV |
| egk | -90 | - | mV |
| egNa | 55 | - | mV |
| v Shift | -12 | -12 | mV |
| gk dend | 300 | 300 | pS µm-2 |
| gNa dend | 200 | 100 | pS µm-2 |
| gNa Soma | 2000 | - | pS µm-2 |
model neuron parameters for network modeling.
https://doi.org/10.7554/eLife.49872.021| Parameter | FS PV+ | Pyramidal | Units |
|---|---|---|---|
| 90 | 115 | pF | |
| 1.7 | 0.1 | nS/mV | |
| 14 | 3.3 | nS/mV | |
| −60.6 | −65.8 | mV | |
| 2.5 | 22.6 | mV | |
| −43.1 | −57 | mV | |
| 0.1 | 0.0012 | ms−1 | |
| −0.1 | 3 | nS | |
| −67 | −65.8 | mV | |
| 0.1 | 10 | pA | |
| 0.25 | 0.2 | ms | |
| 0.77 | 1.7 | ms | |
| 0.27 | 0.3 | ms | |
| 1.7 | 3.5 | ms | |
| 2 | - | ms | |
| 60 | - | ms | |
| 0 | 0 | mV | |
| −70 | −70 | mV | |
| 0.015/(npyr2) | - | - | |
| 9 | - | pF | |
| 3/npyr | - | - | |
| 5 | - | nS | |
| −60.6 | - | mV | |
| 28 | - | - | |
| 212 | - | - | |
| 28 | 27 | - | |
| 5 | 1 | - |
-
Izhikevich Parameter values as in Ferguson et al. (2014) and Ferguson et al. (2013). Synaptic time constants: (Bartos et al., 2002; Geiger et al., 1997; Roth and van Rossum, 2009). Remaining parameters where adjusted to allow the network to generate a gamma rhythm.
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.49872.022





